Abstract
The identification of small molecules that target specific CFTR variants has ushered in a new era of treatment for cystic fibrosis (CF), yet optimal, individualized treatment of CF will require identification and targeting of disease modifiers. Here we use genome-wide association analysis to identify genetic modifiers of CF lung disease, the primary cause of mortality. Meta-analysis of 6,365 CF patients identifies five loci that display significant association with variation in lung disease. Regions on chr3q29 (MUC4/MUC20; P=3.3 × 10−11), chr5p15.3 (SLC9A3; P=6.8 × 10−12), chr6p21.3 (HLA Class II; P=1.2 × 10−8) and chrXq22-q23 (AGTR2/SLC6A14; P=1.8 × 10−9) contain genes of high biological relevance to CF pathophysiology. The fifth locus, on chr11p12-p13 (EHF/APIP; P=1.9 × 10−10), was previously shown to be associated with lung disease. These results provide new insights into potential targets for modulating lung disease severity in CF.
 Cystic fibrosis imposes a decline in quality of life but new treatments are being developed that target specific CFTR variants. Here the authors identify five genome loci significantly associated with variation in disease severity in a meta-analysis, which may provide targets for individualized treatment of cystic fibrosis.
Cystic fibrosis imposes a decline in quality of life but new treatments are being developed that target specific CFTR variants. Here the authors identify five genome loci significantly associated with variation in disease severity in a meta-analysis, which may provide targets for individualized treatment of cystic fibrosis.
Cystic fibrosis (CF) affects ∼70,000 individuals worldwide and is caused by loss-of-function variants in CFTR. Although CF is regarded as a single-gene disorder, patients who have the same variants in CFTR exhibit substantial variation in severity of lung disease, of which >50% is explained by non-CFTR genetic variation1. The identification of small molecules that target specific CFTR variants has ushered in a new era of treatment for cystic fibrosis (CF)2, but optimal individualized treatment will require identification and targeting of disease modifiers.
The advent of large-scale genome-wide association studies (GWAS) and capability for imputation has made it possible to explore millions of polymorphisms in search of genetic determinants of phenotypic variation. Our previously reported GWAS in 3,444 CF patients led to identification of genome-wide significant single-nucleotide polymorphism (SNP) associations with lung disease severity in an intergenic region between EHF and APIP (chr11p13), as well as several additional suggestive loci (chr6p21.3 and chrXq22-q23)3. In other published candidate gene studies4, additional loci/genes have been reported to reach significance thresholds for the individual study, but many of these studies were based on relatively small sample sizes and/or limited phenotyping, and most have not been replicated, generating uncertainty as to their pathophysiological relevance.
In this manuscript, we have extended our study of CF gene modifiers by testing 2,921 additional patients from North America (n=1,699) and France (n=1,222). We use the same lung phenotype as in the previous GWAS (Consortium lung phenotype (KNoRMA))5, which allows for direct comparison of the lung function of CF patients irrespective of age and gender. A meta-analysis of both imputed and genotyped variants is reported that combines data from the new subjects with the previously reported GWAS, allowing for an unprecedented sample size of 6,365 CF patients and analysis of over 8 million variants. To maximize power, linear mixed models are used to allow for inclusion of CF-affected siblings. The combined analysis confirms a previous genome-wide association and identifies four new loci that contain genes with high biological relevance for pathophysiology of CF lung disease.
Results
Characteristics of patients in GWAS2 and GWAS1
The cohort study design and the demographic and clinical characteristics of GWAS2 subjects are detailed in Table 1 and Methods. In the combined GWAS1+2 data set, 99.8% of subjects were pancreatic exocrine insufficient (primarily defined by CFTR genotypes); 65.0% were p.Phe508del homozygotes; 95.5% were of European ancestry; and only 5.4% were diagnosed by newborn screening. GWAS1 subjects from three cohorts were genotyped on the same Illumina platform3, while GWAS2 included 10 subgroups defined by different combinations of site and Illumina genotyping platforms. These 13 subgroups had similar distributions of the lung disease phenotype (Supplementary Fig. 1).
Table 1. Characteristics of patients enrolled in GWAS2 and GWAS1 by the International Cystic Fibrosis Gene Modifier Consortium.
| Age (years) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lead Institution(s) | Design | Subjects n | Mean (±s.d.) | Range | Male n (%) | European* n (%) | p.Phe508del/p.Phe508del n (%) | Pancreatic exocrine insufficient n (%) | Subjects identified by NBS n (%) | |
| GWAS2 | ||||||||||
| French CF Gene Modifier Consortium (FrGMC) | University of Pierre and Marie Curie, Inserm U938 | Population based | 1,222 | 21.0 (9.2) | 6.0–57.6 | 627 (51.3) | 1,211 (99.1) | 716 (58.6) | 1,222 (100.0) | 63 (5.2) |
| Genetic Modifier Study (GMS) | University of North Carolina/ Case Western Reserve University | Extremes of phenotype | 469 | 25.8 (10.9) | 7.9–62.2 | 256 (54.6) | 407 (86.8) | 191 (40.7) | 467 (99.6) | 3 (0.01) |
| Population-based† | 357 | 20.3 (10.0) | 6.6–60.2 | 191 (53.5) | 336 (94.1) | 214 (59.9) | 357 (100.0) | 137 (38.4) | ||
| Canadian Consortium for Genetic Studies (CGS) | Hospital for Sick Children | Population based‡ | 285 | 13.0 (7.6) | 6.4–40.0 | 150 (52.6) | 268 (94.0) | 189 (66.3) | 282 (98.9) | 0 (0.0) |
| Twins and Sibs Study (TSS) | Johns Hopkins University | Family based and population based§ | 588 | 15.8 (10.3) | 6.0–56.0 | 305 (51.9) | 533 (90.6) | 315 (53.6) | 583 (99.1) | 54 (9.2) |
| Summary GWAS2 | 2,921 | 19.9 (10.4) | 6.0–62.2 | 1,529 (52.3) | 2,755 (94.3) | 1,625 (55.6) | 2,911 (99.7) | 257 (8.8) | ||
| GWAS1 | ||||||||||
| Summary GWAS1|| | 3,444 | 19.2 (8.5) | 6.0–56.0 | 1,839 (53.4) | 3,324 (96.5) | 2,514 (73.0) | 3,444 (100.0) | 84 (2.4) | ||
| GWAS1+2 | ||||||||||
| Summary GWAS1+2 | 6,365 | 19.5 (9.4) | 6.0–62.2 | 3,368 (52.9) | 6,079 (95.5) | 4,139 (65.0) | 6,355 (99.8) | 341 (5.4) | ||
GWAS, genome-wide association study; NBS, newborn screening.
