Abstract
Acute kidney injury (AKI) is a common, serious complication of cardiac surgery. Since prior studies have supported a genetic basis for postoperative AKI, we conducted a genome-wide association study (GWAS) for AKI following coronary bypass graft (CABG) surgery. The discovery dataset consisted of 873 non-emergent CABG surgery patients with cardiopulmonary bypass (PEGASUS), while a replication dataset had 380 cardiac surgical patients (CATHGEN). Single nucleotide polymorphism (SNP) data were based on Illumina Human610-Quad (PEGASUS) and OMNI1-Quad (CATHGEN) BeadChips. We used linear regression with adjustment for a clinical AKI risk score to test SNP associations with the postoperative peak rise relative to preoperative serum creatinine concentration as a quantitative AKI trait. Nine SNPs meeting significance in the discovery set were detected. The rs13317787 in GRM7|LMCD1-AS1 intergenic region (3p21.6) and rs10262995 in BBS9 (7p14.3) were replicated with significance in the CATHGEN data set and exhibited significantly strong overall association following meta-analysis. Additional fine-mapping using imputed SNPs across these two regions and meta-analysis found genome wide significance at the GRM7|LMCD1-AS1 locus and a significantly strong association at BBS9. Thus, through an unbiased GWAS approach, we found two new loci associated with post-CABG AKI providing new insights into the pathogenesis of perioperative AKI.
Keywords: acute kidney injury, coronary artery bypass graft surgery, GWAS, BBS9
Introduction
Acute kidney injury (AKI), as reflected by systemic accumulation of nitrogenous waste products due to impaired plasma filtration (e.g., creatinine and blood urea nitrogen), occurs in a variety of clinical scenarios where it is consistently associated with poor outcome.1 The postoperative period represents an ideal setting for epidemiological investigation of AKI, since up to 47% of all in-hospital episodes follow surgery.2 Particularly, cardiac surgery is the most common etiology of postoperative AKI,3 with an incidence ranging between 5 and 30% following coronary artery bypass graft (CABG) surgery.4–6 Escalating degrees of AKI are closely associated with more complicated postoperative courses, including increased in-hospital mortality rates, and (for survivors) needs for intensive and post-discharge supportive care, hospital readmissions, and poorer subsequent quality of life and long-term survival.7–10
Numerous AKI risk factors have been identified in cardiac surgery cohorts, including advanced age, obesity, chronic kidney disease (CKD), diabetes, poor ventricular function, hypertension, embolic and inflammatory processes, and specific surgery-related interventions (e.g., intra-aortic balloon counter pulsation or the use of cardiopulmonary bypass).11–17 Nonetheless, current risk models poorly explain observed variability in AKI occurrence.18
Beyond traditional clinical risk factors, a genetic predisposition for postoperative AKI has been suggested by previous candidate gene studies.19 To date, association studies of post-CABG AKI have mostly focused on selected candidate genes that modulate inflammatory and vasomotor responses to injury, including functional alleles influencing cytokine production that can cause renal tubular and microvascular damage,20–27 but are limited by marked heterogeneity of AKI phenotype definitions, lack of power, and poor reproducibility. Family and linkage studies, although impractical as tools for the study of perioperative AKI, demonstrate impaired glomerular filtration (GFR) to be a heritable trait,28, 29 supporting the heritability of renal dysfunction in general. However, a heritability index has not been specifically assessed for AKI. Similarly, although several genome-wide association studies (GWAS) have identified susceptibility loci for indices of renal function (estimated GFR) and CKD,30–32 comparable studies are lacking for AKI in general and following cardiac surgery in particular. We therefore conducted a GWAS among participants from Perioperative Genetics and Safety Outcome Study (PEGASUS), followed by independent replication using data from CATHeterization GENetics (CATHGEN) study at Duke Heart Center to identify common genetic variants that show association with risk of developing AKI following cardiac surgery with cardiopulmonary bypass (CPB).
Results
Descriptive statistics for demographic and clinical variables and comparisons between the two datasets are presented in Table 1. The discovery (PEGASUS) and replication (CATHGEN) datasets consisted of 873 and 380 subjects of self-reported European ancestry, respectively; CATHGEN had more females than PEGASUS (38.4% vs. 23.6%, p<0.001). Postoperative AKI was common and occurred at similar rates in both cohorts, as reflected by the relative increase in serum creatinine concentration from baseline (preoperative) to peak values within the first ten days after surgery expressed as a percentage rise (%ΔCr),14 which averaged 22.5 (standard deviation, SD=35.9) for PEGASUS and 23.6 (SD=37.0) for CATHGEN, respectively (p=0.6). This is further supported by similar AKI case rates as defined by AKIN, RIFLE, and KDIGO criteria. Finally, severe AKI (KIDGO stage 3) complicated the postoperative courses of 16 (1.2%) patients in the PEGASUS and 6 (1.6%) in the CATHGEN cohort.33 Although the prevalence of baseline CKD was marginally higher in the PEGASUS cohort, serum creatinine concentrations and estimated glomerular filtration rates (eGFR) were similar between groups both at baseline (preoperative) and postoperatively. Notably, the types of cardiac surgical procedures were different between the two datasets (p<0.001), with all patients in the discovery cohort undergoing isolated CABG, whereas 29% of patients in the replication cohort had concomitant valve surgery. Additional differences in comorbidities between the two cohorts included higher prevalence of congestive heart failure, hypertension, and hypercholesterolemia in CATHGEN. Consequently, the average clinical AKI risk score was significantly higher in the replication than discovery cohort (32.1±6.8 vs. 26.3±12.6, p< 0.001, Table 1).
Table 1.
