Abstract
Nucleases containing programmable DNA-binding domains can alter the genomes of model organisms and have the potential to become human therapeutics. Here we present DNA-binding phage-assisted continuous evolution (DB-PACE) as a general approach for the laboratory evolution of DNA-binding activity and specificity. We used this system to generate TALE nucleases with broadly improved DNA cleavage specificity, establishing DB-PACE as a versatile approach for improving the accuracy of genome-editing agents.
Genome-editing tools are revolutionizing our understanding of how genotype influences phenotype and have the potential to serve as treatments for genetic diseases1, 2. These tools include fusions of programmable DNA-binding domains (DBDs) such as zinc fingers and transcription activator-like effectors (TALEs) to functional domains including nucleases, recombinases, and transposases2, 3. Zinc fingers (ZFs) are DBDs of approximately 30 amino acids that typically bind three DNA nucleotides along the major groove2, 4. Several methods have been developed to generate zinc-finger arrays with tailor-made DNA specificities5, 6. TALE proteins consist of an N-terminal domain followed by a series of tandem repeats each of 33 to 35 amino acids, a nuclear localization sequence, a transcription activation domain, and a C-terminal domain7, 8. Two repeat variable diresidues (RVDs) at positions 12 and 13 within each repeat recognize and bind to a specific DNA base9, 10, and altering the RVDs allows TALE repeats to be programmed using a simple code7, 11.
A limitation of TALEs is that the 5′ nucleotide of the target is specified to be T9. Although promiscuous TALEs with no specificity at the 5′ position have been described12, 13, no TALE variants that preferentially recognize 5′ A or 5′ C have been described12, 13. Moreover, TALEs bind appreciably to off-target sites within the genome, limiting their potential for use as human therapeutics14. A method to enhance the specificity of a particular TALE array by decreasing its ability to bind to specific off-target DNA sequences found in a genome has not been reported.
We recently developed a system, phage-assisted continuous evolution (PACE), that allows proteins to continuously evolve in the laboratory at a rate ~100-fold faster than conventional methods (Fig. 1a and Supplementary Results)15. PACE has been used to rapidly evolve RNA polymerases and proteases with tailor-made properties15–17. We speculated that PACE could be adapted to continuously evolve DNA-binding domains with altered or improved DNA-binding specificity.
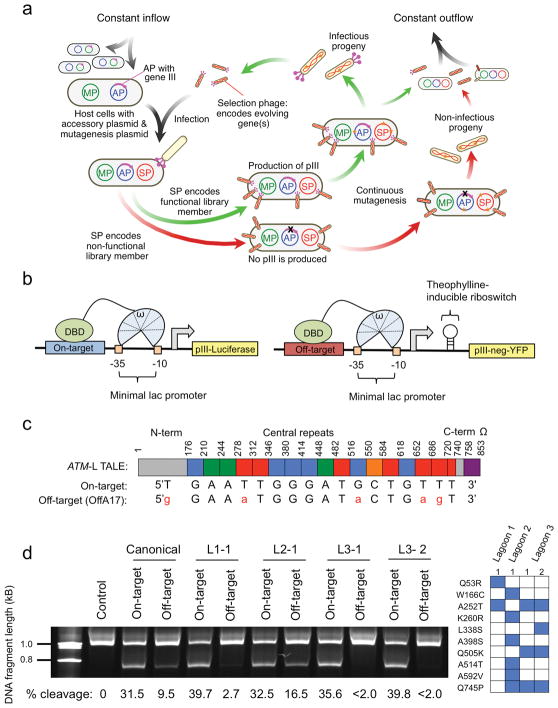
Figure 1. Development of DB-PACE and its application to TALEs.