*On the basis of Eigenstrat principal components analysis and closeness to CEU.
†Includes patients enrolled into studies at Children's Hospitals in Boston, Colorado and Wisconsin, and through UNC/CWRU; includes 3 two-sibling families.
‡Includes 13 two-sibling families and 1 three-sibling family, plus 256 singletons.
§148 two-sibling families, 4 three-sibling families, plus 280 singletons.
||Wright et al.3.
Genome-wide significance in five regions by meta-analysis
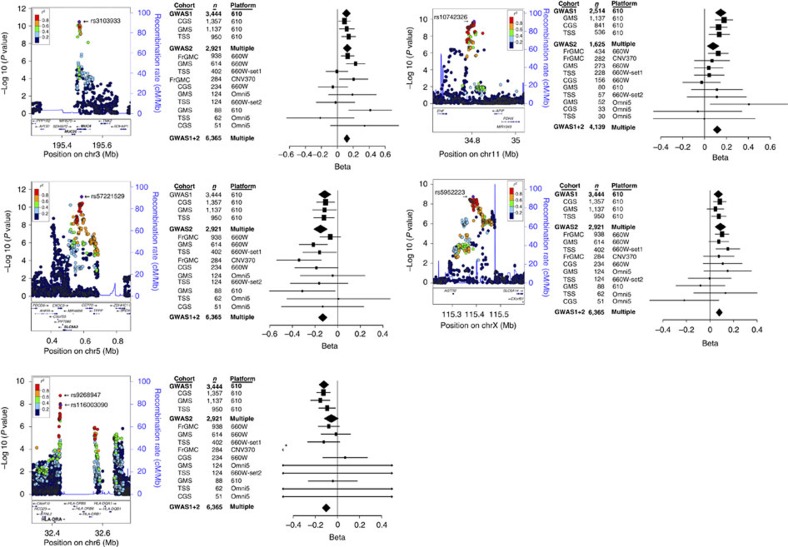
To account for potential effect size heterogeneity, a random effects meta-analysis association model6 (Methods) was applied across the 13 subgroups. Acknowledging that the power of fixed effects meta-analysis can be greater than that of random effects, even under heterogeneity7, we also performed fixed effect meta-analysis, and report loci exceeding significance thresholds by either random or fixed effects analysis. Analysis included 8,520,458 genotyped and imputed SNPs with minor allele frequency >0.005 and markers with imputation r2>0.3. Principal component-based stratification control was used (genomic control lambda=0.95) (Methods and Supplementary Figs 2 and 3). We considered loci with P<1.25 × 10−8 to be genome-wide significant, based on the standard for genome-wide significance (P=5 × 10−8)8 and multiplicity correction for CFTR genotype (all genotypes versus p.Phe508del homozygotes) and model (random versus fixed effects). Associations in five regions (chr3q29, chr5p15, chr6p21, chr11p12-p13 and chrXq22-q23) exceeded genome-wide significance (Fig. 1). Only the chr11p12-p13 locus provides significant evidence of interaction with CFTR (P=0.048, from a Wald test of the interaction term in the linear mixed model that is then meta-combined using the inverse variance-based weighting). This result corroborates our prior observation that association at this locus achieves a lower P value in p.Phe508del homozygous subjects than in all subjects3.
Figure 1. Genome-wide Manhattan plot of associations with the Consortium lung phenotype.
Evidence from GWAS1+2 for all patients (closed circles) and for p.Phe508del homozygotes (open triangles). The horizontal dashed line represents the threshold for genome-wide significance (P<1.25 × 10−8). Genome-wide significance was achieved in five regions. The results from regions on chr5p15, chr11p12-p13 and chrXq22-q23 are from meta-analysis using a random effects model, and for chr3q29 and chr6p21 using a fixed effects model.
Each of the regions contained at least one genotyped SNP achieving significance (Table 2), except for chr6p21.3 near HLA-DRA. However, five imputed SNPs that exceeded genome-wide significance at the HLA-DRA locus were directly genotyped in a subset of the sample and provided independent genotype confirmation (Methods). LocusZoom9 and effect size forest plots show the regional association evidence and the relative contribution from each of the 13 cohort by platform subgroups (Fig. 2). A formal test (Cochran's Q test) revealed that only the MUC4 locus has significant evidence of heterogeneity (P=0.04); the inconsistent association among subgroups may be due to the small sample size of some of these cohorts. All five regions all remained significant when restricted to 6,079 individuals determined by principal components to be of European ancestry (Supplementary Table 1, Supplementary Fig. 4). Complementary results from sub-setting the sample into a North American discovery (n=5,143) and French replication set (n=1,222) show genome-wide significance and independent replication in four of the five loci reported in Table 2. The remaining locus (chrX; AGTR2/SLC6A14) achieved suggestive evidence of association in the North American cohort with compelling evidence of replication in the French subjects (Methods and Supplementary Table 2).
