Abstract
Heliobacterium modesticaldum is an anaerobic photosynthetic bacterium that grows optimally at pH 6–7 and 52°C and is the only phototrophic member of the Firmicutes phylum family (gram-positive bacteria with low GC content). The ATP synthase of H. modesticaldum was isolated and characterized at the biochemical and biophysical levels. The isolated holoenzyme exhibited the subunit patterns of F-type ATP synthases containing a 5-subunit hydrophilic F1 subcomplex and a 3-subunit hydrophobic FO subcomplex. ATP hydrolysis by the isolated HF1FO ATP synthase was successfully detected after pretreatment with different detergents by an in-gel ATPase activity assay, which showed that the highest activity was detected in the presence of mild detergents such as LDAO; moreover, high catalytic activity in the gel was already detected after the initial incubation period of 0.5 h. In contrast, HF1FO showed extremely low ATPase activity in harsher detergents such as TODC. The isolated fully functional enzyme will form the basis for future structural studies.
Keywords: ATP synthase, Heliobacterium modesticaldum, ATP hydrolysis, in-gel function assay
Introduction
Heliobacterium modesticaldum is a thermophilic anoxygenic phototrophic bacterium that grows either photoheterotrophically or chemotrophically in the dark by fermentation but does not grow photoautotrophically due to the lack of several genes encoding key enzymes required for the known autotrophic carbon fixation pathways [1, 2]. Heliobacterium modesticaldum may have played a key role in the evolution of phototrophic bacteria. The electron transport chain contains a type I photosynthetic reaction center (RC) that employs bacteriochlorophyll (Bchl g) as the primary electron donor. The majority of recent studies have focused on characterization of the photosynthetic reaction center and electron transport chain. The energy metabolism of H. modesticaldum has been studied to a much lesser extend, and no isolation or biochemical studies have been reported thus far for the H. modesticaldum ATP synthase, which is one of the key enzymes involved in bioenergetic energy conversion. Based on the genomic information of H. modesticaldum, it is known that this organism contains a gene cluster for an F-type ATP synthase in which all eight subunits are encoded in a single conserved operon [2]. However, due to the lack of any biochemical data, the structure, function and regulation of the ATP synthase of H. modesticaldum are still poorly understood.
The membrane-bound F-type ATP synthase is a multi-subunit membrane protein complex that functions as a universal rotary nano-machine in the energy-transducing membranes of bacteria, mitochondria, and chloroplasts [3]. The chloroplast ATP synthase is a multi-subunit complex comprising 9 different subunits (α3β3γδεI1II1III12–14IV1) located in two major domains (CF1 and CF0). The hydrophilic CF1 domain comprises 5 different subunits (α3β3γδε). In most bacteria, the hydrophobic membrane-bound CF0 domain comprises 3 different subunits (a, b2, and c10–15), which correspond to the chloroplast subunits (a = IV1, b2 = I1 and II1, and c10–15 = III10–15 IV1). The CF1 and CF0 domains are connected in the CF1F0 protein complex by two peripheral stalks: a central interior stalk comprising γ and ε subunits, and the peripheral exterior stalk comprising the I and II subunits as well as the δ subunit [4]. Three ATP molecules are synthesized in the three catalytic sites in CF1 during one full rotation of the central stalk and the c-ring, driven by an electrochemical proton gradient. Up to 400 ATP molecules per second are synthesized in the natural environment [5–8]. F-type ATP synthases can act in both synthesis and hydrolysis directions; in the synthesis direction, the enzyme synthesizes ATP, which is driven by an electrochemical proton gradient. In the reverse direction, ATP hydrolysis drives the translocation of protons across the photosynthetic membrane.
However, photosynthetic organisms must avoid ATP hydrolysis in the absence of an electrochemical gradient; therefore, the hydrolytic function of CF1FO is inactivated at night by redox modulation of a disulfide bridge on its γ subunit via thioredoxin [9–15].
This study is, to the best of our knowledge, the first report of the successful isolation and biochemical characterization of the ATP synthase from Heliobacterium modesticaldum. We used an in-gel ATPase activity assay, which is well established and has previously been successfully applied to the detection of the activity of mitochondrial ATP synthases and spinach chloroplast ATP synthases, as well as the ATP synthases from the cyanobacterium T. elongatus and the green algae C. reinhardtii [16], to study the activity of the ATP synthase in the presence of different detergents. In contrast to mitochondrial ATP synthases, for which high ATP hydrolysis activities have been reported [16], the chloroplast ATP synthase has low ATPase activity due to the inhibition mechanism that prevents wasteful ATP hydrolysis in the dark. This inactivation is mediated by a conformational change of the ε subunit of CF1 [17, 18], which occurs induced by the lack of proton gradient, the binding of Mg2+-ATP [19–22], and the oxidation of cysteine residues in the γ subunits to a disulfide bond [23, 24]. Several studies used harsh pre-treatments of the chloroplast enzyme, including heating, organic solvents, and trypsin, to stimulate the ATPase activity of CF1, which is the hydrophilic part of the enzyme that contains the nucleotide binding sites [25–28]. The interaction of some harsher detergents with the solubilized CF1 part of the chloroplast ATP synthases led to enhanced ATPase activity [16, 29, 30]. The stimulation of CF1 ATPase activity by harsh detergents that form small micelles may be induced by the dissociation of the ε subunit from the chloroplast CF1 subcomplex. Various detergents, including alkylglucosides, LDAO, and TDOC, have previously been tested to study the effect of the dissociation of the CF1- ε subunit [16, 29, 30]. With an increase of the micelle size by increasing the chain length, the interaction of alkylglucosides with CF1 becomes less and less effective in removing the ε subunit, resulting in lower ATPase activity [30].
In contrast to the chloroplast enzyme, the hydrolytic function of ATP synthase is a vitally important process in anaerobic bacteria that lack a membrane-bound respiratory chain to establish a gradient of electrochemical potential for protons or sodium ions [31]. The F1FO ATP synthase of the alkaliphilic bacterium Natranaerobius thermophilus is one example that it does not function in ATP synthesis [31] but the enzyme catalyzes ATP hydrolysis to expel cytoplasmic Na+ to avoid Na toxicity. H. modesticaldum lacks the internal membranes present in cyanobacteria; the photosynthetic electron transport chain is instead embedded into the cytoplasmic membrane. Therefore, it is commonly assumed that an electrochemical gradient across the cytoplasmic membrane that is established during the photosynthetic process, drives ATP synthesis. However, whether ATP hydrolysis might also be an essential function of the heliobacterial HF1FO remains unknown. It is also unknown whether regulation of the reverse direction of the H. modesticaldum ATP synthase is similar to the inactivation mechanism of CF1FO at night.
In this manuscript, we report on the isolation and biochemical characterization of the functional ATP synthase from H. modesticaldum. The identities of all subcomplexes and individual subunits of the ATP synthase were determined. We also discovered that the isolated ATP synthase remains active during native gel electrophoresis using an in-gel ATPase activity assay. In contrast to the chloroplast ATP-synthase, no activation is required for the ATPase activity of the ATP synthase from H. modesticaldum. The H. modesticaldum enzyme is fully active in the presence of mild detergents, while it is denatured in harsh detergents. Our studies show that the isolated ATP synthase from H. modesticaldum is fully functional and is suitable for further functional and structural studies.