(a) Overview of phage-assisted continuous evolution (PACE). (b) Left: Reporter system used to couple DNA binding to production of pIII-luciferase, and Right: Reporter system used to couple DNA binding to an off-target sequence to production of pIII-neg-YFP. (c) Schematic of the ATM-targeting TALE–ω fusion and the relationship between individual TALE repeats and the nucleotides they recognize for the on-target sequence (ATM: 5′-TGAATTGGGATGCTGTTT-3′), or the most highly cleaved human genomic off-target sequence (OffA17: 5′-GGAAATGGGATACTGAGT-3′). (d) Left: relative cleavage efficiencies of the canonical ATM TALEN pair or four ATM TALEN pairs containing the canonical ATM-right half site TALEN and an evolved ATM-left half site TALEN (L1-2, L2-1, L3-1, or L3-2) on a linear 6-kb DNA fragment containing either the ATM on-target sequence or the OffA17 off-target sequence. The top band is non-cleaved DNA, while the bottom band is a cleavage product. The gel has been cropped to simplify presentation. Cleavage percentages were determined using densitometry analysis (GelEval), and are included below each lane. Right: mutations in the evolved ATM-left half site TALEs used in the left panel.
To develop a PACE-compatible DNA-binding selection, we linked a DBD of interest to a subunit of bacterial RNA polymerase III (RNAP). Based on previous one-hybrid systems18, we envisioned that binding of this fusion protein to operator sequences upstream of a minimal lac promoter would induce transcription of a downstream gene III-luciferase reporter through recruitment or stabilization of the RNAP holoenzyme (Fig. 1b). Using the DBD of Zif268 (residues 333–420)19 fused to the ω subunit of RNAP, we established sequence-specific and binding-dependent induction of gene III and production of pIII, the phage protein that enables propagation during PACE (Supplementary Fig. 1 and Supplementary Results).
To port this selection system into PACE, we moved the DNA operator-gene III cassette to an accessory plasmid (AP), and moved the RNAP ω-Zif268 protein to a selection phage (SP) construct. We then performed plaque assays to establish activity-dependent phage propagation in vitro (Supplementary Figs. 2–4 and 5a, and Supplementary Results), and validated continuous propagation of Zif268-SP phage in DNA-binding PACE (DB-PACE) (Supplementary Fig. 5b and Supplementary Results). Next, we performed a mock evolution to evolve DNA-binding activity starting from an inactive mutant Zif268 protein (Supplementary Fig. 5c,d and Supplementary Results).
To apply this system to continuously evolve TALE proteins, we optimized the one-hybrid fusion architecture, and verified activity-dependent phage propagation in vitro and in PACE (Supplementary Figs. 6 and 7a,b, and Supplementary Results). Next, we used DB-PACE to evolve a canonical CBX8-targeting TALE towards recognition of non-canonical 5′ nucleotides (5′ A, C, and G). Following 48 h of DB-PACE we isolated phage with up to 6-fold increased activity on 5′ A relative to the canonical TALE protein, 5-fold increased activity on 5′ C, and 5-fold higher activity on 5′ G target sequences (Supplementary Figs. 8 and 9, and Supplementary Results). Analysis of the mutations in these clones revealed a number of neutral and beneficial amino acid substitutions that have not previously been described (Supplementary Figs. 8–11 and Supplementary Results).
As expected given the absence of a counter-selection, the activity of the evolved CBX8-targeting TALEs was increased in a promiscuous manner on all 5′ bases (Supplementary Fig. 12a). To evolve selective recognition of non-T 5′ nucleotides, we adapted our recently described PACE negative selection that links undesired activities to the production of pIII-neg, a dominant negative pIII that poisons phage propagation16. We constructed a series of negative selection APs (APNegs) in which binding of a TALE–ω fusion protein to an off-target DNA sequence induces expression of gene III-neg from a minimal lac promoter (Fig. 1b). To enable tuning of negative selection stringency, we placed a theophylline-inducible riboswitch upstream of gene III-neg. After validating the system (Supplementary Fig. 12b,c and Supplementary Results), we applied it to evolve TALE domains that preferentially bind a 5′ A target site over a 5′ T site using simultaneous positive and negative selection (Supplementary Results). All clones resulting from two different PACE lagoons displayed a substantial (> 2-fold) increase in DNA-binding activity on sequences beginning with 5′ A, 5′ C, and 5′ G, and clones from lagoon 2 (L2) displayed a two-fold reduction in binding affinity for the canonical 5′ T site, resulting in a ~4-fold 5′ A vs. T specificity change relative to the canonical TALE protein (Supplementary Fig. 13a and Supplementary Results). Analysis of mutations in these clones revealed context-dependent amino acid substitutions that alter TALE 5′ specificity (Supplementary Figs. 13 and 14, and Supplementary Results).