Table 2. Genome-wide significant association results for GWAS1+2.
| Chr | 3 | 5 | 6 | 11 | X |
|---|---|---|---|---|---|
| Nearby gene(s) | MUC4/MUC20 | SLC9A3 | HLA-DRA | EHF/APIP | AGTR2/SLC6A14 |
| SNP* | rs3103933 | rs57221529 | rs116003090† | rs10742326 | rs5952223 |
| Base pair‡ | 195,485,440 | 586,624 | 32,434,850 | 34,810,010 | 115,386,565 |
| Minor allele§ | A | G | C | A | T |
| Major allele§ | G | A | G | G | C |
| Minor allele frequency|| | 0.37 | 0.2 | 0.31 | 0.42 | 0.28 |
| GWAS1+2 all beta coefficient¶ | 0.12 | −0.13 | −0.1 | 0.09 | 0.08 |
| GWAS1+2 p.Phe508del/p.Phe508del beta coefficient¶ | 0.12 | −0.13 | −0.09 | 0.12 | 0.09 |
| P value, GWAS1+2 all# | 3.3 × 10−11 | 6.8 × 10−12 | 1.2 × 10−8 | 4.8 × 10−9 | 1.8 × 10−9 |
| P value, GWAS1+2 p.Phe508del/p.Phe508del# | 7.6 × 10−8 | 3.4 × 10−8 | 3.6 × 10−5 | 1.9 × 10−10 | 1.3 × 10−5 |
| Analysis with maximum significance | GWAS1+2 all** | GWAS1+2 all | GWAS1+2 all** | GWAS1+2 p.Phe508del/p.Phe508del | GWAS1+2 all |
| SNP, genotyped†† | rs2246901 | rs3749615 | rs2395185 | rs10466455 | rs5905376 |
| P value, genotyped†† | 1.3 × 10−10 | 2.2 × 10−9 | 1.5 × 10−7 | 1.3 × 10−9 | 3.3 × 10−9 |
GWAS, genome-wide association study; SNP, single-nucleotide polymorphism.
*SNP IDs are from 1000 Genomes Project (Phase I, Version 3). The proportion of variability in Consortium lung phenotype explained by the five SNPs is 0.05.
†rs116003090 is an imputed SNP that was genotyped on a subset of subjects (n=374) for independent genotype confirmation.
‡Genome Reference Consortium Human Build 37 (GRCh37).
§Major/minor alleles indexed to 1000 Genomes Project.
||Minor allele frequencies are listed for all GWAS1+2.
¶Beta-coefficients refer to the average change in Consortium lung phenotype for each copy of the minor allele.
#Meta-analysis P values based on a random effects model,
**with the exceptions of rs3103933 and rs116003090, which were based on a fixed effects model.
††Most significant SNP genotyped on all platforms; P values based on called genotypes.
Figure 2. LocusZoom and forest plots for five regions with significant association in GWAS1+2.
On the left side of the five panels are plots of the association evidence (build GRCh37, LocusZoom viewer) in the five genome-wide significant regions for all patients, except that chr11p12-p13 shows only p.Phe508del homozygotes. Colours represent 1000 Genomes EUR linkage disequilibrium r2 values with each SNP in column three of Table 2 (shown as purple diamonds and labelled with dbSNP ID). The purple diamond in the chr6 region denotes the SNP that has independent genotype confirmation, but the top imputed SNP is also indicated by a dbSNP ID (rs number). On the right side of the five panels are forest plots of the relative effect sizes for the most significant SNP in each of the 13 subgroups, ordered by size. Beta (coefficient) refers to the average change in Consortium lung phenotype for each copy of the minor allele. The size and shape of the squares are proportional to the weights used in the meta-analysis, and the line segments are 95% confidence intervals of each beta. The black diamonds represent summary data for GWAS1, GWAS2, and GWAS1+2. The asterisk on chr6 (HLA region) forest plot illustrates a beta (and confidence interval) for the FrGMC CNV370 subgroup of −19.9 (−35.4, −4.4). In addition, the beta (and confidence interval) for four other subgroups in the chr6 region are as follows: GMS Omni5, 0.87 (−19.5, 21.2); TSS 660W-set 2, −1.67 (−19.6, 16.3); TSS Omni5, 15.4 (−14.1, 44.8); and CGS Omni5, 1.7 (−23.8, 27.2).
Conditioning on significant SNPs
Conditioning on the most significant SNP in the five regions that had genome-wide significance eliminated significant association in three regions (chr3q29; chr6p21; chrXq22-q23), but not for chr5p15 or chr11p12-p13. On chr5p15 (Supplementary Fig. 5a), conditioning on rs57221529 (located 5′ of SLC9A3 and CEP72) revealed significantly associated SNPs 3′ of SLC9A3 (near AHRR), suggesting a complex contribution by genes and/or regulatory domains in the region. Similar complex mechanisms may be at play on chr11p12-p13, as conditioning on rs10742326 (in EHF/APIP intragenic region) revealed significantly associated SNPs 3′ of APIP (Supplementary Fig. 5b).
CFTR variants and lung function
As this study has the largest assembly of CF patients for modifier identification, we employed a combined test for association between CFTR variants and lung function10, using the maximal unrelated sample of patients from GWAS1+2 (n=5,762). Summing the association test statistics (the square of the association Z-statistic) for each SNP in/near CFTR enabled capture of the combined effect of disease-causing mutations (Methods). Overall, this gene-based (CFTR) statistic showed that CFTR and lung disease severity are associated (P=0.0043, from a permutation-based test that is then meta-combined using Stouffer's Z-score method), but the evidence is very modest compared to the evidence for multiple modifier loci reported here. As expected, this association with lung disease was not present when we restricted analysis to the p.Phe508del homozygotes (n=3,815; P=0.1604, from a permutation-based test that is then meta-combined using Stouffer's Z-score method).
eQTLs for significant SNPs
To determine whether the most significantly associated GWAS SNPs show evidence as expression quantitative trait loci (eQTLs) in lung or immune cells, we examined three available databases for eQTLs (Genotype-Tissue Expression, GTEx, http://www.gtexportal.org/home/; University of Chicago SNP and CNV Annotation Database, SCAN, http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/; University of North Carolina at Chapel Hill seeQTL, http://www.bios.unc.edu/research/genomic_software/seeQTL/). We considered eQTL evidence for each of the top-ranked GWAS SNPs at each locus, and for other local SNPs with r2 (linkage disequlibrium, LD)>0.6 with the top-ranked GWAS SNPs. The most significant eQTLs were noted at the chr6 (human leukocyte antigen (HLA) Class II) locus (DRB1 and DRB5) and the chr11 locus (APIP). The nominal P values from association testing in a linear regression model for eQTLs at other loci were modest (P>10−5), and of uncertain biological significance (Supplementary Table 3).