2. Materials and methods
Purification of the ATP synthase complex from Heliobacterium modesticaldum
The HF1FO ATP synthase complex was isolated from H. modesticaldum as previously described [3, 32, 33] for the chloroplast ATP synthase. Liquid cultures of H. modesticaldum were grown anaerobically using pyruvate-yeast extract medium (pH 6.9) at 52°C under 4,500–6,000-lux incandescent illumination until the stationary phase is reached [34]. The cells were harvested by centrifugation at 10,000× gmax for 15 min at 4°C and then re-suspended in 200 ml of harvesting buffer containing 50 mM MOPS (pH 7.0) and 5 mM magnesium chloride. The cells were disrupted by sonication with a 100 % duty cycle for 15 cycles at 50 % power using an ultrasonic homogenizer, Model 300 V/T (Biologics Inc., Manassas, Virginia). The lysate was subjected to high-speed centrifugation at 200,000× gmax for 1 h at 4°C, and the membrane pellets were suspended in the harvesting buffer described above. Prior to detergent solubilization, DL-Dithiothreitol (DTT) was added to reach a final concentration of 50 mM and incubated with agitation for 15 min at 4°C. The membrane proteins were then solubilized in a buffer containing 20 mM Tricine (pH 8.0), 200 mM sucrose, 5 mM magnesium chloride, 400 mM ammonium sulfate, 2 mM Na2-ATP, 6.25 mM sodium cholate (Sigma, USA), 12 mM n-octyl-β-D-glucoside (OG) (GLYCON, Germany), and 50 mM DTT for 15 min at 4°C and then subjected to centrifugation at 200,000× gmax for 1 h at 4°C to separate solubilized protein-detergent micelles from non-solubilized membranes. The solubilized protein-detergent micelles were precipitated with ammonium sulfate (45 %, v/v) at 4°C. The ammonium sulfate precipitation pellet containing the HF1FO ATP synthase was re-suspended in buffer containing 30 mM monobasic sodium phosphate (pH 7.2), 200 mM sucrose, 2 mM magnesium chloride, 0.5 mM Na2-EDTA, and 4 mM n-dodecyl-β-D-maltopyranoside (β-DDM) (GLYCON, Germany) and was further purified by sucrose density gradient centrifugation at 242,000× g for 17 h at 44°C in gradient buffer containing 30 mM monobasic sodium phosphate (pH 7.2), 2 mM magnesium chloride, 0.5 mM Na2-EDTA, 8 mM β-DDM, 1 mg/ml asolectin and sucrose at a concentration range from 20–60 % (w/v). The fractions from the sucrose density gradient were collected from the bottom to the top of the gradient. The sucrose was removed by using Sephadex G-25 desalting columns at 4°C previously equilibrated in buffer containing 10 mM Tricine (pH 8.0), 5 mM magnesium chloride, and 8 mM β-DDM and the ATP synthase was concentrated with a 100-kDa cutoff concentrators membrane filter devices (Millipore, Billerica, MA) prior to further experiments. The protein concentration was determined according to the modified Lowry method [35, 36].
Protein analysis by electrophoresis techniques
The subunit composition of the HF1FO ATP synthase was analyzed using 15% Tricine-SDS–PAGE polyacrylamide gels, according to Schagger [37] with a few modifications: protein samples were diluted at a 1:2 ratio with sample loading buffer (62.5 mM Tris-HCl (pH 6.8), 2 % SDS, 25 % glycerol, 0.01 % bromophenol blue), and incubated at 80°C for 30 min to completely disassemble the c-ring of ATP synthase. The gels were run at 4°C for 3 h.
The intact HF1FO ATP synthase was analyzed using 4–16 % native polyacrylamide gels as previously described [38, 39] with modifications: protein samples, in the presence of β-DDM at 8 mM, were mixed with 2× native sample buffer (100 mM sodium chloride, 100 mM imidazole-HCl, 4 mM 6-aminohexanoic acid, 10% glycerol, 2 mM EDTA (pH 7.0)). Subsequently, protein samples for blue native gel electrophoresis (BN-PAGE) and high-resolution clear native gel electrophoresis (hrCN-PAGE) were additionally supplemented with Coomassie Blue G-250 dye (a detergent/Coomassie ratio of 8) and Ponceau S (0.01 %), respectively. The native electrophoretic buffers were identical except that the BN-PAGE cathode buffer contained the anionic Coomassie Blue G-250 dye (0.02 %), and the hrCN-PAGE cathode buffer contained two detergents (0.05 %) sodium deoxycholate (Sigma, USA) and 0.01 % non-ionic β-DDM). Native gel electrophoresis was performed at 4°C to maintain protein-complex integrity.
Protein bands were visualized with either the Coomassie Blue staining method or the Silver staining method [40]. Heme-containing proteins were detected with the heme staining method [41]. For immunoblot analysis, performed in principle as previously described [42], the proteins were transferred onto a PVDF membrane in a buffer optimized for the transfer of ATP synthase subunits (25 mM Tris base, 192 mM glycine, 0.1 % SDS, and 20 % methanol). The membrane was blocked in 5% non-fat milk containing 50 mM Tris-Base, 150 mM sodium chloride, and 0.05% Tween-20 for 1 h and then incubated with primary antibodies (AptA and AptH, 1:10,000, Agrisera, Sweden) in 5 %, non-fat dry milk in 1× Tris-buffered saline with Tween-20 (TBST buffer), containing 50 mM Tris–HCl pH 8.0, 150 mM sodium chloride, 0.05 % Tween-20), washed three times with 1× TBST, incubated with goat anti-rabbit IgG-HRP-conjugated secondary antibodies (1:5,000), washed again, and stained with the Immun-Star HRP substrate reagent.
ATP hydrolysis activity in-gel assay
The in-gel assay of ATP hydrolysis activity was performed as previously described [16] with modifications, as follows: hrCN-PAGE gel strips were pre-equilibrated in 40 mM Tris-HCl, 4 mM ATP, and 1.5 mM magnesium chloride (pH 8.0) supplemented with 30 mM of various detergents, including beta-dodecylmaltoside (β-DDM) (Glycon), β-octylglycoside (OG) (Glycon), N,N-dimethyl-1-dodecanamine-N-oxide (LDAO) (Affymetrix/Anatrace), or tauro-deoxycholate (TDOC) (Sigma-Aldrich), for 3 hours at room temperature. Following removal of the incubation solution and a 1-min brief rinse with deionized water, the gel strips were incubated in an assay buffer comprising 35 mM Tris-HCl, 270 mM glycine, 14 mM magnesium sulfate, 0.075 % lead nitrate, and 0.8 mM ATP (pH 7.8) with 20 % methanol at room temperature for 30 min, 60 min and for 24 h. Increasing ATP hydrolysis activity leads to increasing lead phosphate precipitate formation. For imaging, the reaction was stopped with 50 % methanol. The gel strips were incubated for 30 min and then rinsed with deionized water. The transparent gel strips were imaged on top of a blue background to improve the visibility of the lead nitrate bands.
3. Results and discussion
Purification and subunit composition of the F1FO-ATPase from H. modesticaldum
Since the discovery of heliobacteria, remarkable progress has been made in the understanding of their physiology and energy metabolism, as well as in the characterization of their photosynthetic reaction center, including the pigment composition and electron transfer reactions [1, 33, 43–47]. Heliobacteria differ from many other photosynthetic organisms as they lack differentiated internal membranes or membrane-bound vesicles. Therefore, the cytoplasmic membrane is the only membrane system found in Heliobacteria [48]. All membrane-bound complexes, including all photosynthetic complexes and a large number of various transporters for transporting essential substrates into the cells, are confined to this cytoplasmic membrane [2, 48]. According to the complete genome sequence analysis of H. modesticaldum [49], a gene cluster was identified for the ATP synthase that contains all eight subunits and it is assumed that the enzyme is present in the cytoplasmic membrane [2, 49].
The primary aim of this study was to establish a method to purify the intact ATP synthase complex from H. modesticaldum for further bioenergetic and structural studies.
Studies aimed to isolate the photosynthetic reaction center of H. modesticaldum used a low percentage of either β-DDM or β-DM to extract the photosynthetic reaction center from the membrane [33, 50]. We modified the protocol developed for the photosynthetic RC by varying the nature of the detergents as well as their concentrations, taking into account procedures established for the chloroplast enzyme from plants and the ATP synthase from cyanobacteria. The maximal yield of ATP synthase extracted from cytoplasmic membranes was obtained using a detergent mixture of 6.25 mM cholate and 15 mM OG, as described in the materials and methods section. Subsequently, the ATP synthase complex was precipitated by ammonium sulfate fractionation at 45 % (v/v) saturated ammonium sulfate solution at 4°C (Figure 1, lane 2). The fraction also contained a lower amount of heme-containing proteins (cyt c533), as shown in Figure 2, lane 7. However, these contaminating heme-proteins were removed by the subsequent sucrose gradient centrifugation step (Figure 2, lane 9). To increase enzymatic stability and activity [32], the two detergents used in the initial solubilization step (cholate and OG) were replaced by 8 mM β-DDM during the sucrose density gradient centrifugation.