Patient, Renal and Procedural Characteristics
| Discovery cohort (PEGASUS N=873) |
Replication cohort (CATHGEN N=380) |
p-value | |
|---|---|---|---|
| Patient characteristics | |||
| Demographic variables | |||
| Age (years) | 63.8 (10.1) | 62.9 (11.0) | 0.175 |
| Body weight (kg) | 86.7 (18.7) | 87.2 (20.2) | 0.689 |
| Female sex | 206 (23.6%) | 146 (38.4%) | <0.001 |
| Comorbidities | |||
| Left ventricular ejection fraction (%) | 55.3 (14.3) | 55.6 (17.7) | 0.776 |
| Angina | 495 (58.0%) | 138 (42.6%) | <0.001 |
| Arrhythmia | 189 (22.0%) | 52 (16.1%) | 0.023 |
| Congestive heart failure | 97 (11.3%) | 111 (29.5%) | <0.001 |
| Chronic obstructive lung disease | 45 (6.6%) | 31 (8.2%) | 0.341 |
| Diabetes | 266 (30.7%) | 136 (36.0%) | 0.078 |
| History of hypertension | 438 (50.2%) | 288(75.8%) | <0.001 |
| Hypercholesterolemia | 488 (57.2%) | 267 (70.8%) | <0.001 |
| Previous myocardial infarction | 381 (43.9% | 135 (36.2%) | 0.011 |
| Peripheral vascular disease | 87 (10.2%) | 38 (10.0%) | 0.915 |
| Smoking history | 394(46.2%) | 220 (58.1%) | <0.001 |
| Chronic kidney diseasea,b | 199 (22.8%) | 107 (28.2%) | 0.042 |
| Procedural variables | |||
| Duration of CPB (min) | 114.7 (36.3) | 141.5 (51.8) | <0.001 |
| Duration of aortic cross-clamping (min) | 64.0 (26.7) | 79 (34.2) | <0.001 |
| Blood transfusionc | 368 (42.2%) | 241 (63.4%) | <0.001 |
| Intraoperative balloon counterpulsation | 32 (3.8%) | 31 (9.6%) | <0.001 |
| Concomitant valve surgery | 0 | 110 (29.0%) | <0.001 |
| Clinical AKI risk scored | 26.3 (12.6) | 32.1 (6.8) | <0.001 |
| Markers of renal function | |||
| Serum creatinine concentrations (mg/dL) | |||
| baseline preoperative | 1.06 (0.46 | 1.06 (0.32) | 0.893 |
| peak postoperative | 1.28 (0.57) | 1.30 (0.51) | 0.711 |
| eGFRcrea (ml/min/1.73m2)b | |||
| baseline preoperative | 73.7(18.1) | 71.9 (19.9) | 0.142 |
| nadir postoperative | 62.0 (19.6) | 60.2 (20.7) | 0.144 |
| AKI criteriae | |||
| Peak relative to baseline | 22.5 (35.9) | 23.6 (37.0) | 0.621 |
| creatinine %ΔCr (%) | |||
| KDIGO | 294 (33.7%) | 119(381.3%) | 0.139 |
| AKIN | 290 (33.2%) | 115 (30.3%) | 0.221 |
| RIFLE | 149 (17.1%) | 64(16.8%) | 0.333 |
| KDIGO AKI Stage | 0.612 | ||
| Stage 1 | 69 (7.9%) | 29 (7.6%) | |
| Stage 2 | 29 (3.3%) | 11 (2.9%) | |
| Stage 3 | 10 (1.2%) | 6 (1.6%) |
Results presented as mean (standard deviation) for continuous variables, and frequency (percentage) for categorical variables.
Abbreviations: CPB – cardiopulmonary bypass; CABG – coronary artery bypass grafting; AKI – acute kidney injury; eGFRcrea – estimated glomerular filtration rate (creatinine) based on the CKD-EPI equation; AKIN – Acute Kidney Injury Network;37 RIFLE - risk, injury, failure, loss, end-stage kidney disease;38 KDIGO - Kidney Disease: Improving Global Outcomes.39
Chronic kidney disease defined as baseline eGFR < 60 ml/min/1.73m2 (modified KDIGO criteria, lacking the 3 month preoperative window)
eGFR was computed based on CKD-EPI equation
Transfusion defined as receipt of any blood transfusion perioperatively.
- −2.59207 −7.72486 (Preop creatinine) + 0.30737(weight) + 0.14174 (cross-clamp time) + 16.35924 (transfusion) −9.06373 (hypertension).
Reflect only serum creatinine criteria (i.e., lack oliguria criteria).
Association results
The genome wide association results from the discovery cohort are depicted as Manhattan and quantile-quantile (QQ) plots, which showed good adherence to null expectations (Figure S1-A and -B). Nine single nucleotide polymorphisms (SNPs), located in seven loci, showed promising association with %ΔCr (p<10−5) from the GWAS and were brought forward for replication (Table 2). Two of these SNPs showed nominal significant associations with %ΔCr in the replication cohort - rs13317787 (p=0.02) at 3p21.6 (intergenic region between GRM7|LMCD1-AS1), and rs10262995 (p=0.03) at 7p14.3 (located in BBS9), with allelic effects on %ΔCr in the same direction as observed in the discovery cohort. The overall association results derived from meta-analysis of both datasets revealed strong association with AKI for both rs13317787 (meta-p=5.35×10−7) and rs10262995 (meta-p= 2.24×10−7), close to commonly accepted genome wide significance levels (p< 5×10−8). The heterogeneity I2 between the two datasets at these two SNPs was not significant (Table 2: I2=0).
Table 2.
Summary of SNPs Selected in the Discovery Cohort for Replication
| Discovery cohort: PEGASUS | Replication cohort: CATHGEN | Combined dataset | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Base Pair | Gene * | allele | MAF | Effect | SE | P | MAF | Effect | SE | P | Effect | SE | I2 | meta-P |
| 1 | rs2352039 | 78819945 | MGC27382 | A | 0.156 | 10.07 | 2.26 | 9.78×10−6 | 0.168 | 6.23 | 3.43 | 0.070 | 8.90 | 1.89 | 0 | 2.45×10−6 |
| 3 | rs13317787 | 8141952 | GRM7 | LMCD1-AS1 | A | 0.028 | 21.56 | 4.84 | 9.67×10−6 | 0.02 | 22.04 | 9.56 | 0.022 | 21.66 | 4.32 | 0 | 5.35×10−7 |
| 7 | rs10262995 | 33550041 | BBS9 | A | 0.087 | 14.33 | 2.98 | 1.83×10−6 | 0.099 | 9.51 | 4.47 | 0.034 | 12.84 | 2.48 | 0 | 2.24×10−7 |
| 12 | rs2248098 ** | 48253356 | VDR | A | 0.488 | −7.48 | 1.60 | 3.60×10−6 | 0.474 | −0.21 | 2.83 | 0.942 | −5.71 | 1.40 | 80 | 4.22×10−5 |
| 16 | rs1109836 | 57660346 | GPR56 | A | 0.013 | 37.57 | 7.46 | 5.67×10−7 | 0.012 | −11.32 | 12.33 | 0.36 | 24.48 | 6.38 | 91.3 | 0.0001 |
| 18 | rs8086030 ** | 9750395 | RAB31 | A | 0.420 | 7.64 | 1.69 | 7.33×10−6 | 0.438 | −1.07 | 2.83 | 0.705 | 5.35 | 1.45 | 85.7 | 0.0002 |
| 18 | rs8099036 ** | 9756056 | G | 0.389 | 8.47 | 1.71 | 9.01×10−7 | 0.397 | −0.44 | 2.92 | 0.880 | −6.19 | 1.48 | 85.5 | 2.74×10−5 | |
| 21 | rs2831026 | 28969040 | LOC100288252 | G | 0.289 | 8.95 | 1.82 | 1.11×10−6 | 0.263 | −5.39 | 3.08 | 0.081 | −5.22 | 1.57 | 93.8 | 0.0009 |
| 21 | rs1551588 | 28982407 | | NCRNA00113 | A | 0.165 | 11.86 | 2.20 | 9.14×10−8 | 0.147 | −7.52 | 3.73 | 0.044 | 6.85 | 1.90 | 95 | 0.0003 |
Abbreviations: SNP - single nucleotide polymorphism; Chr – chromosome; MAF – minor allele frequency; Effect – the beta coefficient of linear regression; SE – standard error; I2 – heterogeneity; PEGASUS, CATHGEN – see text.
Intergenic regions are identified by the two flanking genes separated by “|”.
Imputed SNPs in the CATHGEN cohort.