TALE arrays are frequently used in the context of TALE nucleases (TALENs) to initiate genome editing2. We hypothesized that DB-PACE could be used to improve TALEN specificity by decreasing TALE domain recognition of specific off-target sequences while maintaining on-target recognition. We used a TALEN pair that targets a 36-bp sequence within the human ATM locus (Supplementary Table 1) for which we previously identified off-target cleavage sites in human cells14. We generated an SP encoding the TALE specifying recognition of the 18-bp left half-site (ATM-L) fused to the ω RNAP subunit, an AP containing the corresponding ATM on-target binding sequence, and an APNeg containing a sequence corresponding to the left half site of OffA17, the most frequently cleaved genomic off-target sequence of this TALEN14 (Fig. 1c).
We performed an initial DB-PACE experiment on the ATM-L TALE in duplicate lagoons (L1 and L2) by incrementally increasing negative selection stringency against OffA17 binding (Supplementary Results). Next, we pooled phage from L1 and L2 and subjected the mixture to an additional 24 h of PACE (L3). Using an in vitro DNA cleavage specificity profiling assay14, we found that TALEN pairs containing evolved ATM-L TALEs from L1 or L3 retained on-target DNA cleavage activity comparable to that of the canonical TALEN (~32%), but exhibited virtually no detectable cleavage of OffA17 (compared with 9.5% cleavage for the canonical TALEN) (Fig. 1d). The L3-evolved clones assayed showed ≥ 16-fold higher on-target:OffA17 off-target cleavage specificity in vitro than the canonical TALEN (Fig. 1d). Importantly, evolved TALEs displayed wild-type or improved activity on the on-target sequence (Supplementary Figs. 15 and 16a). Analysis of individual amino acid mutations in evolved clones uncovered A252T as a key mutation and L338S as a potential accessory mutation that alter the on-target:off-target cleavage propensity of the ATM-targeting TALE (Supplementary Figs. 16b–d and 17a–d, and Supplementary Results).
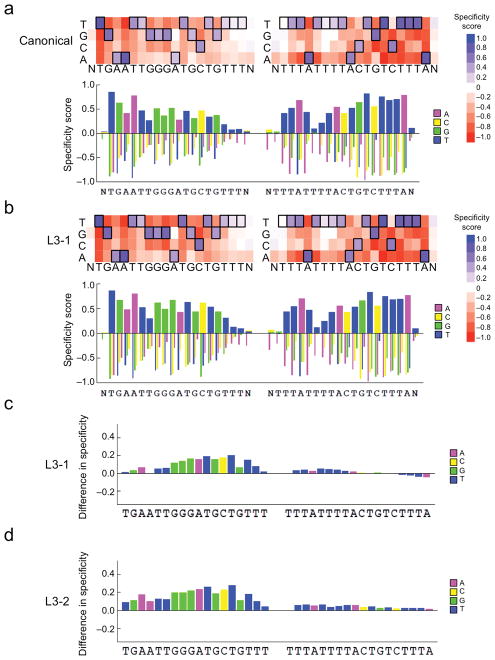
To investigate if the specificity enhancements of the evolved TALENs are limited to the OffA17 sequence or if they also improve specificity against other sequences, we tested their activity on derivatives of the OffA17 sequence (Supplementary Fig. 17e,f), and used our previously described TALEN specificity profiling method14 which measures the ability of a TALEN to cleave any of >1012 DNA sequences that are related to the on-target site (Supplementary Results). We found that TALEN pairs containing the evolved TALEs L3-1 and L3-2 showed a substantially decreased ability to cleave a wide range of off-target sequences containing four to nine mutations relative to the canonical TALEN (Supplementary Fig. 18 and Supplementary Table 2), indicating a broad specificity improvement. Moreover, TALEN pairs incorporating the evolved TALEs L3-1 or L3-2 displayed increased specificity relative to the canonical TALEN at nearly all positions in the left half-site of the ATM binding sequence, but no substantial change in specificity in the right half-site that was not used during DB-PACE (Fig. 2a–d and Supplementary Figs. 19–24 and Supplementary Results).