Comparison of GWAS1+2 with prior association studies
We also compared the results of the GWAS1+2 analysis with variants in 30 candidate genes or loci previously reported to associate (nominal P<0.05) with some aspect of CF pulmonary disease phenotype (Supplementary Table 4). For previously reported candidate gene SNP associations, only those in SLC9A3 demonstrated significance (after correction for multiplicity of replication testing) in GWAS1+2, although others trended towards nominal significance. It is important to recognize that none of these studies used precisely the same lung phenotype as GWAS1+2, that each study involved substantially fewer subjects than the 6,365 individuals studied here, and that other SNPs in these candidate regions were not assessed here. Likewise, homogeneity of non-genetic factors such as healthcare delivery might have enabled discovery of modifier associations in some studies that are not detectable in the heterogeneous populations aggregated for this study.
Discussion
Given the rarity of Mendelian disorders such as CF, it is not possible to accrue the large population of samples achieved for common complex traits11,12. The uniform genetic aetiology of Mendelian disorders can minimize phenotypic and genetic heterogeneity, and we take advantage of this uniformity to identify five genome-wide significant loci associated with the severity of lung disease in CF. Despite minimizing genetic heterogeneity, some residual effect of CFTR sequence variation on variation in lung function remained evident, suggesting a contributing but modest role compared to the modifiers. On the basis of the calculated beta-coefficients, the potential effect sizes of the SNPs of interest are estimated to be clinically relevant. The largest absolute beta-coefficient value (0.13) for SLC9A3/rs57221529 is equivalent to a change in forced expiratory volume at one second (FEV1) of 190 ml (4.64% predicted) for males and 130 ml (3.98% predicted) for females based on extrapolation from 18-year-old White subjects with lung function and height both at the 50th percentile for individuals with CF13,14. Likewise, the smallest absolute beta-coefficient value (0.08) for AGTR2/SLC6A14/rs5952223 is equivalent to a change in FEV1 of 110 ml (2.68% predicted) for males and 80 ml (2.45% predicted) for females.
The five associated loci contain genes with pathophysiological relevance for lung function in CF, and genes in these regions are expressed in lung3,15,16,17,18,19. Identification of the gene or genes responsible for the modifying effect of each locus will require functional study; however, each locus contains genes of compelling biologic plausibility based on extensive understanding of CF pathophysiology. MUC4 and MUC20 located at chr3q29 encode membrane-spanning ‘tethered' mucins on ciliated airway mucosal surfaces18,20,21 that contribute to the periciliary brush layer, and prevent mucus penetration into the periciliary space22. Further, MUC4 and MUC20 are present in airway mucus, reflecting secretion and/or shedding from the airway epithelium18, and likely play a role in mucociliary host defense. SLC9A3 on chr5 is the cation proton antiporter 3 (NHE3) involved in pH regulation and epithelial ion transport23,24. Knockout of SLC9A3 alleviated intestinal obstruction commonly observed in the mouse model of CF25, and a subsequent hypothesis-driven genome-wide association study identified SLC9A3 as a modifier of neonatal intestinal obstruction in humans with CF26. Candidate gene studies in the Canadian pediatric CF population have shown SLC9A3 to be pleiotropic for disease severity in multiple affected organs17,27. The broadness of the associated region does not exclude EXOC3 (telomeric, and just downstream of SLC9A3), a component of the exocyst complex that is involved in post Golgi trafficking and specification of membrane surfaces, including those in epithelial cells28. Further, the two more centromeric genes, CEP72 and TPPP, have been implicated in microtubule function. Microtubule disturbance has been reported in CF cells29 and a microtubule-associated gene, DCTN4 has been implicated in a CF-lung related phenotype of onset of Pseudomonas aeruginosa (P. aeruginosa) infection30. The HLA Class II region on chr6 has been associated with asthma, variation in lung function in the non-CF populations, and CF lung disease19,31,32, along with susceptibility to allergic bronchopulmonary aspergillosis33,34,35. Moreover, HLA Class II pathways have been recently associated with CF lung disease and age-of-onset of persistent P. aeruginosa in a large gene expression association study36. The chrX locus contains AGTR2 and SLC6A14 and either gene, or possibly both, could modify lung disease. AGTR2, the angiotension type II receptor, has been implicated in a variety of pulmonary functions including mediating signalling in lung fibrosis15, regulating nitric oxide synthase expression in pulmonary endothelium37, and has recently been described as a therapeutic target for lung inflammation38. SLC6A14 encodes an amino-acid transporter and variants in its 5′-regulatory region have been reported to modify risk for neonatal intestinal obstruction26, lung disease severity, and age at first P. aeruginosa infection in individuals with CF under 18 years of age17. The chr11 locus containing EHF and APIP was reported to be associated with CF lung disease in p.Phe508del homozygotes in an earlier GWAS3 and this association persists in this report. This EHF transcription factor is reported to modify the CF phenotype by influencing p.Phe508del processing16, and to modulate epithelial tight junctions and wound repair39. APIP is a methionine salvage pathway enzyme known to be associated with both apoptosis and systemic inflammatory responses40,41. Examination of the most significantly associated GWAS SNPs revealed eQTLs for genes at the HLA Class II locus (DRB1 and DRB5) and APIP (chr11). These findings are congruent with lung disease severity being mediated through differential gene expression (eQTLs), but additional studies will be required to clarify this possibility for each locus.