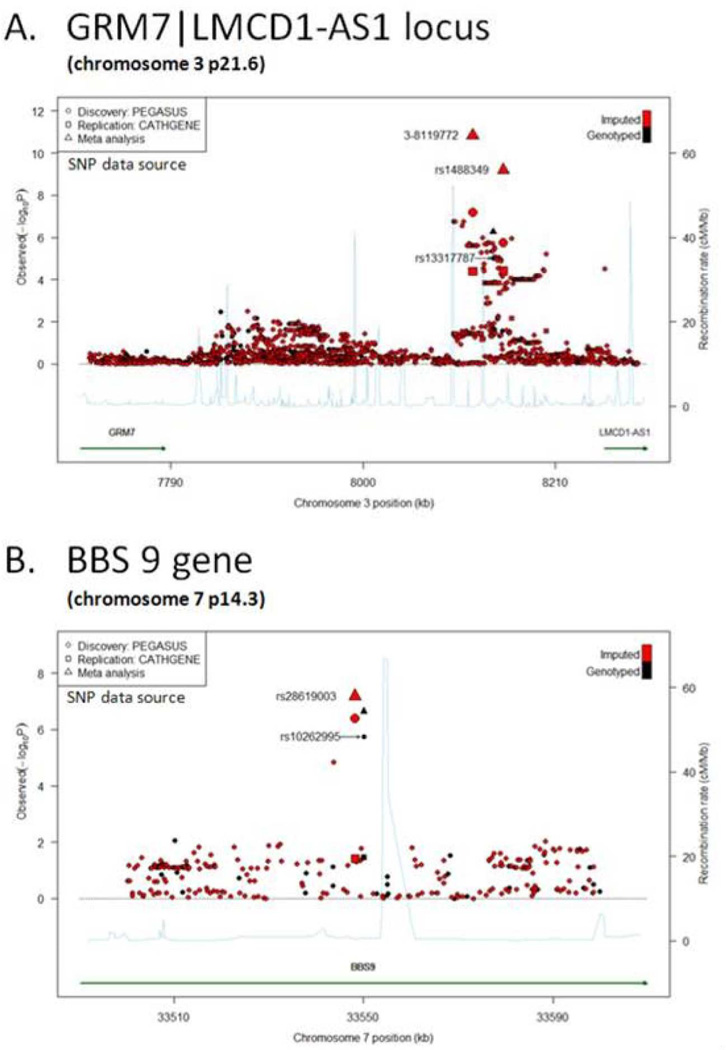
To provide a high-resolution overview of the association signal across the 3p31.6 and 7p14.3 loci, we performed in silico fine-mapping by imputing the untyped SNPs on chr3: 6,907,193–8,537,944 (for the GRM7 to LMCD1-AS1 region), and chr7: 33,173,404–33,639,870 (for the BBS9 region), respectively. Among 2029 genotyped and imputed SNPs at 3p31.6, 44 including the initially identified rs13317787 (spanning from chr3: 8,099,146–8,161,987) met discovery criteria (p<10−5), and 17 of these reached genome-wide significance in meta-analysis (meta-p < 5×10−8). The most significant SNP (meta-p=2.49×10−11) is an un-named SNP located at chr3:8,119,772 (SNP 3-8119772; Figure 1A; Table S1), which is also in strong linkage disequilibrium (LD) (r2=0.97) with rs1488349 (chr3: 8,153,260), the second most significant SNP in meta-analysis (meta-p=5.41×10−10) (Table S1). Since minor allele frequencies for 3-8119772 and rs1488349 are relatively low (between 1% and 3%), we also conducted permutation tests with 106 repeats to obtain empirical p-values (min empirical p=4.07×10−5 for 3-8119772, Table S1). Furthermore, all other top SNPs (43 SNPs) at 3p31.6 were highly correlated with SNP 3-8119772 (r2=0.52–0.77), including rs13317787 (r2=0.65) the initial SNP identified from GWAS (Table S1). Fine-mapping of the BBS9 region identified one additional imputed SNP (rs28619003; chr7:33548225), in complete LD with the original top SNP rs10262995 (r2=1), which also approached genome-wide significance after meta-analysis (meta-p=6.51×10−8) (Figure 1B, Table S1).
Figure 1.
Regional association plot for (A) chr3p21.6 locus (GRM7|LMCD1-AS1) and (B) BBS9 gene, presenting – log10(p-values) from the discovery (PEGASUS) and replication (CATHGEN) datasets, as well as the meta-analysis. Directly genotyped SNPs are plotted in black; imputed markers are plotted in red.
Further analysis of the relationship of identified loci with AKI
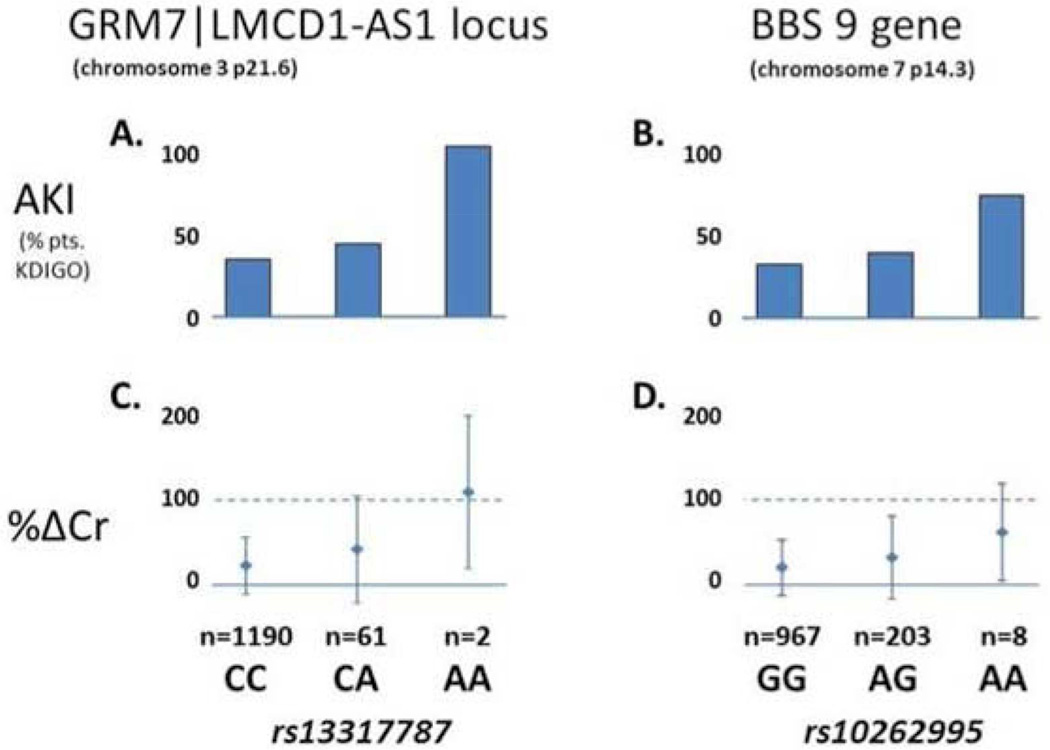
To further assess the clinical relevance of the identified loci, we estimated the AKI incidence and severity observed with variation in the chromosomal regions of interest using the original genotyped SNPs rs13317787 and rs10262995 as representative tag SNPs in the combined dataset (N=1,253). For both SNPs, AKI incidence increased with each additional copy of the minor allele (Figure 2). Average %ΔCr (SD) for rs13317787 was 21.8% (0.34) for the CC genotype, 40.5% (0.63) for CA, and 108.0% (0.90) for AA. Similarly, for rs10262995, average %ΔCr (SD) was 20.6% (0.32), 32.4% (0.49), and 62.1% (0.57) for CC, CA, and AA genotypes, respectively.