Figure 2. High-throughput specificity profiling of canonical and evolved TALENs.
Top: Heat map showing DNA cleavage specificity scores across > 1012 off-target sequences for either (a) canonical or (b) L3-1 evolved TALENs targeting the ATM locus at each position in the left and right half-sites plus a single flanking position (N). Bottom: Bar graph showing the quantitative specificity score for each nucleotide position. A score of zero indicates no specificity, while a score of 1.0 corresponds to perfect specificity. (c) Bar graph indicating the quantitative difference in specificity score at each position between the canonical and L3-1 evolved TALENs (scoreL3-1-scorecanonical) at each position in the target half-sites plus a single flanking position (N). A score of zero indicates no change in specificity. For all heat maps, the cognate base for each position in the target sequence is boxed. For the right half-site, data for the sense strand are displayed. (d) Identical to (c), except for the L3-2 evolved TALEN versus the canonical TALEN.
Finally, we tested the behavior of our evolved TALENs in human cells. We found that while cleavage at the on-target ATM site was comparable for the canonical and evolved TALENs (Supplementary Tables 1 and 3), both evolved TALENs exhibited greatly reduced or undetectable off-target activity relative to the canonical TALEN on all four genomic off-target loci assayed (Supplementary Table 3 and Supplementary Results). We confirmed that the improved DNA cleavage specificity of the evolved TALENs was independent of FokI architecture and cell line (Supplementary Table 3).
DB-PACE brings the power of continuous evolution to bear on improving the activity and specificity of a variety of DNA-binding proteins. Because DB-PACE does not require the use of targeted libraries that can constrain or bias evolutionary outcomes, it naturally supports the discovery of evolved solutions with desired properties that could not be rationalized a priori (Supplementary Discussion). Furthermore, DB-PACE coupled with in vitro specificity profiling represents a new systematic approach to removing specific off-target activities of TALENs, and may be used to facilitate generation of highly specific genome engineering tools for therapeutic applications (Supplementary Fig. 25).
Materials and Methods
Cloning and plasmid construction
PCR fragments for pOH, pAP, pAPNeg, pJG, and SP plasmids (see Supplementary Note 1 for plasmid design specifics) were generated using either PfuTurbo Cx Hotstart (Agilent) or VeraSeq Ultra (Enzymatics) DNA polymerases, and assembled by USER cloning (NEB) according to the manufacturer’s instructions. The Q5 Site-Directed Mutagenesis kit (NEB) was used for all site-directed mutagenesis, and to produce minimized pOH plasmids (pTet). DNA encoding TALEN cleavage sites were purchased as gBlocks (IDT) and inserted into pUC19 using XbaI and HindIII restriction enzymes. Representative primer sequences used for cloning are presented in Supplementary Table 4.
Phage-assisted continuous evolution (PACE) of DNA-binding domains
In general, PACE setup was performed as previously described16. E. coli were maintained in chemostats containing 200 mL of Davis’ Rich Media (DRM) 16 using typical flow rates of 1–1.5 vol/h. DRM media was supplemented with appropriate antibiotics to select for transformed plasmids: APs (50 μg/mL carbenicillin), APNegs (75 μg/mL spectinomycin), MPs (25 μg/mL chloramphenicol). Lagoon dilution rates were 1.3–2 vol/h. In all PACE experiments S1030 cells carried an MP, either the previously reported pJC18416, or a variant of this plasmid lacking RecA, pAB086a (see Supplementary Note 1). Mutagenesis was induced by continuously injecting arabinose (500 mM) at a rate of 1 mL/h into each 40-mL lagoon. Typical phage titers during each PACE experiment were 106–108 p.f.u./mL. Specific parameters for each evolution experiment are detailed below.