Collectively, this study suggests new mechanistic insight into ameliorating lung disease progression in individuals with CF. Functional analysis of associated SNPs and genes at each modifier locus could identify novel targets for treating CF. For example, BCL11A, a key transcription factor for fetal haemoglobin expression, is a modifier of beta thalassaemia and sickle cell disease42,43. Identification of a common variant associated with fetal haemoglobin level that alters BCL11A expression provides a therapeutic rationale for targeting this modifier of haemoglobinopathies44. The evolving paradigm for individualized therapy in CF involves small molecules targeting specific CFTR variants2,45. The discovery of modifier loci that are strongly associated with severity of CF lung disease provides an opportunity to enhance individualized treatment in CF.
Methods
Recruitment
The recruitment in North America was performed through three independent groups (GMS, CGS and TSS studies; Table 1)1,46,47. In brief, the majority of the GMS subjects were recruited as unrelated extremes-of-phenotype (lung disease severity, mild versus severe), whereas a subset of patients were recruited as a population-based sample at the Children's Hospital of Denver, Wisconsin, and Boston, and also from the multicenter study UNC/CWRU GMS cohort (Table 1). The CGS patients were recruited from the Canadian population of CF patients. The TSS patients were recruited based on having a surviving affected sibling. Informed consent was obtained from each participant of the study. Studies were approved by institutional review boards at participating sites and include: Committee on Clinical Investigation, Boston Children's Hospital; Institutional Review Board at Children's Hospital of Wisconsin; Colorado Multiple Institutional Review Board; Johns Hopkins School of Medicine eIRB2 (Committee: IRB-3); Research Ethics Board of The Hospital for Sick Children; Biomedical Institutional Review Board, Office of Human Research, University of North Carolina at Chapel Hill; and University Hospitals Case Medical Center, Institutional Review Board for Human Investigation. In France, patients were recruited from 48 CF centers, and phenotypic information was available for 2,898 patients older than 6 years of age; further, only patients with both parents born in European countries were considered (n=2,627). Only patients with documented pancreatic insufficiency or having two severe CFTR mutations were further considered. Written informed consent was obtained from adults, and for patients<18 years old there was consent from parents or guardians for participation in the study. The study was approved by the French ethical committee (CPP n°2004/15) and the information collection was approved by CNIL (n°04.404).
Lung disease severity phenotyping
In CF, FEV1 is recognized as producing the most clinically useful measurements of lung function and a known predictor of survival46,47. However, comparison of disease severity by FEV1 across a broad range of ages is confounded by the decline in FEV1 over time in CF patients, and by mortality attrition. In brief, we calculated average age-specific CF percentile values of FEV1 for each patient using three years of data in patients 6 years or older, using the Kulich-derived US (national)3 or French (national)48 CF percentiles (relative to other CF patients of the same age, sex and height), and adjusted for mortality5. The resulting quantitative phenotypes were distributed as expected, based on ascertainment. This quantitative phenotype was also highly correlated with the Schluchter survival phenotype (r2=0.91), when compared for the GMS patients47.
Genotyping and quality control
Genotyping used Illumina platforms, including: CNV370 (n=284 samples; FrGMC); 610 (n=3,532 samples; primarily GWAS1); 660W (n=2,312 samples); and Omni5 (n=237 samples). Genotype calling was performed using GenomeStudio V2011.1. Position and annotation information is based on hg19.
For the 660W platform, the total number of probes was 655,214 (64,569 were CNV probes) and 55 of the SNPs had no genotype calls for any of the samples. The 2,312 samples were composed of 938 (FrGMC)+614 (GMS)+234 (CGS)+402 (TSS 660W-set1)+124 (TSS 660W-set2) (Supplementary Fig. 1).
For the Omni5 platform, the total number of probes was 4,301,332 (961 of the SNPs had no genotype calls for any of the samples). The 237 samples were composed of 124 (GMS)+51 (CGS)+62 (TSS) (Supplementary Fig. 1).
For GWAS2, some samples (n=24) were genotyped on both 660W and Omni5 platforms and these duplicates showed >98% concordance. There were also 40 quality control duplicates of GWAS2 and GWAS1 samples, and those duplicates also showed high concordance. Samples with a call rate <98%, or sex discordance were excluded. Unintentional duplicates of GWAS2 and GWAS1 samples, and individuals missing a Consortium lung phenotype were also excluded. After exclusions, 2,921 GWAS2 samples remained for the analysis. There were 473,514 SNPs present on both the 660W and Omni5 platforms.
Imputation
MaCH/Minimac software (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) was used for phasing and imputing the genotyped data. Phase I, Version 3 haplotype data from 1000 Genomes project (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/) was used as the reference. Approximately 5% of our patients were deemed as not of European ancestry by principal components (Supplementary Fig. 4), so all 1000 Genomes reference samples were included. Samples were imputed separately by genotyping platform and site. Genotyped SNPs with a low minor allele frequency and low call rate were excluded prior to imputation. Imputed SNPs with a MaCH quality score r2<0.30 were excluded from the analysis.