Figure 2.
Comparative graphical representation of genotypic effects of rs13317787 at the chr3p21.6 locus (A&C) and rs10262995 in BBS9 (B&D) on incident post-cardiac surgery acute kidney injury (AKI) – defined using either the KDIGO criteria56 or peak postoperative serum creatinine increase (%ΔCr). The dashed line represents a 2-fold (100%) increase in serum creatinine from baseline, approximately equivalent to a 50% reduction in glomerular filtration rate.
We also evaluated the ability of two SNPs with strongest association signals (rs1488349 in GRM7|LMCD1-AS1 and rs28619003 in BBS9 regions) to predict inter-individual variability in %ΔCr. When jointly added to the patient-specific clinical AKI risk score, the two loci explain roughly double the %ΔCr variance (r2: 9.7% vs. 4.9% in the discovery cohort, and 9% vs 3.6% in the replication cohort, Table 3). The improved r2, corroborated by two commonly used global measures of relative model fit like the Akaike information criterion (AIC) and Bayesian information criterion (BIC), both demonstrating reduced (albeit modestly) values (differences of 39.5 and 39.5 for AIC and BIC, respectively, Table 3) support the superior performance of the clinical-genomic model as a postoperative AKI risk stratification tool to potentially individualize reno-protective interventions.
Table 3.
Comparison of Clinical and Clinico-genomic AKI Predictive Models with and without Inclusion of the top SNP in each region (rs1488349 and rs28619003) for two datasets
| Dataset | Model variables | beta(SE) | P | Model p | r2 | AIC | BIC |
|---|---|---|---|---|---|---|---|
| Discovery Cohort: PEGASUS | Clinical Model | <0.0001 | 0.049 | 6029.55 | 6031.37 | ||
| AKI risk score | 0.64 (0.10) | 7.3×10−11 | |||||
| Clinico-Genomic Model | <0.0001 | 0.097 | 5990.02 | 5992.06 | |||
| AKI risk score | 0.60(0.09) | 2.43×10−10 | |||||
| rs1488349 | 28.81(6.24) | 4.50×10−6 | |||||
| rs28619003 | 13.37(3.05) | 1.34×10−5 | |||||
| Replication Cohort: CATHGEN | Clinical Model | 0.001 | 0.036 | 2706.96 | 2708.70 | ||
| AKI risk score | 1.13(0.31) | 3.2×10−4 | |||||
| Valve procedure | 10.83(4.64) | 0.02 | |||||
| Clinico-Genomic Model | <0.0001 | 0.090 | 2691.26 | 2708.77 | |||
| AKI risk score | 1.22(0.30) | 6.76×10−5 | |||||
| Valve procedure | 10.84(4.52) | 0.017 | |||||
| rs1488349 | 50.58(11.98) | 3.06×10−5 | |||||
| rs28619003 | 9.14(4.43) | 0.04 | |||||
Abbreviations: SNP - single nucleotide polymorphism; AKI – acute kidney injury, Chr – chromosome; MAF – minor allele frequency; Beta – regression coefficient; SE – standard error; Model p – p-value for the corresponding model; AIC -- Akaike information criterion; BIC -- Bayesian information criterion; PEGASUS, CATHGEN – see text.
Discussion
In this study, we present a genome-wide analysis to screen genetic variants associated with AKI following CABG surgery with CPB. Using a discovery-replication analysis approach involving independent cardiac surgical cohorts and a continuous variable (%ΔCr) to reflect AKI severity, we describe two novel susceptibility loci: the first with genome-wide significant association is located in the intergenic region GRM7|LMCD-AS1 (chr3p21.6; lowest meta-p= 2.49×10−11), and the second at the boundary of genome wide significance in the Bardet-Biedl syndrome 9 (BBS9) gene (chr7p14.3; min meta-p=6.51×10−8). Patients carrying one or both of the minor alleles at these loci show an incremental increase in risk of incident AKI, even after accounting for currently known clinical AKI risk factors. We believe this is the first such analysis and speculate that our findings have uncovered a novel predictive tool to improve individualized AKI risk stratification, which may also provide pathophysiologic clues to better investigate and prevent postoperative AKI.
Although no previous GWAS for postoperative AKI is available for comparison, a survey of the two risk loci for links with renal disorders is warranted. At the 3p21.6 locus, an intergenic region bounded by GRM7 (glutamate receptor, metabotrophic 7) and LMCD1 (LIM and cysteine rich domains protein 1, dyxin) genes, our study identified a 62.8kb peak region highly associated with AKI (17 genome wide significant SNPs in Table S1, Figure 1). No direct functional roles are currently attributed to this intergenic region. However, SNPs in this region were located within active regulatory elements based on ENCODE ChIP-Seq and DNase-Seq data in RegulomeDB34. Additionally, HaploReg35 lists rs1488349 as located within a hypothetical gene, AC018832.1 (based on GENCODE data), or at the 3’ end of LMCD1 antisense RNA 1 (LMCD1-AS1), a non-coding RNA (based on the RefSeq data). LMCD1 is a member of the LIM-domain family of zinc finger proteins, abundantly expressed in kidney tissue, and functionally involved in protein-protein interactions with transcriptional co-repressor activity (MIM*604859) (http://www.ncbi.nlm.nih.gov/omim), including regulation of the calcineurin-NFAT signaling cascade known to play a critical role in recovery from AKI.36 Our literature review did not identify direct functional links between the GRM7|LMCD-AS1 intergenic region and AKI pathophysiology, thus future studies are needed to uncover its potential regulatory roles.
Most interestingly, our second risk locus involves a peak 1.8kb region in BBS9 (MIM*607968), also known as parathyroid hormone-responsive B1 gene (PTHB1), named for its relationship with Bardet-Biedl syndrome (BBS, MIM*209900). Kidney disease is a key feature and major source of early mortality with BBS.37, 38 Approximately 10% of children and adolescents with BBS have end-stage renal disease, and 25% of surviving patients have CKD by their fifth decade. Almost all BBS patients have renal structural defects, and while renal glomerular abnormalities are rare, one-third of patients have vasopressin-resistant urinary concentration defects. A BBS9 translocation is also associated with the most common pediatric renal malignancy, Wilm’s tumor.39
BBS is a genetically heterogeneous multiorgan ciliopathy of non-motile cilia that includes mutant variants of their anchoring structure or “BBSome” (also known as the basal body).40 In the kidney, BBS9 proteins are expressed in focal adhesions and play a central role in controlling cilia length through regulation of actin cytoskeleton polymerization.41 While the exact role of BBS9 within the BBSome remains unknown, the protein is conserved across species, highly expressed in adult human kidney,38 and approximately 6% of BBS cases involve BBS9 mutations.42–44 Although these mutations are not available in our SNP panels, the two BBS9 intronic variants identified in this study (rs10262995 and rs28619003) were part of an LD block located immediately upstream of a recombination hotspot in intron 20 (Figure S2).