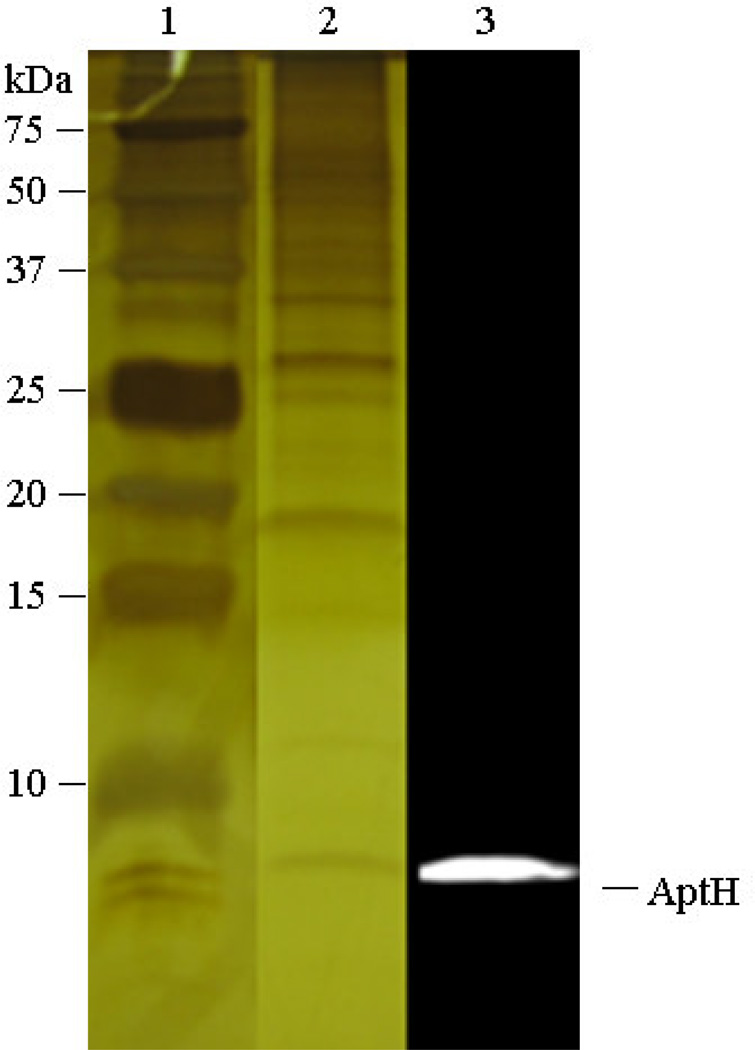
Figure 1. Tricine-SDS polyacrylamide gel and immunoblot analysis of the 45% (v/v) ammonium sulfate precipitate fraction of solubilized membrane proteins of H. modesticaldum.
The silver-stained 15% Tricine-SDS polyacrylamide gel and immunoblotting detection of the 8-kDa c-subunit (AptH), shows HF1FO-ATP synthase complex was enriched in the 45% (v/v) saturated ammonium sulfate ((NH4)2SO4) fraction (lanes 2 and 3, respectively). [lane 1, kDa, protein dual color standards (Bio-Rad, USA); lane 2, 45% (v/v) ammonium sulfate precipitate fraction in 15% Tricine-SDS-PAGE; lane 3, 45% (v/v) Western-blot of the ammonium sulfate precipitate fraction using the AptH antibody against the C-subunit of the H. modesticaldum ATP synthase].
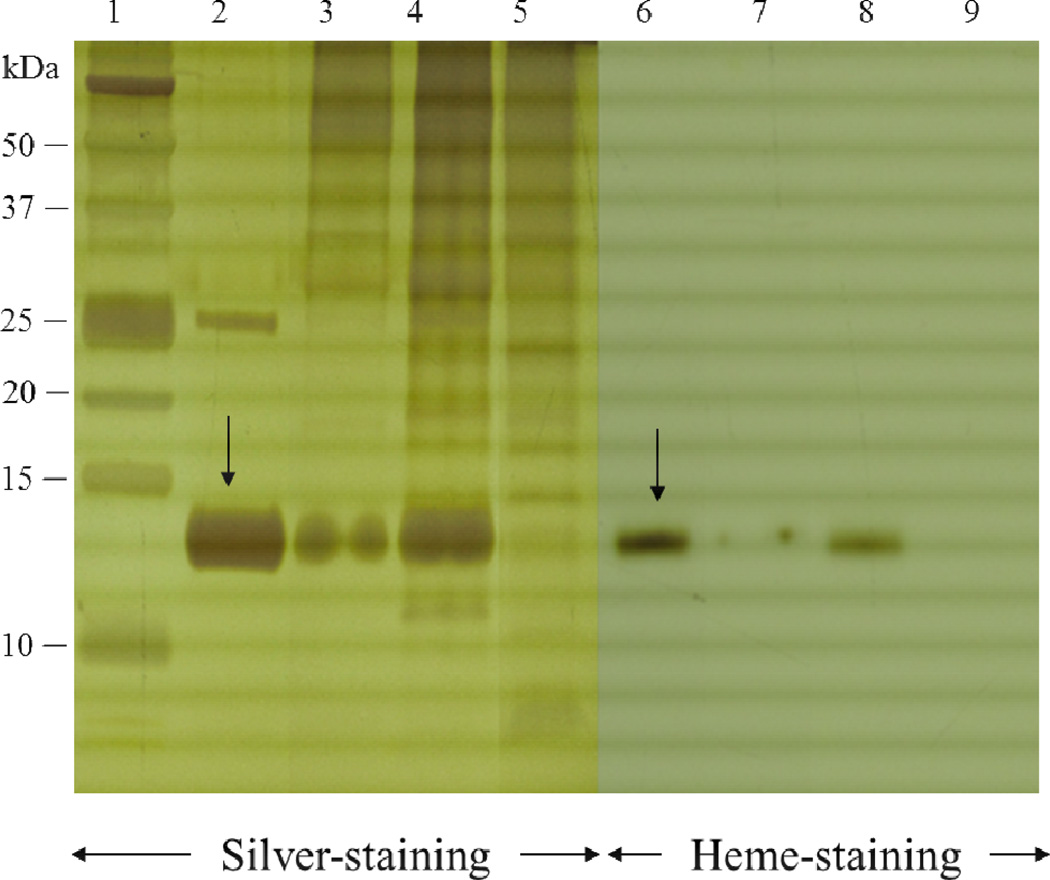
Figure 2. Analysis of the ammonium sulfate fractions of the ATP synthase extract by denaturing electrophoresis.
Silver-stained and heme-stained Tricine-SDS polyacrylamide gel analysis of different fractions collected from the ammonium sulfate precipitation steps and the discontinuous sucrose density gradient centrifugation of H. modesticaldum. The fractions45%(4P), 60%(6P), and 44%(SG) were analyzed by 15 % Tricine-SDS-PAGE electrophoresis with silver staining (lanes 3, 4 and 5) and heme staining (lanes 7, 8, and 9). The heme staining indicated that fraction45%(4P) (lanes 3 and 7) and fraction60%(6P) (lanes 4 and 8) are contaminated by 14-kDa cytochrome c according to the molecular mass of the positive control (lanes 1 and 6) cytochrome c (see arrow) [33]. The majority of cyt. c was precipitated by the 60% (v/v) saturated ammonium sulfate solution (fraction60%(6P), lanes 4 and 8). The fraction45% (4P) enriched the intact HF1FO ATP synthase still contained minimal amount of cyt. c, which was removed via discontinuous sucrose density gradient centrifugation. Lane 9 shows the 44% sucrose fraction from the density gradient, which contained intact HF1FO ATP synthase without cyt. c contamination.
[lane 1, protein dual color standards (Bio-Rad, USA); lanes 2 and 6, positive control, cytochrome c (provided by Dr. Sarrou); lanes 3 and 7, fraction45%(4P), proteins fractionated by 45% (v/v) saturated ammonium sulfate solution; lanes 4 and 8, fraction60%(6P), proteins fractionated by 60% (v/v) saturated ammonium sulfate solution; lanes 5 and 9, fraction44%(SG), fractions collected between 44% from the sucrose gradient. Abbreviations: P, pellets; SG, sucrose density gradient].