Reversion of Zif268-V24R
A lagoon receiving host cell culture from a chemostat containing S1059 cells transformed with an MP was inoculated with Zif268-V24R phage. The lagoon flow rate during drift was 2 vol/h. After 24 h of drift, phage were isolated and used to inoculate a PACE experiment with S1030 host cells carrying pAPZif268 (see Supplementary Notes 1 and 2) and an MP. Evolved phage were isolated after 24 h and characterized using plaque assays.
Positive selection of TALEs with altered 5′ preference (5′ A, C, G)
Three parallel evolution experiments were performed to evolve phage with higher affinity for 5′ A, 5′ C, or 5′ G target sequences. For each experiment, two separate lagoons receiving culture from a chemostat containing S1030 cells transformed with the appropriate AP (pAPCBXTAL:5A, pAPCBXTAL:5C, pAPCBXTAL:5G) (see Supplementary Note 1) and an MP were inoculated with SPCBXTAL. PACE proceeded for 48 h at a lagoon dilution rate of 1.3 vol/h prior to harvest and analysis of the resultant phage pools.
Negative selection to generate TALEs with 5′ A specificity
Two separate lagoons receiving culture from a chemostat containing a mixed population of S1030 cells were inoculated with evolved 5′ A phage from the positive selection experiment. This E. coli population consisted of a 1:1:1 mixture of host cells carrying an APNeg plasmid (pAPNegCBXTAL:5C, pAPNegCBXTAL:5G, or pAPNegCBXTAL:5T) together with pAPCBXTAL:5A and an MP. Over the course of a six-day PACE experiment, an increasing dose of theophylline was added to each lagoon at a rate of 1 mL/h to yield increasing final theophylline lagoon concentrations of 0.1 mM, 0.2 mM, and 0.3 mM (+0.1 mM theophylline every 48 h).
Positive selection and negative selection (OffA17) of ATM-L TALE
Two separate lagoons receiving culture from a chemostat containing a S1030 cells transformed with pAPATMLTAL, pAPNegATMTAL:OffA17, and an MP were inoculated with SPATMTAL phage (see Supplementary Note 1). The lagoon flow rate was 1.3 vol/h. Theophylline was added to each lagoon at increasing quantities (+0.1 mM every 24 h), from a starting dose of 0 mM to a final concentration of 0.4 mM; the injection rate into each lagoon was 1 mL/h. After 120 h of PACE, phage from both lagoons were pooled and subjected to an additional 24 h of PACE at a lagoon flow rate of 2 vol/h in the presence of 0.4 mM theophylline.
Luciferase assay
pOH plasmids were transformed by electroporation into S1030 cells (see Supplementary Notes 1 and 2), and grown overnight at 37 °C on LB-agar plates supplemented with 50 μg/mL carbenicillin. Single colonies were used to inoculate cultures which were allowed to grow for ~12 h at 37 °C in DRM supplemented with 50 μg/mL carbenicillin in a shaker. Cultures were diluted to an OD600 of ~0.3 and allowed to grow for an additional 2 h at 37 °C. Next, each culture was diluted 1:15 into 300 μL of DRM supplemented with 50 μg/mL carbenicillin in the presence or absence of 200 ng/mL anhydrotetracycline and incubated in a 96-well plate for an additional 4–6 h (shaking). 200 μL aliquots of each sample were then transferred to 96-well opaque plates and luminescence and OD600 readings were taken using a Tecan Infinite Pro instrument. Luminescence data were normalized to cell density by dividing by the OD600 value.