Non-motile or primary cilia (containing BBS9) are solitary apical appendages found on most cells in the body that function as signal transduction antennae. Renal primary cilia act as mechanosensors that protrude into the nephron lumen from tubule and collecting duct epithelial cells, and are necessary for water absorption in the kidney.45 Critical drops in tubular flow, such as occur with AKI, are sensed by the cilia and activate cell proliferation, presumably to promote renal recovery.46, 47 Following ischemia/reperfusion induced AKI, renal primary cilia undergo predictable morphologic changes, as observed in kidney transplant patients and animal experiments; these include a doubling in length over the first 7 days, and return to normal size over weeks as recovery occurs.48 Collectively, these observations suggest that further investigation of the potential mechanistic involvement of BBS9 in postoperative AKI pathogenesis is warranted.
While this report expands the investigation of cardiac surgery-associated AKI from candidate gene to an unbiased GWAS approach, several limitations remain. First, although power estimates indicate that our sample sizes (N=873 in discovery alone or N=1,253 for the combined two datasets) can reach 80% power to detect SNPs in similar ranges of MAF (0.03–0.09) and proportion of %ΔCr variation ( to 4.4%) as observed in this study (see Supplementary Materials), we may still miss potential susceptibility variants with smaller effect sizes. A more powerful study could also have been achieved through refinements to our AKI phenotype and clinical risk score incorporating additional variables such as preoperative albuminuria, acute decline in renal function over the months prior to and following surgery, and perioperative use renin-angiotensin system inhibitors, data which was not available for all patients. Further, our results would have been bolstered if a validation cohort from another institution were included in the analysis. Nonetheless, baseline to peak postoperative serum creatinine concentration increase is highly validated as a marker of adverse outcome following cardiac surgery in studies from numerous institutions, and variables identified for inclusion in our clinical renal risk score are similar to those from previous studies.23 Of note, sensitivity analyses revealed that the top two variants identified (rs13317787 and rs10262995, Table 2) remain associated with postoperative AKI as defined using the standard KDIGO criteria, albeit at nominal significance levels (p=0.05 and 0.03, respectively). The weaker association signal, likely reflecting a limited number of cases and controls (294 vs. 579) available for association analysis using the dichotomous KDIGO AKI phenotype, supports however our primary findings using %ΔCr as a continuous phenotype.
Second, markers at the 3p21.6 locus are rare (minor allele frequencies between 1–3%) and, although supported by empirical p-values, a larger sample size would increase confidence in this finding. Combining strict genotype QC criteria with visual inspection of cluster plots for rs13317787 revealed well-separated genotype clusters (Figure S3), thus confirming accurate genotype calling for this rare maker. Although we used an imputation method to refine the two most significant association loci, this strategy is not designed toward rare functional variants. These concerns notwithstanding, our findings could form the basis of a genetic pre-operative risk stratification tool which, by individually assessing risk alleles of rs13317787 and rs10262995 in the current samples, would have identified 2.3% and 7.4% of cardiac surgery patients, respectively, to have considerably elevated AKI risk (1.5–5 fold greater rise in serum creatinine) relative to non-carriers. As preliminary evidence for increased predictive ability, genotype information at the two loci improves the performance of a patient-specific clinical risk score for postoperative AKI, with the clinico-genomic model explaining a higher proportion of variability in the primary AKI phenotype (%ΔCr) and showing improved (albeit modestly) relative fit as evidenced by lower AIC and BIC. Such a genetic susceptibility biomarker for postoperative AKI would be useful not only to assist clinical decision-making, but also to aid researchers in identifying candidates for evaluation of promising reno-protective interventions.
Finally, although we provide intriguing indirect evidence for possible mechanistic roles of the observed risk loci, our study offers no direct functional analysis to validate the GWAS hits. Such interpretation of these putative noncoding regulatory variants could entail gene expression, eQTL, and allelic imbalance analyses in animal models. The limited availability of well-characterized animal models of post-cardiac surgery AKI represents a possible obstacle, and may require the use of experimental models of acute renal ischemia-reperfusion injury, which in addition to the significant pathobiological differences reported between common forms of human AKI and rodent models, would limit clinical relevance and translatability to a cardiac surgical population.
As AKI is a significant complication of cardiac surgery that contributes to perioperative mortality and medical cost, a better understanding of its risk factors is important. Our comprehensive study design presented here has pinpointed two novel susceptibility loci (chr3p21.6 and BBS9) for AKI after cardiac surgery with CPB. The conclusion of this report will provide candidate regions for future genetic research on cardiac surgery-associated AKI, and may eventually lead to improved preoperative screening, novel prevention and intervention options to reduce AKI and associated morbidity and mortality.
Materials and Methods
Study design
We utilized two independent cohorts, from the PEGASUS and the CATHGEN studies at Duke Heart Center, to conduct initial common variant discovery by GWAS followed by replication analysis of top candidate single nucleotide polymorphisms (SNPs), respectively. Both PEGASUS and CATHGEN studies were approved by the Duke University School of Medicine Institutional Review Board, and all patients provided informed consent. Our study was performed in accordance with the Declaration of Helsinki and followed the “Strengthening the Reporting of Genetic Association Studies” (STREGA) recommendations.49 For this AKI substudy, patients in the discovery cohort were participants in the ongoing PEGASUS longitudinal study, and underwent isolated non-emergent CABG surgery with CPB between 1997 and 2006.50 Patients were excluded from enrollment in PEGASUS if they had a history of end-stage renal disease, active liver disease, bleeding disorders, autoimmune diseases, or immunosuppressive therapy. An a priori decision was made to limit the analyses to subjects of self-reported European ancestry, justified by the limited number of non-Caucasian patients in the PEGASUS GWAS dataset and to avoid potential confounding from population admixture given previous reports identifying self-reported race as an independent predictor of postoperative AKI.7, 23 After applying quality control criteria (see below) and excluding patients with missing genotypes or phenotypic information, 873 subjects of self-reported European ancestry met eligibility for genome-wide association analysis.
The replication dataset consists of 380 subjects of self-reported European ancestry from the CATHGEN study,51, 52 who underwent non-emergent CABG with or without concomitant valve surgery using CPB between 2006 and 2010. Similarly, CATHGEN enrollees with end-stage renal disease prior to surgery were excluded from this substudy. Patient and procedural characteristics for both cohorts were collected and curated from the Duke Information System for Cardiovascular Care, an integral part of the Duke Databank for Cardiovascular Disease.