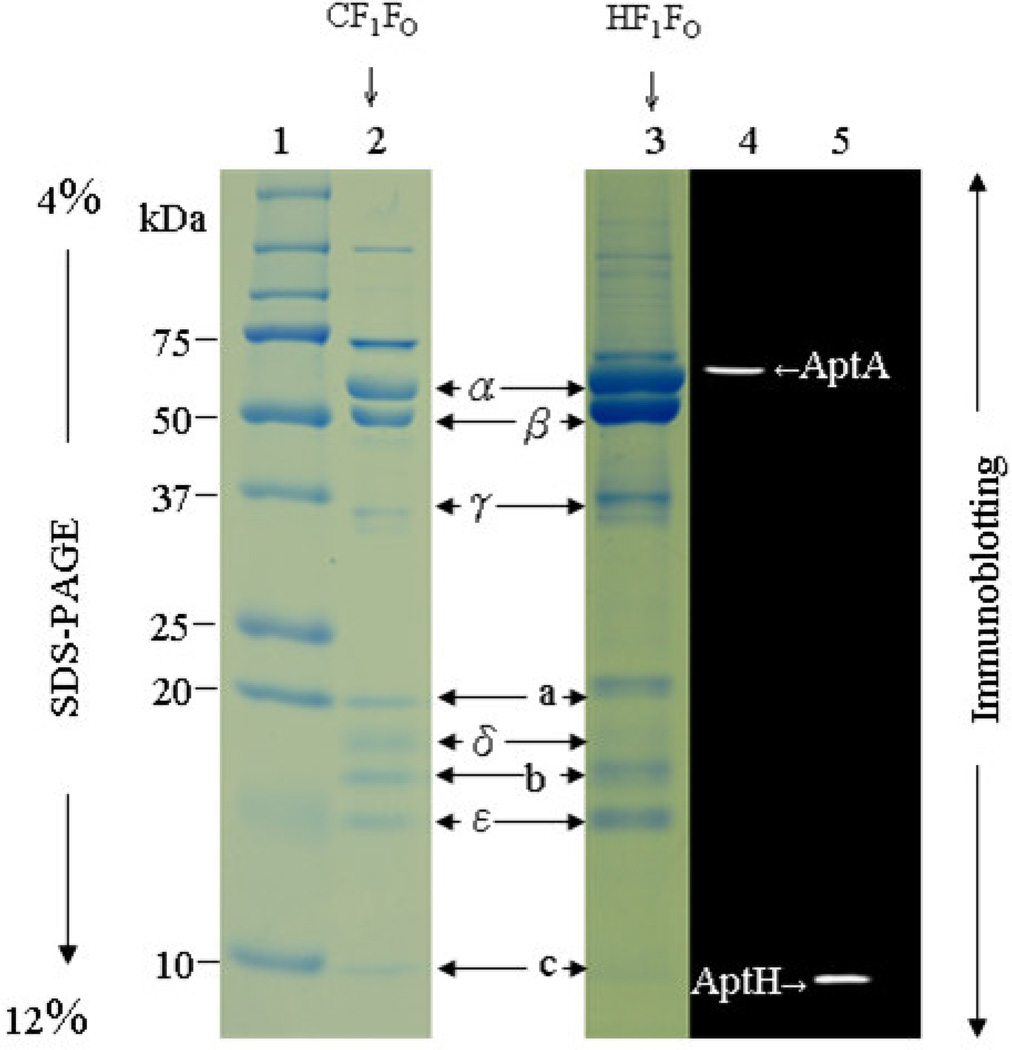
As determined by silver-stained SDS-PAGE, the ATP synthase complex is essential in the interfacial fractions between 44 % and 52 % of the sucrose gradient (Figures 3 and 4). Tricine SDS-PAGE electrophoresis of the isolated ATP synthase complex showed the typical subunit pattern of an F-ATP synthase (Figure 5, lane 3), with five F1 subunits (α, β, γ, δ and ε), and three FO subunits (a, b and c), according to the molecular mass pattern of the chloroplast enzyme CF1FO, which was used as a positive control in the SDS-PAGE analysis (Figure 5, lane 2). The identities of the α-subunit (AtpA) and the c-subunit (AtpH) of HF1FO were confirmed by western blotting, as shown in Figure 5, lanes 4 and 5.
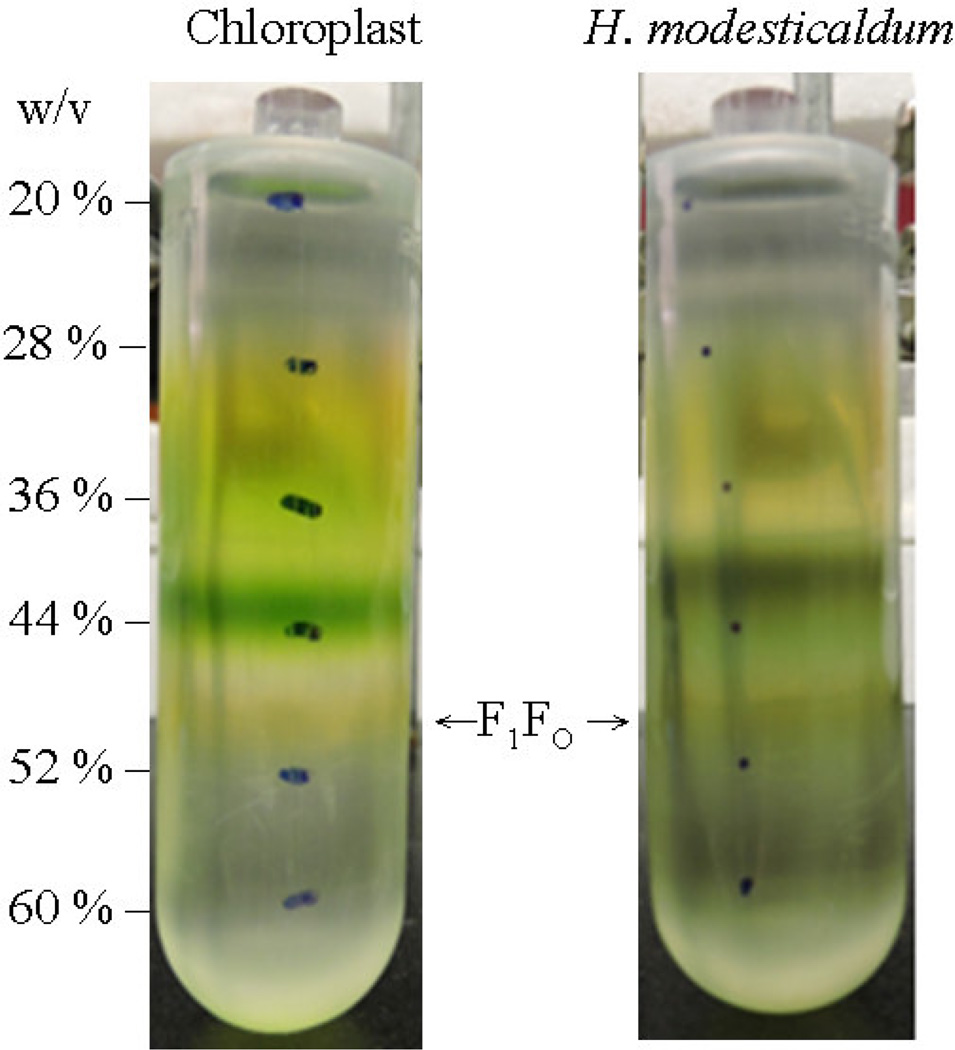
Figure 3. Discontinuous sucrose density gradient centrifugation of the ATP synthase form chloroplast (left) and H. modesticaldum (right).
The intact ATP synthase was separated from other membrane proteins using discontinuous sucrose density gradient centrifugation. The interface fraction between 44 % and 52 % contains the ATP synthases. The yellowish fraction from the isolated fraction form chloroplast (left) contains chloroplast CF1FO ATP synthase, which were isolated according to the method previously described [32]. The olive greenish fraction of the isolated fraction (right) of the H. modesticaldum contains the HF1FO ATP synthase. The indicated numbers on the right represent the concentration of sucrose (w/v) in each fraction of the step gradient.