Genotype verification of imputed SNPs
At the HLA locus, six SNPs (rs140348826, rs143609473, rs145621799, rs145182993, rs116003090 and rs115367740) in high linkage disequilibrium with the SNP (rs9268947) with the lowest P value (in the meta-analysis using inverse variance-based weighting) reached genome-wide significance in the meta-analysis. The allele assignments of all seven SNPs were imputed, although some of these SNPs were genotyped on one or more platforms. To evaluate the accuracy of the imputation of these seven SNPs we performed two analyses. First, we compared the genotyping results of one of these SNPs (rs116003090 included on the Omni5 platform) in 374 individuals. Concordance between the imputed allele assignments for this SNP and the 374 genotypes of the SNP was 100%. Second, we obtained whole-genome sequencing results from 94 Canadian CF individuals who were subjects in the meta-GWAS analysis. These 94 subjects had all seven SNPs phased by imputation based on genotyping on the 610 platform. Five of the seven SNPs were called with high confidence in the whole-genome sequence. Of these five SNPs, the concordance rate between the sequencing results and the imputation (rounded to the nearest integer) was 98% (rs143609473), 99% (rs140348826 and rs145182993) and 100% (rs116003090 and rs115367740). These results independently verify the quality of the imputation of five of the seven SNPs at the HLA locus that exceeded genome-wide significance. The remaining imputed SNP (rs9268947) has the lowest P value in the region, but lacks independent genotype confirmation. Thus, rs116003090, the SNP with the most extensive and best concordance with independent genotyping, was selected as the representative associated SNP from the HLA region.
Estimating population structure with principal components
Eigensoft (http://www.hsph.harvard.edu/alkes-price/software/) was utilized to obtain principal components to serve as covariates to adjust for population stratification. Genotyped SNPs common in all platforms with a high minor allele frequency and low linkage disequilibrium were included. Principal components were generated separately for each subgroup. To account for relatives, a maximal set of unrelated individuals were first identified by choosing a random member from each family for principal component analysis. Principal components for the held-out members were then inferred from the loadings. The Tracy–Widom statistic, as computed in Eigensoft, was used to evaluate the statistical significance of each principal component. Individuals were judged to have European ancestry if principal components 1 and 2 fell within a mean±6 s.d. rectangle formed using principal components from the HapMap3 European data set (Supplementary Fig. 4).
Association testing
Genome-wide association was conducted in each of the 13 subgroups. Linear regression was used for the GMS and FrGMC samples; linear mixed modelling was used in the TSS and CGS subgroups to allow the inclusion of siblings in the analysis, accounting for familial correlation by including a random effect for each family. For each subgroup, we tested for interaction between CFTR and modifier loci by adding an indicator variable for p.Phe508del homozygotes versus others then performing a meta-analysis on the interaction term. Within each subgroup analysis, we included sex and significant principal components (based on the Tracy–Widom statistic P<0.05) as covariates in the association models for each SNP. The SNP was coded additively in the model. The SNP-specific beta coefficient and s.e. from each subgroup analysis were used as input for the meta-analysis. Genome-wide significance was defined (P≤1.25 × 10−8)49.
Alternate study design
Subjects were divided into two pools to evaluate our results using a replication-based study design. The North American subjects (n=5,143) constituted a Discovery sample and the French subjects (n=1,222) served as a Replication sample, a strategy employed in a prior publication3. Under this design, four of the five loci (chr3q29, chr5p15, chr6p21, and chr11p12-p13) achieved genome-wide significance (P<5 × 10−8)49 in the North American sample with independent replication in the French subjects (P=8.7 × 10−5, 0.003, 0.054 and 0.003, respectively; Supplementary Table 2). The AGTR2/SLC6A14 locus achieved suggestive evidence of association in the North American cohort (P=9.8 × 10−7) with compelling evidence of replication in the French subjects (1.8 × 10−4). Note that one locus (chr11p12-p13) had previously achieved genome-wide significance in the discovery sample and replication sample in a study of North American subjects3. In the current study, the EHF/APIP locus retains genome-wide significance in subjects from North America and the association is replicated in the 1,222 French subjects (P=0.003).
CFTR gene-based testing
To assess whether variability in lung disease is associated with CFTR genotype within our sample of individuals with severe (pancreatic insufficient) mutations, we used gene-based association analysis. From our imputed SNP set, there were 539 SNPs annotated to CFTR (+/− 10 kb). We constructed a gene-based test statistic in the maximal set of unrelated individuals in each of the 13 subgroups (n=5,762), and restricted to this unrelated set for ease of permutation. To compute the gene-based test statistic, we obtained residuals by regressing the Consortium lung phenotype (KNoRMA) and each SNP genotype on the principal components and sex. The association test statistic between the residualized phenotype and residualized SNP was computed, squared and then summed across the 539 SNPs. We used permutation to obtain subgroup-specific permuted statistics, permuting the residualized phenotype 10,000 times to obtain 10,000 gene-based sum statistics under the null hypothesis of no association to obtain P values for each subgroup, which preserves the linkage disequilibrium structure in the region. The P value was calculated as the proportion of sum statistics from the 10,000 permutations that were more extreme than the observed CFTR sum statistic. Stouffer's Z-score method was used to combine P values from each subgroup and weight them by their respective sample sizes.
Phenotype variation attributable to association
To estimate the proportion of variability in Consortium lung phenotype that is explained by our five significant regions, we conducted an association analysis using the maximum set of unrelated individuals in each subgroup, using the five SNPs from Table 2 in the regression model. Then we calculated an average r2, weighted by sample size.
eQTL assessment for most significantly associated GWAS SNPs
We searched for eQTLs for the top-ranked SNP in each of five loci, and for local SNPs in r2 (LD)>0.6 with the top-ranked GWAS SNPs, using three publicly available databases (Genotype-Tissue Expression, GTEx, lung and whole blood; University of North Carolina at Chapel Hill seeQTL, HapMap LCL and Zeller monocyte; University of Chicago SNP and CNV Annotation Database, SCAN, HapMap CEU LCL). Genes and transcripts within 100 kb of the top-ranked SNPs in five loci were included. Results are presented as nominal P values for each region, except Q values are given for the results from UNC seeQTL (Supplementary Table 3).