Primary phenotype and clinical data
Daily serum creatinine concentrations considered in this study were those routinely measured at baseline (preoperative) and up to 10 postoperative days at a single core hospital laboratory for both the discovery and validation datasets. The primary study outcome was a continuous AKI endophenotype, the percentage change of the highest postoperative serum creatinine from the baseline preoperative concentration (%ΔCr).14 For each patient, %ΔCr reflects a gross approximation of the maximum relative loss of renal function. For example, a postoperative serum creatinine doubling (100% rise) approximates a 50% acute functional nephron loss. Notably, in this study we selected %ΔCr (as with our previous studies23, 53) in preference to standard dichotomous definitions of AKI (e.g., KDIGO, AKIN and RIFLE criteria54–56), which include thresholds for relative creatinine rise (e.g., 50%) that closely resemble %ΔCr. Considering that dichotomous outcomes are known not to be as informative as continuous outcomes, the rationale for using %ΔCr as a quantitative AKI trait was to enhance the ability and power to identify risk variants,57 as evidenced by previously reported GWAS of other continuous renal traits such as eGFR30, 58 and serum creatinine59 in ambulatory populations. Importantly, %ΔCr reflects a spectrum of injury to the kidneys that often does not meet the dichotomous AKI threshold criteria; even small relative rises in serum creatinine are associated with substantial reductions in post-operative event-free survival.60, 61
Multiple preoperative and postoperative clinical measures were collected including patient characteristics, procedural variables, and information related to renal function (Table 1). Descriptive statistics for serum creatinine concentrations, estimated glomerular filtration rate (eGFR) based on the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) equation62, as well as alternative definitions of AKI (KDIGO, AKIN, RIFLE) are provided for both baseline (preoperative) and peak postoperative measures. Further, a patient-specific clinical AKI risk score was computed based on a previously developed multivariable model that used traditional clinical and procedural risk factor in a large contemporaneous consecutive cohort (N=10,708) of non-emergent CABG surgery with CPB procedures between July 2000–July 2010, as (−2.59207 − 7.72486 × (preoperative creatinine) + 0.30737 × (weight) + 0.14174 × (aortic cross − clamp time) + 16.35924 × (transfusion) − 9.06373 ×(hypertension)). The clinical risk score was a priori confirmed to be independently predictive of AKI in the PEGASUS study cohort, and thus incorporated as a covariate in regression models to adjust the SNP associations with %ΔCr.
Genotyping and quality controls (QCs)
Genomic DNA was isolated from whole blood using standard procedures. Genotyping the PEGASUS discovery cohort used the Illumina Human610-Quad BeadChip at the Duke Genomic Analysis Facility for a total of 1004 samples. The Illumina GenomeStudio program (Illumina Inc., San Diego CA) was used for genotype calling. Markers with low GenCall score (≤ 0.15) or call frequency < 98% were filtered out. Samples with call rate < 98% or gender specification errors were also excluded in this initial QC. Additional QCs, conducted using PLINK software,63 included checks for cryptic relatedness and duplications. For a pair of samples with identity by descent (IBD) >0.1875 (between 2nd and 3rd degree relative), one sample was excluded from further analysis. Population structure was investigated using EigenSoft program64. All 15 principal components (PCs) were computed for each sample, and multiple PC plots were generated to determine whether any obvious outliers deviated from the main cluster and hence should be excluded. In total, we filtered out 44 samples (14 with call rate < 0.98, 3 with gender errors, 21 due to their relatedness with other samples, and 6 outliers from PC analysis). The QC’ed genotype dataset consisted of 960 subjects with 561,091 markers. Additionally, 86 subjects missing %ΔCr data, and one outlier with extreme high %ΔCr (outside of 3 SD from the mean %ΔCr) were excluded. Therefore, the final PEGASUS analysis dataset consists of 873 patients, all of European descent, with both genotype and phenotype data available.
All CATHGEN samples were genotyped using Illumina OMNI1-Quad BeadChip, and subjected to the same marker and sample QC criteria described above for PEGASUS. Following QC, a subset of CATHGEN samples was selected based on availability of %ΔCr data. Only SNPs identified in the discovery dataset were tested in the CATHGEN cohort for replication purposes.
Imputation of untyped markers
To increase overlap in SNP coverage between the genotyping platforms used for PEGASUS and CATHGEN cohorts for both replication analysis and meta-analysis, as well as to improve coverage of top candidate regions for fine-mapping associations, imputation of untyped autosomal SNPs was conducted in the post-QCed PEGASUS genotype dataset (960 samples with 561,091 markers) using a hidden Markov Model algorithm implemented in IMPUTE v2 software65 and phased haplotypes from the 1000 Genomes CEU reference panel. The best-guess imputed genotype for any untyped SNP per sample was chosen as the genotype with the highest imputation probability (imputation score). If the highest imputation score for an imputed SNP of a sample was less than 90%, a missing imputed genotype was assigned.
Statistical Analysis
Descriptive statistics of clinical variables are presented as frequency (percentage) for categorical variables and mean (standard deviation) for continuous variables. None of the principal components (PCs) derived from population structure analysis were significantly associated with %ΔCr in univariate linear regression tests, suggesting a lack of population stratification in our ethnically homogenous patient cohorts. As such, no ancestry covariates were included in the final multivariable linear regression model, which therefore tested SNP allelic association with %ΔCr adjusting only for the patient-specific AKI clinical risk score. An additive genetic model was used for coding each SNP genotype (0 for common homozygous, 1 for heterozygous, and 2 for rare homozygous).
Association analyses were carried out in the discovery cohort for all variants that passed QCs. Additional marker exclusion criteria – significant deviations from Hardy-Weinberg equilibrium (HWE) (P<10−6), genotype missingness rate < 10%, heterozygous haploid, minor allele frequency (MAF) < 1% - were applied to all association tests throughout the study. For the discovery dataset, no markers were excluded based on the 10% missingness threshold due to our prior stringent QC criteria to choose markers with >98% call frequencies. In total, 30,375 SNPs were excluded, leaving 530,716 SNPs to be tested in the final dataset. We used an a priori defined significance threshold of p<10−5 to select candidate SNPs for replication in the CATHGEN cohort, as a balance between the overly conservative Bonferroni correction and type II error, given that we had an a priori defined replication dataset to obviate type I error.
Association tests for the replication dataset were performed based on the same regression model with an additional covariate indicating patient-specific heart valve surgery status; statistically significant replication was defined as nominal significance in the replication cohort (p<0.05) with the same direction of allelic effect. All analyses were conducted using PLINK.
Finally, meta-analyses were performed to assess the overall effect of SNPs tested in both cohorts (meta-p values) using z-scores weighted by the inverse variance of effect size of each study, implemented in METAL66(http://www.sph.umich.edu/csg/abecasis/metal). The weighted z-score method allows for an overall p-value (meta-p) to be computed, by taking into account the beta-estimates and their standard errors from both datasets. The top genomic regions with SNPs meeting meta-p value <10−6 were further investigated (fine-mapped) by adding all imputed markers within the region in discovery dataset, followed by replication and meta-analysis as described above. Same marker exclusion criteria described for HWE, genotype missingness rate, and MAF were also applied to imputed markers prior to association analysis.
Linkage disequilibrium (LD) block information was computed and displayed using HaploView67(version 4.2) for top candidate regions. To evaluate whether genetic information independently adds prognostic value for postoperative AKI above traditional risk factors, we contrasted multiple regression models of AKI clinical risk score alone (clinical model) and with the addition of most significant independent SNPs (defined by LD) in top associated regions (clinical-genomic model). To facilitate model performance comparison, we present p-values, r2, and two commonly used information criteria (AIC and BIC) for each model.