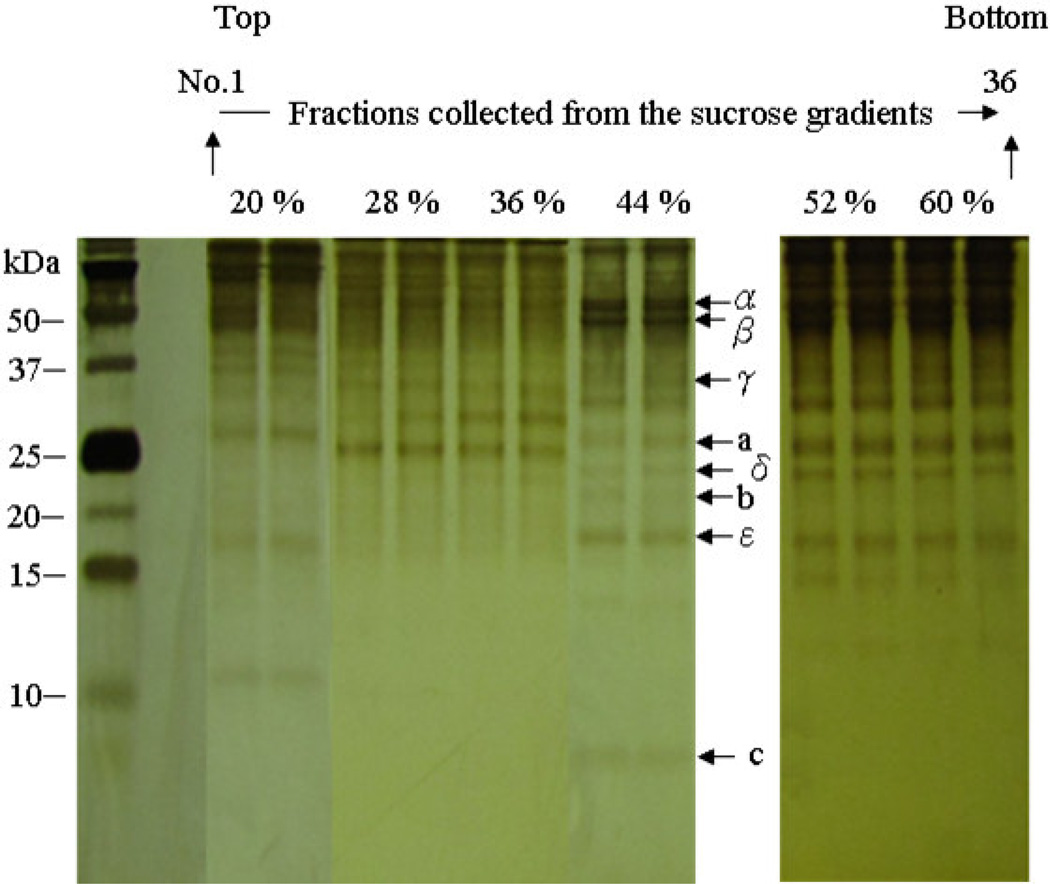
Figure 4. Silver-stained 15% Tricine-SDS polyacrylamide gel of the total proteins in the collected sucrose gradient fractions.
The analysis showed that the interface fractions between 44 % and 52 % were enriched in H. modesticaldum HF1FO ATP synthase, indicated by arrow that represents the c-subunit of HF1FO ATP synthase.
Figure 5. Tricine-SDS polyacrylamide gel electrophoresis and immunoblot analysis of the H. modesticaldum HF1FO ATP synthase.
The 5 subunits (α, β, γ, δ, and ε) of the HF1 subcomplex and the 3 subunits (a, b, and c) of the HFO subcomplex of the H. modesticaldum HF1FO ATP synthase complex were identified (lane 3) according to the molecular mass of each subunit and a compared to the positive control chloroplast CF1FO ATP synthase (lane 2). The immunoblot detects the ~52-kDa α subunit of the HF1 subcomplex (lane 4) and the ~8-kDa c subunit of the HFO subcomplex (lane 5). [lane 1, protein dual color standards (Bio-Rad, USA); lane 2, positive control, chloroplast CF1FO ATP synthase complex; lanes 3 H. modesticaldum HF1FO ATP synthase complexes collected from the interface gradient between 44 % and 52 % (w/v) of the sucrose gradient; Lanes 4 and 5, Western blot analysis with antibodies against the α-subunit (AptA) ans c-subunit (AtpH)].
Native gel electrophoresis
The quaternary structure of HF1FO was analyzed in more detail by native gel electrophoresis. This technique has been previously used to analyze ATP synthase complexes of mitochondria and chloroplasts. It has also been applied to the analysis of enzymatically active oligomeric states [51–55] or in advanced structural studies using 2D-crystallization and electron microscopic single particle analysis of proteins extracted from BN-PAGE [53, 56–58].
The isolated HF1FO-ATP synthase complex was analyzed by native electrophoresis techniques to investigate whether it forms oligomers. In-gel activity assays were performed to test if the complex still retains its physiologically functional state after the isolation procedure.
Under the conditions of native gel electrophoresis, the integral HF1FO-ATP synthase complexes migrated as a band with an apparent molecular mass of 600 kDa in both BN-PAGE and hrCN-PAGE gels and was identified as a monomeric complex according to the native electrophoretic mobility of the CF1FO complex in BN-PAGE, as previously described [59–61] (Figure 6, panel A). The heliobacterial ATP synthase appears to be more stable than the chloroplast enzyme under the stress of the native gel electrophoresis: a significant band for the chloroplast CF1 subcomplex was identified just below 480 kDa in both BN-PAGE and hrCN-PAGE gels (Figure 6, panel A, lanes 2 and 4); however, in the region of the identified CF1 subcomplex, only a weak band for the HF1 is identified with Coomassie Blue staining. The gel strips of hrCN-PAGE containing HF1FO-ATP synthase complexes were further analyzed using two-dimensional electrophoresis (1-D hrCN-PAGE, 2-D SDS-PAGE), which identified the individual subunits of the HF1FO complex on 2-D SDS-PAGE according to the molecular mass shown in Figure 6, panel B. The HF1FO complex contains all HF1 and HFO subunits.
Figure 6. Native gel electrophoresis analysis and 2D-electrophoresis analysis of the β-DDM-solubilized intact ATP synthase complex of H. modesticaldum.
(A) The intact F1FO complexes and F1 subcomplexes were identified on the gels of blue native electrophoresis (BN-PAGE) (lanes 2 and 3) and high-resolution clear native electrophoresis (hrCN-PAGE) (lane 4 and 5). Lanes 4 and 5 which represent the clear native gel were stained with Coomassie Blue after completion of the electrophoresis run. (B) A hrCN-PAGE gel strip of the H. modesticaldum HF1FO ATP synthase complex was analyzed by 2D electrophoresis, which showed all individual subunits of the H. modesticaldum HF1FO ATP synthase complex. [lane 1, unstained native protein standard (Life Technologies, USA); lane 2, BN-PAGE of the chloroplast CF1FO ATP synthase complex; lane 3, BN-PAGE of the H. modesticaldum HF1FO ATP synthase complex; lane 4, hrCN-PAGE of the chloroplast CF1FO ATP synthase complex; lane 5, hrCN-PAGE of the H. modesticaldum HF1FO ATP synthase complex].
Biochemical properties of the isolated ATP synthase from H. modesticaldum
The ATP synthesis and hydrolysis functions of ATP synthases isolated from different sources has been studied most frequently via liposome reconstitution and chemiosmotic pH jump experiments. However, the techniques involved are complex and time consuming as they study the function of the enzyme after detergent removal in lipid bilayers. However, these methods cannot analyze the catalytic function of the ATP synthase in form of a protein-detergent complex.
Therefore, the combination of native gel electrophoresis techniques and an in-gel functional activity assay has evolved into a powerful tool for protein-protein interaction analysis in mitochondrial respiratory complexes [62]. One of the established in-gel assays, named here the “in-gel ATP hydrolysis assay,” was remarkably useful in both qualitative and quantitative analyses of the catalytic activity of the mitochondrial ATP synthase (complex V) [38]. Because ATP hydrolysis activity can be monitored in native gels via an in-gel ATPase assay, it can be used to rapidly screen for enzymatic activity of the isolated ATP synthase complex in detergents directly in the gel without involving reconstitution into liposomes. We used this assay to screen for the best detergent to stabilize the structure and function of the heliobacterial ATP synthase in different protein-detergent complexes. This is not possible using ATP synthesis assays, as they all require removal of the detergent during reconstitution into lipid membranes.
The functional properties of the isolated HF1FO-ATP synthase complex were examined by monitoring the ATP hydrolysis activity by observing lead phosphate precipitate formation using the in-gel ATPase activity assay at room temperature, as previously described [16].
For the in-gel ATPase activity assay, hrCN-PAGE is highly preferred compared with BN-PAGE because the dye Coomassie Blue applied on BN-PAGE can dissociate detergent-labile subunits from the super-complex or disassemble the super-complex into sub-complexes. Furthermore, lead precipitate detection is very difficult in the presence of the Coomassie Blue dye. The hrCN-PAGE gels are colorless; therefore, the in-gel assays can be directly performed without any interference with the Coomassie Blue dye [38, 55, 62, 63].