Plaque assays
S1030 cells were transformed with the appropriate plasmids via electroporation and grown in LB media to an OD600 of 0.8–1.0. Diluted phage stock samples were prepared (10−4, 10−5, 10−6, or 10−7-fold dilution) by adding purified phage stock to 250 μL of cells in Eppendorf tubes. Next, 750 μL of warm top agar (0.75% agar in LB, maintained at 55 °C until use) was added to each tube. Following mixing by pipette, each 1 mL mixture was pipetted onto one quadrant of a quartered petri plate that had previously been prepared with 2 mL of bottom agar (1.5% agar in LB). Following solidification of the top agar, plates were incubated overnight at 37 °C prior to analysis. Colorimetric plaque assays were performed in parallel with regular plaque assays using S2060 cells instead of S1030 cells, and used S-Gal/LB agar blend (Sigma) in place of regular LB-agar.
High-throughput analysis of TALE mutations
PCR fragments containing evolved phage with ~500 bp of flanking sequence on either end were amplified from minipreps (Qiagen) of cells infected with evolved phage pools using the following primers: HTSFwd – 5′-TGAAAATATTGTTGATGCGCTGGCAGTGTTC-′3, HTSRev – 5′-TAGCAGCCTTTACAGAGAGAATAACATAAAA-′3. HTS preparation was performed as previously reported20 using a Nextera kit (Illumina). Briefly, 4 μL of amplified DNA (2.5 ng/μL), 5 μL TD buffer, and 1 μL TDE1 were mixed together and heated at 55 °C for 5 min to perform “tagmentation”. Following DNA clean up using a Zymo-Spin column (Zymo), samples were amplified with Illumina-supplied primers according to the manufacturer’s instructions. The resulting products were purified using AMPure XP beads (Agencourt), and the final concentration of DNA was quantified by qPCR using PicoGreen (Invitrogen). Samples were sequenced on a MiSeq Sequencer (Illumina) using 2×150 paired-end runs according to the manufacturer’s protocols. Analysis of mutation frequency was performed using MATLAB as previously described20. Observed background mutation frequencies were subtracted from the mutation frequencies of each experimental sample to account for DNA sequencing errors20.
YFP assay
pTet plasmids were co-transformed with pAPNeg plasmids by electroporation into S1030 cells (see Supplementary Notes 1 and 2), and grown overnight at 37 °C on LB-agar plates supplemented with 50 μg/mL carbenicillin and 100 μg/mL spectinomycin. Single colonies were used to inoculate cultures which were allowed to grow for ~12 h in antibiotic-supplemented DRM in a bacterial shaker. Cultures were diluted to an OD600 of ~0.3 and allowed to grow for an additional 2 h at 37 °C. Next, each culture was diluted 1:15 into 300 μL of DRM supplemented with antibiotics and 5 mM theophylline in the presence or absence of 50 ng/mL anhydrotetracycline and incubated in a 96-well deep well plate for an additional 4–6 h (shaking). 200 μL aliquots of each sample were then transferred to 96-well opaque plates and YFP fluorescence (λex = 514 nm, λem = 527 nm) and OD600 readings were taken using a Tecan Infinite Pro instrument. Fluorescence data were normalized to cell density by dividing by the OD600 value.
In vitro TALEN cleavage assay
In vitro TALEN cleavage assays were performed as previously described with slight modifications to the procedure14. Briefly, 1 μg of each TALEN-encoding plasmid (pJG) was added individually to 20 μL of methionine-supplemented T7-TnT Coupled Transcription/Translation System (Promega) lysate and incubated for 1.5 h at 30 °C. Determination of protein concentrations and preparation of linear DNA for TALEN cleavage was performed as previously reported14. Each reaction consisted of 50 ng of amplified DNA, 12 μL NEB Buffer 3, 3 μL of each in vitro transcribed/translated TALEN left and right monomers (corresponding to ~15 nM final TALEN concentration), and 6 μL of empty lysate brought up to a final volume of 120 μL in distilled water. The digestion reaction was allowed to proceed for 30 min at 37 °C (or 1 h where indicated), and then incubated with 1 μg/uL RNase A (Qiagen) for 2 minutes prior to being purified using a Minielute column (Qiagen). Reactions were subsequently run in a 5% TBE Criterion PAGE gel (Bio-rad), and stained with 1X SYBR Gold (Invitrogen) for 10 minutes. Gels were imaged using a Syngene G:BOX Chemi XRQ, and densitometry was performed using GelEval 1.37 software.