GWAS1+2 comparison with prior candidate associations
A literature search was conducted to identify previously published reports of variants associated with some aspect of pulmonary function with a significance value of P<0.05, from association testing in a regression model, (Supplementary Table 4). The gene suggested as likely responsible for the associations was identified, and the chromosome on which it resides. For SNPs, the rs number was identified, along with other nomenclature (aliases) for the variant. A variety of phenotyping methods had been used in these studies, ranging from X-ray and clinical scores to objective measures of lung function, both cross-sectional and longitudinal. The associating phenotypes are summarized under ‘Phenotypes Tested' and the reported P value. The number of subjects analysed and the publication were defined and the P values for any of these SNPs that were tested in this study are given for all subjects (All) and for p.Phe508del homozygotes alone.
Additional information
How to cite this article: Corvol, H. et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 6:8382 doi: 10.1038/ncomms9382 (2015).
Supplementary Material
Supplementary Figures 1-5, Supplementary Tables 1-4 and Supplementary References.
Acknowledgments
National Heart, Lung and Blood Institute 1DP2OD007031, R01HL068890, R01HL095396, R01HL68927; National Institute of Diabetes and Digestive and Kidney Diseases 1R01DK61886-01, K23DK083551, P30DK079637; National Human Genome Research Institute R21HG007840; Cystic Fibrosis Foundation SONTAG07A0, KNOWLE00A0, DRUMM0A00, CUTT00AO, CUTT06PO, COLLAC13IO; PES Clinical Scholars Award, Schwachman (COLLACO09A0); Canadian Institutes of Health Research (MOP 258916), Cystic Fibrosis Canada (CFC; #2626), Genome Canada through the Ontario Genomics Institute (2004-OGI-3-05); Institut National de la Santé et de la Recherche Médicale, Assistance Publique-Hôpitaux de Paris, Université Pierre et Marie Curie Paris, Agence Nationale de la Recherche (R09186DS), DGS, Association Vaincre La Mucoviscidose, Chancellerie des Universités (Legs Poix), Association Agir Informer Contre la Mucoviscidose, GIS-Institut des Maladies Rares. Genome-wide genotyping of subjects in North America provided by the US and CFC Foundations. We thank the US CF Foundation for the use of CF Foundation Patient Registry data. We thank the patients, care providers and clinic coordinators at CF Centers throughout the US and Canada (see list in Supplementary Information) for their contributions to the CF Foundation Patient Registry and Canadian Gene Modifier Study. We thank the following: P-F. Busson, J-F. Vibert, A. Blondel, P. Touche (French study design/patient recruitment); R. Bersie, M. Reske (Children's Hospitals of Wisconsin and Boston data collection); M. Anthony (Denver study design/patient recruitment); P. Diaz, S. Norris (UNC recruitment); A. Infanzon (UNC editorial assistance); H. Kelkar, A. Xu (UNC bioinformatics); W. Wolf (UNC genotyping); Canadian Genome Sequencing Resource for CF; M. Corey, R. Dorfman, A. Sandford, P. Pare, Y. Berthiaume (CGS recruitment); P. Hu (genotype calling/quality control, The Centre for Applied Genomics, The Hospital for Sick Children); K. Naughton, P. Cornwall, B. Vecchio (TSS recruitment/data analysis). We also acknowledge the contributions of our dear colleague, the late Dr. Julian Zielenski.
Footnotes
Author contributions S.M.B., P.Y.B., A.C., G.R.C., H.C., M.L.D., P.R.D., L.G., M.R.K., R.G.P., J.M.R., L.J.S., L.S., M.K.S., J.R.S. and F.A.W. worked on the study design. S.M.B., P.Y.B., G.R.C., H.C., H.L., M.L.D., P.J.G., M.R.K., W.K.O., R.G.P., J.M.R., J.R.S., L.J.S. and F.A.W. were involved in the manuscript preparation. S.M.B., P.Y.B., G.R.C., H.D., M.L.D., P.J.G., J.G., M.R.K., H.L., W.L., J.M.R., L.J.S., F.A.W., and Y.-H.Z. performed the data analysis. F.J.A., S.M.B., A.C., G.R.C., H.C., J.M.C., A.T.D., A.F., K.K., M.R.K., H.L., F.L., W.L., R.G.P., M.V.P., K.S.R., M.K.S., J.R.S. and L.J.S. participated in patient recruitment, sample collection and phenotyping. P.Y.B., H.D., A.T.D., M.V.P., J.M.R. and L.J.S. applied bioinformatics. P.Y.B., S.M.B., H.D., A.F., P.J.G., R.G.P., J.R.S., L.J.S. and F.A.W. performed the genotyping and data cleaning.