We compared the CF1FO isolated from spinach chloroplast with the HF1FO complex isolated from H. modesticaldum. Previous studies provided that ATP hydrolysis activity of the chloroplast CF1FO-ATP synthase complex is extremely low, but can be improved by activating the CF1FO-ATP synthase complex via detergent treatments prior to the in-gel ATPase activity assay [16, 29, 30]. One interesting question was if the heliobacterial ATP synthase would require activation or if the intact enzyme was fully functional for ATP hydrolysis without any activation such as harsh detergent treatment. hrCN-PAGE gel strips were treated with selected detergents as described in detail in materials and methods section. We tested the ATP hydrolysis activity of the chloroplast and H. modesticaldum ATP synthase in the presence of the following detergents: OG (octyl-glucoside) a non-ionic short length detergent, β-DDM (beta-dodecylmaltoside) a mild non-ionic long chain detergent, LDAO (N,N-dimethyl-1-dodecanamine-N-oxide) an ionic mild long chain detergent, and TDOC (tauro-deoxycholate) a harsh detergent.
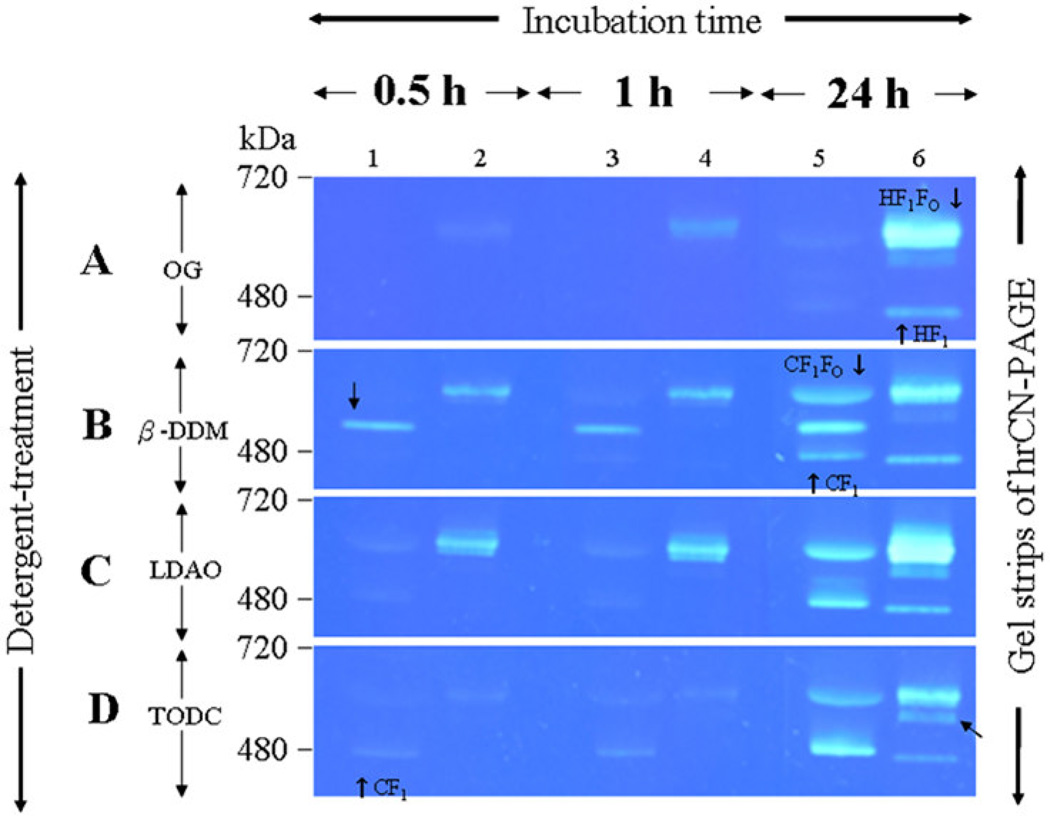
Using OG treatment, ATP hydrolysis could not be detected for either CF1FO or CF1 within the 0.5 h or 1 h incubation periods (Figure 7, A panel, lane 1 and lane 3). After 24 hours of incubation, extremely weak bands indicating hydrolysis activity were visible (Figure 7, A panel, lane 5). For ATP HF1FO hydrolysis of the intact enzyme was already detectable as a weak band after an incubation period of 0.5 h (Figure 7, A panel, lane 2). The intensity of the band increased after 1 hour, and saturated activity was observed after an incubation period of 24 h (Figure 7, A panel, lane 6). Interestingly, weak HF1 hydrolysis activity was only visible after an incubation period of 24 h.
Figure 7. In-gel ATPase hydrolysis activity of ATP synthase complexes isolated from spinach chloroplasts and H. modesticaldum.
The hrCN-PAGE gel strips, each containing CF1FO and HF1FO, were incubated in buffers with different detergents to detect whether ATPase activity was dependent on the detergent. The gel strips were preincubated with buffer containing OG (A), β-DDM (B), LDAO (C) (Affymetrix/Anatrace, USA), or TDOC (D) (Sigma, USA). In the in-gel ATPase activity assay, ATP hydrolysis can be visualized as the white band comprising precipitated lead phosphate. Images of the in-gel assays were taken under a blue background after 0.5 h, 1 h and 24 h incubations, respectively.
Notably, under some detergent conditions, multiple subcomplex bands appear, as indicated by black arrows. These bands are observed, for example, for CF1FO in β-DDM and LDAO and for HF1FO in TODC. They might comprise CF1FO and HF1FO subcomplexes that have lost subunit a and/or b of the FO domain during hrCN-PAGE electrophoresis. [lanes 1, 3, and 5, spinach chloroplast CF1FO ATP synthase complex; lanes 2, 3, and 6, H. modesticaldum HF1FO synthase complex].
Using β-DDM, no ATP hydrolysis of the chloroplast ATP synthase, CF1FO, was detected after incubation times of 0.5 h and 1 h (Figure 7, B panel, lane l and lane 3), whereas a subcomplex of the chloroplast enzyme showed strong ATP-hydrolysis activity already after 0.5 h and 1 h. In contrast, strong HF1FO ATP hydrolysis activity was already observed after an incubation time of 0.5 h (Figure 7, B panel, lane 2), and the band was even more pronounced after the 1 h incubation period. (Figure 7, B panel, lane 4). In contrast to the chloroplast CF1FO subcomplex, the HF1 subcomplex showed no hydrolysis activity after 0.5 h and 1 h; a weak band became visible only after the 24 h incubation period. This is a very interesting result because it indicates that the intact HF1FO is fully active in the ATP hydrolysis direction in the presence of very mild detergents such as β-DDM without requiring activation. Furthermore, the results indicate that removal of the proton-translocating HFO domain blocks ATP hydrolysis, as only very weak ATP hydrolysis was detected after 24 h for HF1 in β-DDM.
Using LDAO, only very weak ATP hydrolysis activity of the chloroplast CF1FO and CF1 was detected after 0.5 and 1 h, whereas HF1FO showed very high activity (Figure 7, C panel, lanes 1 and lane 2). Interestingly, also using LDAO, no HF1 hydrolysis activity was detected after 0.5 and 1 h, and only a weak band appeared after an incubation period of 24 h (Figure 7, C panel, lane 6).
In contrast to the results of the ATP synthase complexes treated with the detergents OG, β-DDM and LDAO, both CF1FO and HF1FO treated with the harsh detergent TDOC showed extremely low ATP hydrolysis activities after the 0.5 h and 1 h incubation periods (Figure 7, D panels, lanes 1, 2, 3, and 4), and significant activity could only be detected after 24 hours of incubation (Figure 7, D panel, lanes 5 and lane 6).
In summary, these results indicate that HF1FO shows high ATPase activity in mild detergents such as LDAO and β-DDM, whereas extremely low ATP hydrolysis activity was detected in harsher detergents such as OG and TDOC.
The fastest and highest ATP hydrolysis activity observed was detected when HF1FO was treated with LDAO. This result may indicate that long-chain detergents such as β-DDM and especially LDAO, which are known to stabilize membrane protein complexes, stabilize HF1FO and maintain its full catalytic activity. In contrast, short-chain detergents such as OG and TDOC may destabilize the heliobacterial ATP synthase and thereby inactivate it. In contrast to the chloroplast enzyme, for which CF1 shows much higher ATP hydrolysis activity that CF1FO, the detachment of the head of the heliobacterial ATP synthase (HF1) from the intact enzyme does not enhance but rather inhibits ATP hydrolysis activity. Such behavior has thus far not been reported for F-type ATP synthases but has been reported for vacuolar type V ATPases, which are enzymes that function to establish an H+ gradient across the membrane [64].