Supplementary Material
Acknowledgement
We thank all patients who participated in this study. This study was funded in part by National Institutes of Health grants HL075273 and HL092071 (MVP); AG09663, HL054316, and HL069081 (MFN); HL096978, HL108280, and HL109971 (JPM); HL095987 (SS), and HL101621 (WK); American Heart Association grants #0120492U (MVP), #0256342U (JPM), #9970128N (MFN);, and the Duke Clinical Research Centers Program (NIH grant #M01-RR-30).
Appendix: Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team Members
Davis RD, Funk B, Gaca JG, Ginsburg GS, Glower DD, Hall RL, Hauser E, Kertai MD, Laskowitz DT, Li YJ, Lodge AJ, Mathew JP, Milano CA, Newman MF, Phillips-Bute B, Podgoreanu MV, Smith MP, Smith PK, Stafford-Smith M, Swaminathan M, Welsby IJ, White WD.
Footnotes
Disclosure
The authors declare that they have no competing interest.
References
- 1.Cruz DN, Ricci Z, Ronco C. Clinical review: RIFLE and AKIN--time for reappraisal. Crit Care. 2009;13:211. doi: 10.1186/cc7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmichael P, Carmichael AR. Acute renal failure in the surgical setting. ANZ J Surg. 2003;73:144–153. doi: 10.1046/j.1445-2197.2003.02640.x. [DOI] [PubMed] [Google Scholar]
- 3.Uchino S, Kellum JA, Bellomo R, et al. Acute renal failure in critically ill patients: a multinational, multicenter study. Jama. 2005;294:813–818. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- 4.Loef BG, Epema AH, Smilde TD, et al. Immediate postoperative renal function deterioration in cardiac surgical patients predicts in-hospital mortality and long-term survival. J Am Soc Nephrol. 2005;16:195–200. doi: 10.1681/ASN.2003100875. [DOI] [PubMed] [Google Scholar]
- 5.Swaminathan M, Hudson CC, Phillips-Bute BG, et al. Impact of early renal recovery on survival after cardiac surgery-associated acute kidney injury. Ann Thorac Surg. 2010;89:1098–1104. doi: 10.1016/j.athoracsur.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Lassnigg A, Schmidlin D, Mouhieddine M, et al. Minimal changes of serum creatinine predict prognosis in patients after cardiothoracic surgery: a prospective cohort study. J Am Soc Nephrol. 2004;15:1597–1605. doi: 10.1097/01.asn.0000130340.93930.dd. [DOI] [PubMed] [Google Scholar]
- 7.Conlon PJ, Stafford-Smith M, White WD, et al. Acute renal failure following cardiac surgery. Nephrol Dial Transplant. 1999;14:1158–1162. doi: 10.1093/ndt/14.5.1158. [DOI] [PubMed] [Google Scholar]
- 8.Mangano CM, Diamondstone LS, Ramsay JG, et al. Renal dysfunction after myocardial revascularization: risk factors, adverse outcomes, and hospital resource utilization. The Multicenter Study of Perioperative Ischemia Research Group. Annals of internal medicine. 1998;128:194–203. doi: 10.7326/0003-4819-128-3-199802010-00005. [DOI] [PubMed] [Google Scholar]
- 9.Brown JR, Parikh CR, Ross CS, et al. Impact of perioperative acute kidney injury as a severity index for thirty-day readmission after cardiac surgery. Ann Thorac Surg. 2014;97:111–117. doi: 10.1016/j.athoracsur.2013.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rewa O, Bagshaw SM. Acute kidney injury-epidemiology, outcomes and economics. Nat Rev Nephrol. 2014;10:193–207. doi: 10.1038/nrneph.2013.282. [DOI] [PubMed] [Google Scholar]
- 11.Aronson S, Fontes ML, Miao Y, et al. Risk index for perioperative renal dysfunction/failure: critical dependence on pulse pressure hypertension. Circulation. 2007;115:733–742. doi: 10.1161/CIRCULATIONAHA.106.623538. [DOI] [PubMed] [Google Scholar]
- 12.Janssen DP, Noyez L, van Druten JA, et al. Predictors of nephrological morbidity after coronary artery bypass surgery. Cardiovasc Surg. 2002;10:222–227. doi: 10.1016/s0967-2109(01)00134-x. [DOI] [PubMed] [Google Scholar]
- 13.Zakeri R, Freemantle N, Barnett V, et al. Relation between mild renal dysfunction and outcomes after coronary artery bypass grafting. Circulation. 2005;112:I270–I275. doi: 10.1161/CIRCULATIONAHA.104.522623. [DOI] [PubMed] [Google Scholar]
- 14.Shaw AD, Stafford-Smith M, White WD, et al. The effect of aprotinin on outcome after coronary-artery bypass grafting. N Engl J Med. 2008;358:784–793. doi: 10.1056/NEJMoa0707768. [DOI] [PubMed] [Google Scholar]
- 15.Brown JR, Cochran RP, Leavitt BJ, et al. Multivariable prediction of renal insufficiency developing after cardiac surgery. Circulation. 2007;116:I139–I143. doi: 10.1161/CIRCULATIONAHA.106.677070. [DOI] [PubMed] [Google Scholar]
- 16.Salis S, Mazzanti VV, Merli G, et al. Cardiopulmonary bypass duration is an independent predictor of morbidity and mortality after cardiac surgery. J Cardiothorac Vasc Anesth. 2008;22:814–822. doi: 10.1053/j.jvca.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Garg AX, Devereaux PJ, Yusuf S, et al. Kidney function after off-pump or on-pump coronary artery bypass graft surgery: a randomized clinical trial. Jama. 2014;311:2191–2198. doi: 10.1001/jama.2014.4952. [DOI] [PubMed] [Google Scholar]
- 18.Huen SC, Parikh CR. Predicting acute kidney injury after cardiac surgery: a systematic review. Ann Thorac Surg. 2012;93:337–347. doi: 10.1016/j.athoracsur.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu JC, Coca SG, Patel UD, et al. Searching for genes that matter in acute kidney injury: a systematic review. Clin J Am Soc Nephrol. 2009;4:1020–1031. doi: 10.2215/CJN.05411008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham PN, Dyanov HM, Park P, et al. Acute renal failure in endotoxemia is caused by TNF acting directly on TNF receptor-1 in kidney. J Immunol. 2002;168:5817–5823. doi: 10.4049/jimmunol.168.11.5817. [DOI] [PubMed] [Google Scholar]
- 21.Segerer S, Nelson PJ, Schlondorff D. Chemokines, chemokine receptors, and renal disease: from basic science to pathophysiologic and therapeutic studies. J Am Soc Nephrol. 2000;11:152–176. doi: 10.1681/ASN.V111152. [DOI] [PubMed] [Google Scholar]
- 22.Heyman SN, Darmon D, Goldfarb M, et al. Endotoxin-induced renal failure. I. A role for altered renal microcirculation. Exp Nephrol. 2000;8:266–274. doi: 10.1159/000020678. [DOI] [PubMed] [Google Scholar]
- 23.Stafford-Smith M, Podgoreanu M, Swaminathan M, et al. Association of genetic polymorphisms with risk of renal injury after coronary bypass graft surgery. Am J Kidney Dis. 2005;45:519–530. doi: 10.1053/j.ajkd.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 24.Chew ST, Newman MF, White WD, et al. Preliminary report on the association of apolipoprotein E polymorphisms, with postoperative peak serum creatinine concentrations in cardiac surgical patients. Anesthesiology. 2000;93:325–331. doi: 10.1097/00000542-200008000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Isbir SC, Tekeli A, Ergen A, et al. Genetic polymorphisms contribute to acute kidney injury after coronary artery bypass grafting. Heart Surg Forum. 2007;10:E439–E444. doi: 10.1532/HSF98.20071117. [DOI] [PubMed] [Google Scholar]
- 26.Popov AF, Hinz J, Schulz EG, et al. The eNOS 786C/T polymorphism in cardiac surgical patients with cardiopulmonary bypass is associated with renal dysfunction. Eur J Cardiothorac Surg. 2009;36:651–656. doi: 10.1016/j.ejcts.2009.04.049. [DOI] [PubMed] [Google Scholar]