References
- Vanscoy L. L. et al. Heritability of lung disease severity in cystic fibrosis. Am. J. Respir. Crit. Care Med. 175, 1036–1043 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral M. D. Novel personalized therapies for cystic fibrosis: treating the basic defect in all patients. J. Intern. Med. 277, 155–166 (2015). [DOI] [PubMed] [Google Scholar]
- Wright F. A. et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat. Genet. 43, 539–546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles M. R. & Drumm M. The influence of genetics on cystic fibrosis phenotypes. Cold Spring Harb Perspect. Med. 2, a009548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. et al. A novel lung disease phenotype adjusted for mortality attrition for cystic fibrosis genetic modifier studies. Pediatr. Pulmonol. 46, 857–869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B. & Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88, 586–598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. & Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32, 227–234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnay P. R. et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat. Genet. 45, 1160–1167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson J. L., Odencrantz J. R. & Fedan K. B. Spirometric reference values from a sample of the general U.S. population. Am. J. Respir. Crit. Care Med. 159, 179–187 (1999). [DOI] [PubMed] [Google Scholar]
- Kulich M. et al. Disease-specific reference equations for lung function in patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 172, 885–891 (2005). [DOI] [PubMed] [Google Scholar]
- Konigshoff M. et al. The angiotensin II receptor 2 is expressed and mediates angiotensin II signaling in lung fibrosis. Am. J. Respir. Cell Mol. Biol. 37, 640–650 (2007). [DOI] [PubMed] [Google Scholar]
- Stanke F. et al. The CF-modifying gene EHF promotes p.Phe508del-CFTR residual function by altering protein glycosylation and trafficking in epithelial cells. Eur. J. Hum. Genet. 22, 660–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R. et al. Modulatory effect of the SLC9A3 gene on susceptibility to infections and pulmonary function in children with cystic fibrosis. Pediatr. Pulmonol. 46, 385–392 (2011). [DOI] [PubMed] [Google Scholar]
- Kesimer M. et al. Molecular organization of the mucins and glycocalyx underlying mucus transport over mucosal surfaces of the airways. Mucosal Immunol. 6, 379–392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron Y. et al. HLA class II polymorphism in cystic fibrosis. A possible modifier of pulmonary phenotype. Am. J. Respir. Crit. Care Med. 159, 1464–1468 (1999). [DOI] [PubMed] [Google Scholar]
- Ali M., Lillehoj E. P., Park Y., Kyo Y. & Kim K. C. Analysis of the proteome of human airway epithelial secretions. Proteome Sci. 9, 4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid C. J., Gould S. & Harris A. Developmental expression of mucin genes in the human respiratory tract. Am. J. Respir. Cell Mol. Biol. 17, 592–598 (1997). [DOI] [PubMed] [Google Scholar]
- Button B. et al. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science 337, 937–941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C. M., Brant S. R., Walker M. S., Pouyssegur J. & Donowitz M. Cloning and sequencing of a rabbit cDNA encoding an intestinal and kidney-specific Na+/H+ exchanger isoform (NHE-3). J. Biol. Chem. 267, 9340–9346 (1992). [PubMed] [Google Scholar]
- Orlowski J., Kandasamy R. A. & Shull G. E. Molecular cloning of putative members of the Na/H exchanger gene family. cDNA cloning, deduced amino acid sequence, and mRNA tissue expression of the rat Na/H exchanger NHE-1 and two structurally related proteins. J. Biol. Chem. 267, 9331–9339 (1992). [PubMed] [Google Scholar]
- Bradford E. M., Sartor M. A., Gawenis L. R., Clarke L. L. & Shull G. E. Reduced NHE3-mediated Na+ absorption increases survival and decreases the incidence of intestinal obstructions in cystic fibrosis mice. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G886–G898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. et al. Multiple apical plasma membrane constituents are associated with susceptibility to meconium ileus in individuals with cystic fibrosis. Nat. Genet. 44, 562–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. Unraveling the complex genetic model for cystic fibrosis: pleiotropic effects of modifier genes on early cystic fibrosis-related morbidities. Hum. Genet. 133, 151–161 (2014). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E. & Macara I. G. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15, 225–242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymut S. M. et al. Reduced microtubule acetylation in cystic fibrosis epithelial cells. Am. J. Physiol. Lung. Cell Mol. Physiol. 305, L419–L431 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond M. J. et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat. Genet. 44, 886–889 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock D. B. et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 8, e1003098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontakioti E., Domvri K., Papakosta D. & Daniilidis M. HLA and asthma phenotypes/endotypes: a review. Hum. Immunol. 75, 930–939 (2014). [DOI] [PubMed] [Google Scholar]
- Chauhan B. et al. Evidence for the involvement of two different MHC class II regions in susceptibility or protection in allergic bronchopulmonary aspergillosis. J. Allergy Clin. Immunol. 106, 723–729 (2000). [DOI] [PubMed] [Google Scholar]
- Muro M. et al. HLA-DRB1 and HLA-DQB1 genes on susceptibility to and protection from allergic bronchopulmonary aspergillosis in patients with cystic fibrosis. Microbiol. Immunol. 57, 193–197 (2013). [DOI] [PubMed] [Google Scholar]
- Knutsen A. P. & Slavin R. G. Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin. Dev. Immunol. 2011, 843763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal W. K. et al. Gene expression in transformed lymphocytes reveals variation in endomembrane and HLA pathways modifying cystic fibrosis pulmonary phenotypes. Am. J. Hum. Genet. 96, 318–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao X., Li X., Lerea K. M. & Olson S. C. Angiotensin II type 2 receptor-dependent increases in nitric oxide synthase expression in the pulmonary endothelium is mediated via a G alpha i3/Ras/Raf/MAPK pathway. Am. J. Physiol. Cell Physiol. 292, C2185–C2196 (2007). [DOI] [PubMed] [Google Scholar]
- Wagenaar G. T. et al. Angiotensin II type 2 receptor ligand PD123319 attenuates hyperoxia-induced lung and heart injury at a low dose in newborn rats. Am. J. Physiol. Lung. Cell Mol. Physiol. 307, L261–L272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossum S. L. et al. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 42, 13588–13598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko D. C. et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc. Natl Acad. Sci. USA 109, E2343–E2352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W. et al. Structural and biochemical basis for the inhibition of cell death by APIP, a methionine salvage enzyme. Proc. Natl Acad. Sci. USA 111, E54–E61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda M. et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl Acad. Sci. USA 105, 1620–1625 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G. et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl Acad. Sci. USA 105, 11869–11874 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D. E. et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L. R. & Ramsey B. W. Cystic fibrosis therapeutics: the road ahead. Chest 143, 207–213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R. et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J. Clin. Invest. 118, 1040–1049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm M. L. et al. Gene modifiers of lung disease in cystic fibrosis. N. Engl. J. Med. 353, 1443–1453 (2005). [DOI] [PubMed] [Google Scholar]
- Corvol H. et al. Ancestral haplotype 8.1 and lung disease severity in European cystic fibrosis patients. J. Cyst. Fibros. 11, 63–67 (2012). [DOI] [PubMed] [Google Scholar]
- Li M. X., Yeung J. M., Cherny S. S. & Sham P. C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131, 747–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-5, Supplementary Tables 1-4 and Supplementary References.