Our results may provide preliminary evidence that the heliobacterial ATP synthase may act in vivo in both ATP synthesis and ATP hydrolysis and that a mechanism may exist in Heliobacterium modesticaldum that leads to switching off of the ATP hydrolysis function when the HF1 head is decoupled from the transmembrane HF0 proton inducing channel.
Conclusion
The combination of a modified isolation method and in-gel ATPase activity assay reported here confirmed that isolation of the functionally intact ATP synthase under anaerobic condition from anaerobic Heliobacterium modesticaldum cells can be achieved. We have shown that approximately 20 mg of purified ATP synthase can be routinely obtained from 4 L of anaerobic cell culture. The most critical breakthroughs in achieving these results were modifications in the way the cells were broken down as well as modifications in the isolation of the protein from the membrane. The in-gel ATPase activity assay for the ATP synthase of H. modesticaldum allowed us to detect ATP hydrolysis activity directly in the gel and to identify the stability and activity of the ATP synthase in different detergent micelles. The use of the in-gel activity assay method not only offers an expeditious way to screen for activity of sample fractions in a high-throughput manner but also allows for the assessment of the stability and activity of different ATP synthase subcomplexes in different detergents. The intact heliobacterial HF1FO enzyme shows fully active ATP hydrolysis activity in mild detergents, whereas the ATP hydrolysis activity is inactivated when the HF1 head is removed from the HFO transmembrane part of HF1FO.
Our reported methods provided a primary isolation process under strictly anaerobic conditions, thereby providing heliobacterial ATP synthase samples for further studies that will include in particular bioenergetics studies, crystallization experiments, and may also be applied to ATP synthases from other anaerobic organisms in the future.
Highlights.
The intact ATP-synthase was isolated from Heliobacterium modesticaldum
The subunit composition was compared to the chloroplast enzyme
The oligomeric state and stability were analyzed by native gel electrophoresis
In gel ATPase activity assays show full activity without need for activation
The heliobacterial enzyme evolutionarily links bacterial and plant ATP synthases
Acknowledgments
The work in the laboratory of Petra Fromme was supported by the National Institutes of Health (grant no GM081490 and GM095583) and the BioXFEL Science and Technology Center funded by the National Science Foundation (award 1231306). The work the laboratory of Kevin E. Redding was funded by the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-SC0010575. JH was also supported by the Galvin Chair award to PF from Arizona State University.
We also thank Patricia Baker from Dr. Redding's lab for helpful guidance and discussion concerning of H. modesticaldum cell culture.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Tang KH, Yue H, Blankenship RE. Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Microbiol. 2010;10:150. doi: 10.1186/1471-2180-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sattley WM, Blankenship RE. Insights into heliobacterial photosynthesis and physiology from the genome of Heliobacterium modesticaldum. Photosynthesis Research. 2010;104(2–3):113–122. doi: 10.1007/s11120-010-9529-9. [DOI] [PubMed] [Google Scholar]
- 3.Turina P, Samoray D, Graber P. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1-liposomes. Embo Journal. 2003;22(3):418–426. doi: 10.1093/emboj/cdg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher B, Schwarz L, Graber P. Direct indication for the existence of a double stalk in CF0F1. J Mol Biol. 1998;281(5):757–762. doi: 10.1006/jmbi.1998.1957. [DOI] [PubMed] [Google Scholar]
- 5.Pänke O, Rumberg B. Energy and entropy balance of ATP synthesis. Biochimica et Biophysica Acta. 1997;1322(2–3) [Google Scholar]
- 6.Peter Gräber UJaGHS. Kinetics of Proton-Transport-Coupled ATP Synthesis in Chloroplasts. Activation of the ATPase by an Artificially Generated ΔpH and Δψ. Berichte der Bunsengesellschaft für physikalische Chemie. 1984;88(7):10. [Google Scholar]
- 7.Seelert H, et al. Dye-ligand chromatographic purification of intact multisubunit membrane protein complexes: application to the chloroplast H+-FoF1-ATP synthase. Biochem J. 2000;346(Pt 1):41–44. [PMC free article] [PubMed] [Google Scholar]
- 8.Poetsch A, et al. Detergent effect on anion exchange perfusion chromatography and gel filtration of intact chloroplast H(+)-ATP synthase. Biochem Biophys Res Commun. 1999;265(2):520–524. doi: 10.1006/bbrc.1999.1688. [DOI] [PubMed] [Google Scholar]
- 9.Evron Y, Johnson EA, McCarty RE. Regulation of proton flow and ATP synthesis in chloroplasts. J Bioenerg Biomembr. 2000;32(5):501–506. doi: 10.1023/a:1005669008974. [DOI] [PubMed] [Google Scholar]
- 10.Hisabori T, et al. Molecular devices of chloroplast F(1)-ATP synthase for the regulation. Biochim Biophys Acta. 2002;1555(1–3):140–146. doi: 10.1016/s0005-2728(02)00269-4. [DOI] [PubMed] [Google Scholar]
- 11.Hisabori T, et al. Molecular evolution of the modulator of chloroplast ATP synthase: origin of the conformational change dependent regulation. FEBS Lett. 2003;545(1):71–75. doi: 10.1016/s0014-5793(03)00395-8. [DOI] [PubMed] [Google Scholar]
- 12.Junesch U, Graber P. The rate of ATP-synthesis as a function of delta pH and delta psi catalyzed by the active, reduced H(+)-ATPase from chloroplasts. FEBS Lett. 1991;294(3):275–278. doi: 10.1016/0014-5793(91)81447-g. [DOI] [PubMed] [Google Scholar]
- 13.Kohzuma K, et al. Light- and metabolism-related regulation of the chloroplast ATP synthase has distinct mechanisms and functions. J Biol Chem. 2013;288(18):13156–13163. doi: 10.1074/jbc.M113.453225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter ML. Gamma-epsilon Interactions Regulate the Chloroplast ATP Synthase. Photosynth Res. 2004;79(3):319–329. doi: 10.1023/B:PRES.0000017157.08098.36. [DOI] [PubMed] [Google Scholar]
- 15.Ort DR, Oxborough K. In Situ Regulation of Chloroplast Coupling Factor Activity. Annual Review of Plant Physiology and Plant Molecular Biology. 1992;43:23. [Google Scholar]
- 16.Suhai T, et al. Highly sensitive detection of ATPase activity in native gels. Electrophoresis. 2009;30(20):3622–3625. doi: 10.1002/elps.200900114. [DOI] [PubMed] [Google Scholar]
- 17.Richter ML, Patrie WJ, McCarty RE. Preparation of the epsilon subunit and epsilon subunit-deficient chloroplast coupling factor 1 in reconstitutively active forms. J Biol Chem. 1984;259(12):7371–7373. [PubMed] [Google Scholar]
- 18.Gertz M, et al. Interactions of rotor subunits in the chloroplast ATP synthase modulated by nucleotides and by Mg2+ Biochim Biophys Acta. 2007;1774(5):566–574. doi: 10.1016/j.bbapap.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Du Z, et al. Assembled F1-(alpha beta) and Hybrid F1-alpha 3beta 3gamma -ATPases from Rhodospirillum rubrum alpha, wild type or mutant beta, and chloroplast gamma subunits. Demonstration of Mg2+versus Ca2+-induced differences in catalytic site structure and function. J Biol Chem. 2001;276(15):11517–11523. doi: 10.1074/jbc.M007568200. [DOI] [PubMed] [Google Scholar]
- 20.Digel JG, et al. Differences between two tight ADP binding sites of the chloroplast coupling factor 1 and their effects on ATPase activity. J Biol Chem. 1996;271(33):19976–19982. doi: 10.1074/jbc.271.33.19976. [DOI] [PubMed] [Google Scholar]
- 21.Digel JG, Hightower KE, McCarty RE. Subunit movement during catalysis by F1-F0-ATP synthases. J Bioenerg Biomembr. 1996;28(5):439–442. doi: 10.1007/BF02113986. [DOI] [PubMed] [Google Scholar]
- 22.Du ZY, Boyer PD. On the mechanism of sulfite activation of chloroplast thylakoid ATPase and the relation of ADP tightly bound at a catalytic site to the binding change mechanism. Biochemistry. 1990;29(2):402–407. doi: 10.1021/bi00454a014. [DOI] [PubMed] [Google Scholar]