- 27.Haase-Fielitz A, Haase M, Bellomo R, et al. Decreased catecholamine degradation associates with shock and kidney injury after cardiac surgery. J Am Soc Nephrol. 2009;20:1393–1403. doi: 10.1681/ASN.2008080915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 29.Pasha DN, Davis JT, Rao F, et al. Heritable influence of DBH on adrenergic and renal function: twin and disease studies. PLoS One. 2013;8:e82956. doi: 10.1371/journal.pone.0082956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu CT, Garnaas MK, Tin A, et al. Genetic Association for Renal Traits among Participants of African Ancestry Reveals New Loci for Renal Function. Plos Genet. 2011;7 doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada Y, Sim X, Go MJ, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Section 2: AKI Definition. Kidney international supplements. 2012;2:19–36. doi: 10.1038/kisup.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langworthy M, Zhou B, de Caestecker M, et al. NFATc1 identifies a population of proximal tubule cell progenitors. J Am Soc Nephrol. 2009;20:311–321. doi: 10.1681/ASN.2008010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putoux A, Attie-Bitach T, Martinovic J, et al. Phenotypic variability of Bardet-Biedl syndrome: focusing on the kidney. Pediatric nephrology (Berlin, Germany) 2012;27:7–15. doi: 10.1007/s00467-010-1751-3. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura DY, Swiderski RE, Searby CC, et al. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. American journal of human genetics. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vernon EG, Malik K, Reynolds P, et al. The parathyroid hormone-responsive B1 gene is interrupted by a t(1;7)(q42;p15) breakpoint associated with Wilms' tumour. Oncogene. 2003;22:1371–1380. doi: 10.1038/sj.onc.1206332. [DOI] [PubMed] [Google Scholar]
- 40.Zaghloul NA, Katsanis N. Mechanistic insights into Bardet-Biedl syndrome, a model ciliopathy. The Journal of clinical investigation. 2009;119:428–437. doi: 10.1172/JCI37041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez-Hernandez V, Pravincumar P, Diaz-Font A, et al. Bardet-Biedl syndrome proteins control the cilia length through regulation of actin polymerization. Human molecular genetics. 2013;22:3858–3868. doi: 10.1093/hmg/ddt241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013;21:8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller J, Stoetzel C, Vincent MC, et al. Identification of 28 novel mutations in the Bardet-Biedl syndrome genes: the burden of private mutations in an extensively heterogeneous disease. Human genetics. 2010;127:583–593. doi: 10.1007/s00439-010-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Billingsley G, Bin J, Fieggen KJ, et al. Mutations in chaperonin-like BBS genes are a major contributor to disease development in a multiethnic Bardet-Biedl syndrome patient population. Journal of medical genetics. 2010;47:453–463. doi: 10.1136/jmg.2009.073205. [DOI] [PubMed] [Google Scholar]
- 45.Marion V, Schlicht D, Mockel A, et al. Bardet-Biedl syndrome highlights the major role of the primary cilium in efficient water reabsorption. Kidney international. 2011;79:1013–1025. doi: 10.1038/ki.2010.538. [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Dong Z. Primary cilia and kidney injury: current research status and future perspectives. American journal of physiology Renal physiology. 2013;305:F1085–F1098. doi: 10.1152/ajprenal.00399.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weimbs T. Polycystic kidney disease and renal injury repair: common pathways, fluid flow, and the function of polycystin-1. American journal of physiology Renal physiology. 2007;293:F1423–F1432. doi: 10.1152/ajprenal.00275.2007. [DOI] [PubMed] [Google Scholar]
- 48.Verghese E, Ricardo SD, Weidenfeld R, et al. Renal primary cilia lengthen after acute tubular necrosis. J Am Soc Nephrol. 2009;20:2147–2153. doi: 10.1681/ASN.2008101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Human genetics. 2009;125:131–151. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- 50.Lobato RL, White WD, Mathew JP, et al. Thrombomodulin gene variants are associated with increased mortality after coronary artery bypass surgery in replicated analyses. Circulation. 2011;124:S143–S148. doi: 10.1161/CIRCULATIONAHA.110.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah SH, Freedman NJ, Zhang L, et al. Neuropeptide Y gene polymorphisms confer risk of early-onset atherosclerosis. Plos Genet. 2009;5:e1000318. doi: 10.1371/journal.pgen.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voora D, Horton J, Shah SH, et al. Polymorphisms associated with in vitro aspirin resistance are not associated with clinical outcomes in patients with coronary artery disease who report regular aspirin use. Am Heart J. 2011;162:166–172. e161. doi: 10.1016/j.ahj.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 53.MacKensen GB, Swaminathan M, Ti LK, et al. Preliminary report on the interaction of apolipoprotein E polymorphism with aortic atherosclerosis and acute nephropathy after CABG. Ann Thorac Surg. 2004;78:520–526. doi: 10.1016/j.athoracsur.2004.02.106. [DOI] [PubMed] [Google Scholar]
- 54.Bellomo R, Ronco C, Kellum JA, et al. Acute renal failure - definition, outcome measures, animal models, fluid therapy and information technology needs: the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group. Crit Care. 2004;8:R204–R212. doi: 10.1186/cc2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta RL, Kellum JA, Shah SV, et al. Acute Kidney Injury Network: report of an initiative to improve outcomes in acute kidney injury. Crit Care. 2007;11:R31. doi: 10.1186/cc5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kidney Disease: Improving Global O. KDIGO clinical practice guidelines for the prevention, diagnosis, evaluation, and treatment of hepatitis C in chronic kidney disease. Kidney Int Suppl. 2008:S1–S99. doi: 10.1038/ki.2008.81. [DOI] [PubMed] [Google Scholar]
- 57.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 58.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chambers JC, Zhang W, Lord GM, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42:373–375. doi: 10.1038/ng.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liotta M, Olsson D, Sartipy U, et al. Minimal changes in postoperative creatinine values and early and late mortality and cardiovascular events after coronary artery bypass grafting. Am J Cardiol. 2014;113:70–75. doi: 10.1016/j.amjcard.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 61.Lassnigg A, Schmid ER, Hiesmayr M, et al. Impact of minimal increases in serum creatinine on outcome in patients after cardiothoracic surgery: do we have to revise current definitions of acute renal failure? Crit Care Med. 2008;36:1129–1137. doi: 10.1097/CCM.0b013e318169181a. [DOI] [PubMed] [Google Scholar]
- 62.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Annals of internal medicine. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 65.Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 66.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.