High-throughput specificity profiling assay
High-throughput specificity profiling of canonical and evolved TALEN pairs and subsequent data analysis was performed as previously described14.
TALEN cleavage in HEK 293 and U2OS cells
pJG29 and pJG30 plasmids (see Supplementary Note 1) were transfected into HEK 293 cells (a cell line that has a high transfection efficiency; obtained from ATCC) using Lipoject (Signagen) according to the manufacturer’s instructions. pJG51 and pJG52 plasmids were nucleofected into U2OS cells as previously described14. For both sets of experiments, genomic DNA isolation was performed as previously reported14, 21. Primers for amplifying on and off-target genomic sites are listed in Supplementary Table 4. Illumina adapter ligation, AMPure XP bead cleanup (Agencourt), sequencing and post-analysis were performed as previously described14, 21. The HEK 293 cells used in this study tested negative for mycoplasma at the time of purchase, and the U2OS cell line was previously authenticated and shown to be negative for mycoplasma contamination 14.
Supplementary Material
Acknowledgments
This work was supported by Defense Advanced Research Projects Agency (DARPA) HR0011-11-2-0003, DARPA N66001-12-C-4207, a grant from the U.S. National Institutes of Health (NIH)/NIGMS (R01 GM095501), and the Howard Hughes Medical Institute (HHMI). S.Q.T., J.D.S., and J.K.J. were supported by an NIH Director’s Pioneer Award (DP1 GM105378). S.Q.T. was supported by NIH F32 GM105189. A.H.B. was supported by the Harvard Chemical Biology Program and a National Science Foundation Graduate Research Fellowship.
Footnotes
Accession codes
Sequence Read Archive: SRP055191 and SRP053327.
Author contributions
B.P.H. designed the research, performed experiments, analyzed data, and wrote the manuscript. A.H.B. assisted in the design of the one-hybrid system, and A.H.B. and K.M.D. contributed materials and performed experiments. J.A.Z, J.P.G., and L.C. performed experiments and data analysis. S.Q.T prepared materials for TALEN cleavage analysis in cells. J.D.S. contributed experimentally validated TALE arrays. D.R.L. designed and supervised the research and wrote the manuscript. All of the authors contributed to editing the manuscript.
Competing financial interests
D.R.L. is a consultant for Editas Medicine. J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
References
- 1.Tebas P, et al. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaj T, Gersbach CA, Barbas CF., 3rd Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epinat JC, et al. Nucleic acids research. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klug A. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 5.Maeder ML, et al. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Proc Natl Acad Sci U S A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moscou MJ, Bogdanove AJ. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 8.Boch J, et al. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 9.Deng D, et al. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb BM, Mercer AC, Barbas CF., 3rd Nucleic acids research. 2013;41:9779–9785. doi: 10.1093/nar/gkt754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuji S, Futaki S, Imanishi M. Biochem Biophys Res Commun. 2013;441:262–265. doi: 10.1016/j.bbrc.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 14.Guilinger JP, et al. Nat Methods. 2014;11:429–435. doi: 10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esvelt KM, Carlson JC, Liu DR. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. Nat Chem Biol. 2014;10:216–222. doi: 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson BC, Packer MS, Badran AH, Liu DR. Nat Commun. 2014;5:5352. doi: 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu JC, Kornacker MG, Hochschild A. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 19.Choo Y, Klug A. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickinson BC, Leconte AM, Allen B, Esvelt KM, Liu DR. Proc Natl Acad Sci U S A. 2013;110:9007–9012. doi: 10.1073/pnas.1220670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuris JA, et al. Nature biotechnology. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.