- 23.Ort DR, Oxborough K. In situ Regulation of Chloroplast Coupling Factor Activity. Annual Review of Plant Physiology and Plant Molecular Biology. 1992;43:269–291. [Google Scholar]
- 24.Nalin CM, McCarty RE. Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J Biol Chem. 1984;259(11):7275–7280. [PubMed] [Google Scholar]
- 25.Farron F. Isolation and properties of a chloroplast coupling factor and heat-activated adenosine triphosphatase. Biochemistry. 1970;9(19):3823–3828. doi: 10.1021/bi00821a023. [DOI] [PubMed] [Google Scholar]
- 26.Sakurai H, et al. Enhancement of Adenosine-Triphosphatase Activity of Purified Chloroplast Coupling Factor-I in an Aqueous Organic-Solvent. Journal of Biochemistry. 1981;90(1):95–102. doi: 10.1093/oxfordjournals.jbchem.a133473. [DOI] [PubMed] [Google Scholar]
- 27.Anthon GE, Jagendorf AT. Effect of Methanol on Spinach Thylakoid Atpase. Biochimica Et Biophysica Acta. 1983;723(3):358–365. [Google Scholar]
- 28.McCarty RE. ATP synthase of chloroplast thylakoid membranes: a more in depth characterization of its ATPase activity. J Bioenerg Biomembr. 2005;37(5):289–297. doi: 10.1007/s10863-005-8640-7. [DOI] [PubMed] [Google Scholar]
- 29.Pick U, Bassilian S. Activation of magnesium ion specific adenosinetriphosphatase in chloroplast coupling factor 1 by octyl glucoside. Biochemistry. 1982;21(24):6144–6152. doi: 10.1021/bi00267a019. [DOI] [PubMed] [Google Scholar]
- 30.Yu F, McCarty RE. Detergent activation of the ATPase activity of chloroplast coupling factor 1. Arch Biochem Biophys. 1985;238(1):61–68. doi: 10.1016/0003-9861(85)90140-7. [DOI] [PubMed] [Google Scholar]
- 31.Mesbah NM, Wiegel J. The Na(+)-translocating F(1)F(0)-ATPase from the halophilic, alkalithermophile Natranaerobius thermophilus. Biochim Biophys Acta. 2011;1807(9):1133–1142. doi: 10.1016/j.bbabio.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Varco-Merth B, et al. Crystallization of the c14-rotor of the chloroplast ATP synthase reveals that it contains pigments. Biochim Biophys Acta. 2008;1777(7–8):605–612. doi: 10.1016/j.bbabio.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarrou I, et al. Purification of the photosynthetic reaction center from Heliobacterium modesticaldum. Photosynth Res. 2012;111(3):291–302. doi: 10.1007/s11120-012-9726-9. [DOI] [PubMed] [Google Scholar]
- 34.Kimble LK, M L, Woese CR, Madigan MT. Heliobacterium modesticaldum, sp. nov., a thermophilic heliobacterium of hot springs and volcanic soils. Arch Microbiol. 1995;163(4):8. [Google Scholar]
- 35.Hartree EF. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal Biochem. 1972;48(2):422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- 36.Lowry OH, et al. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- 37.Schagger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1(1):16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 38.Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6(7):1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Wittig I, et al. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis. 2007;28(21):3811–3820. doi: 10.1002/elps.200700367. [DOI] [PubMed] [Google Scholar]
- 40.Merril CR, Dunau ML, Goldman D. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal Biochem. 1981;110(1):201–207. doi: 10.1016/0003-2697(81)90136-6. [DOI] [PubMed] [Google Scholar]
- 41.Francis RT, Becker RR. Specific Indication of Hemoproteins in Polyacrylamide Gels Using a Double-Staining Process. Analytical Biochemistry. 1984;136(2):509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence RM, et al. Recombinant production and purification of the subunit c of chloroplast ATP synthase. Protein Expr Purif. 2011;76(1):15–24. doi: 10.1016/j.pep.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinnickel M, Golbeck JH. Heliobacterial photosynthesis. Photosynthesis Research. 2007;92(1):35–53. doi: 10.1007/s11120-007-9162-4. [DOI] [PubMed] [Google Scholar]
- 44.Chauvet A, et al. Temporal and spectral characterization of the photosynthetic reaction center from Heliobacterium modesticaldum. Photosynth Res. 2013;116(1):1–9. doi: 10.1007/s11120-013-9871-9. [DOI] [PubMed] [Google Scholar]
- 45.Heinnickel M, et al. Identification of FX in the heliobacterial reaction center as a [4Fe-4S] cluster with an S = (3)/(2) ground spin state. Biochemistry. 2006;45(21):6756–6764. doi: 10.1021/bi060031s. [DOI] [PubMed] [Google Scholar]
- 46.Neerken S, Amesz J. The antenna reaction center complex of heliobacteria: composition, energy conversion and electron transfer. Biochim Biophys Acta. 2001;1507(1–3):278–290. doi: 10.1016/s0005-2728(01)00207-9. [DOI] [PubMed] [Google Scholar]
- 47.Oh-oka H. Type 1 reaction center of photosynthetic heliobacteria. Photochem Photobiol. 2007;83(1):177–186. doi: 10.1562/2006-03-29-IR-860. [DOI] [PubMed] [Google Scholar]
- 48.Asao M, Madigan MT. Taxonomy, phylogeny, and ecology of the heliobacteria. Photosynth Res. 2010;104(2–3):103–111. doi: 10.1007/s11120-009-9516-1. [DOI] [PubMed] [Google Scholar]
- 49.Sattley WM, et al. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J Bacteriol. 2008;190(13):4687–4696. doi: 10.1128/JB.00299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kashey TS, et al. Expression and characterization of cytochrome c553 from Heliobacterium modesticaldum. Photosynth Res. 2014;120(3):291–299. doi: 10.1007/s11120-014-9982-y. [DOI] [PubMed] [Google Scholar]
- 51.Krause F, et al. Active oligomeric ATP synthases in mammalian mitochondria. Biochem Biophys Res Commun. 2005;329(2):583–590. doi: 10.1016/j.bbrc.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Meyer B, et al. Identification of two proteins associated with mammalian ATP synthase. Mol Cell Proteomics. 2007;6(10):1690–1699. doi: 10.1074/mcp.M700097-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Poetsch A, et al. Dye removal, catalytic activity and 2D crystallization of chloroplast H+-ATP synthase purified by blue native electrophoresis. Biochimica Et Biophysica Acta-Biomembranes. 2000;1466(1–2):339–349. doi: 10.1016/s0005-2736(00)00191-7. [DOI] [PubMed] [Google Scholar]
- 54.Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19(8):1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wittig I, Schagger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5(17):4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- 56.Schafer E, et al. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46(44):12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 57.Schafer E, et al. Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem. 2006;281(22):15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 58.Seelert H, Dencher NA, Muller DJ. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J Mol Biol. 2003;333(2):337–344. doi: 10.1016/j.jmb.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 59.Fromme P. Die ATP-synthase aus Chloroplast: Biochemische Untersuchungen zur Struktur and Kinetische Messungen zum Mechanismus des Enzyme. Technical University Berlin; 1988. pp. 1–248. [Google Scholar]
- 60.Fromme P, Boekema EJ, Graber P. Isolation and Characterization of a Supramolecular Complex of Subunit-Iii of the Atp-Synthase from Chloroplasts. Zeitschrift Fur Naturforschung C-a Journal of Biosciences. 1987;42(11–12):1239–1245. [Google Scholar]
- 61.Neff D, Dencher NA. Purification of multisubunit membrane protein complexes: isolation of chloroplast FoF1-ATP synthase, CFo and CF1 by blue native electrophoresis. Biochem Biophys Res Commun. 1999;259(3):569–575. doi: 10.1006/bbrc.1999.0820. [DOI] [PubMed] [Google Scholar]
- 62.Wittig I, Schagger H. Native electrophoretic techniques to identify protein-protein interactions. Proteomics. 2009;9(23):5214–5223. doi: 10.1002/pmic.200900151. [DOI] [PubMed] [Google Scholar]
- 63.Wittig I, Schagger H. Features and applications of blue-native and clear-native electrophoresis. Proteomics. 2008;8(19):3974–3990. doi: 10.1002/pmic.200800017. [DOI] [PubMed] [Google Scholar]
- 64.Wang D, Hiesinger PR. The vesicular ATPase: a missing link between acidification and exocytosis. J Cell Biol. 2013;203(2):171–173. doi: 10.1083/jcb.201309130. [DOI] [PMC free article] [PubMed] [Google Scholar]