Abstract
The sensitivity of chromatin immunoprecipitation (ChIP) assays poses a major obstacle for epigenomic studies of low-abundance cells. Here we present a microfluidics-based ChIP-Seq protocol using as few as 100 cells via drastically improved collection of high-quality ChIP-enriched DNA. Using this technology, we uncovered many novel enhancers and super enhancers in hematopoietic stem and progenitor cells from mouse fetal liver, suggesting that enhancer activity is highly dynamic during early hematopoiesis.
Protein-DNA interaction and chromatin modification play critical roles in gene regulation. Chromatin immunoprecipitation coupled with deep sequencing (ChIP-Seq) has become the technology of choice for examining in vivo genome-wide protein-DNA interactions and chromatin modifications1. The assay involves covalently linking the epitope of interest to DNA by a reversible cross-linking reagent, cell lysis, immunoprecipitation of the protein of interest, reversal of the cross-linking, digestion of the protein, amplification and identification of the enriched DNA (i.e. ChIP DNA) by sequencing. A major limitation of conventional ChIP-Seq protocol is the requirement of a large number of cells (~107 cells). Various strategies have been developed to improve the traditional protocol over the past few years. Nano-ChIP-Seq was developed to examine histone modification using 5000 cells2. Single-tube linear DNA amplification (LinDA) was developed to profile the histone 3 lysine 4 tri-methylation (H3K4me3) mark using 10000 cells and oestrogen receptor-α binding using 5000 cells3. Both nano-ChIP-Seq and LinDA exploit novel strategies for amplifying ChIP DNA. Nano-ChIP-Seq uses a random primer with hairpin structure, optimizes conditions for faithful amplification of ChIP DNA by PCR, and uses BciVI restriction sites to allow direct ligation of Illumina sequencing adaptors2. LinDA amplifies ChIP DNA using an optimized T7 phage RNA polymerase linear amplification protocol that reduces amplification bias due to GC content3. Besides improving amplification of ChIP DNA, the use of histone or mRNA carrier has been shown to increase recovery of ChIP DNA and allowed transcription factor ChIP-Seq using 10000 cells4. Indexing-first ChIP (iChIP) was recently developed to index and pool many chromatin samples before ChIP5. The ChIP DNA from pooled samples (containing DNA prepared from >40000 cells) was then sequenced and the data were demultiplexed based on sample-specific bar codes to yield a sensitivity of 500 cells per individual sample.
Microfluidics provides the platform for conducting molecular assays with drastic reduction in the volume, high level of integration and automation, and effective manipulation of cells and particles. Several microfluidic ChIP protocols were reported recently for studying specific loci using ChIP coupled with qPCR6–8. However, no effective strategies have been developed for high-efficiency collection of ChIP DNA and suppressed nonspecific adsorption at the same time. Meeting both requirements is critical for genome-wide studies (i.e. ChIP-Seq) using a small number of cells.
The sensitivity of ChIP-Seq assays is largely limited by the collection efficiency of ChIP DNA. A diploid mammalian cell contains 4–8 pg of DNA yet previous ChIP-Seq protocols could only obtain tens of picograms of DNA from 10000 cells2,3,5. Here we introduce a simple microfluidics-based protocol, microfluidic-oscillatory-washing-based ChIP-Seq (MOWChIP-Seq). It provides high collection efficiency of ChIP DNA and allows genome-wide analysis of histone modifications using as few as 100 cells. The combined use of a packed bed of beads for ChIP and effective oscillatory washing for removing nonspecific adsorption and trapping is the key to extremely high yield of highly enriched DNA.
We used multilayer soft lithography to design and fabricate a polydimethylsiloxane (PDMS) device, featuring a simple microfluidic chamber (~710 nl in volume) for high-efficiency ChIP. The microfluidic chamber has one inlet 1 and one outlet 2 and the outlet has an on-chip pneumatic microvalve that can be partially closed by exerting a pressure at 37,9 (Fig. 1a, Supplementary Fig. 1 and 2). First, magnetic beads (~2.8 μm in diameter and coated with a ChIP antibody) are flowed into the microfluidic chamber and form a packed bed while the pneumatic microvalve is partially closed. Sonicated chromatin fragments (~200–600 bp) are then flowed through the packed bed of IP beads and adsorbed onto the bead surface. When closely packed, the gaps among the IP beads are smaller than 2 μm and facilitate rapid and high-efficiency adsorption of targeted chromatin fragments under the small diffusion length. The IP beads are then washed by oscillatory washing (Supplementary Video 1) in two different washing buffers to remove nonspecifically adsorbed chromatin fragments. Finally, the IP beads (with adsorbed chromatin fragments) are flowed out of the chamber and collected for off-chip processing. The entire on-chip process takes ~1.5 h.
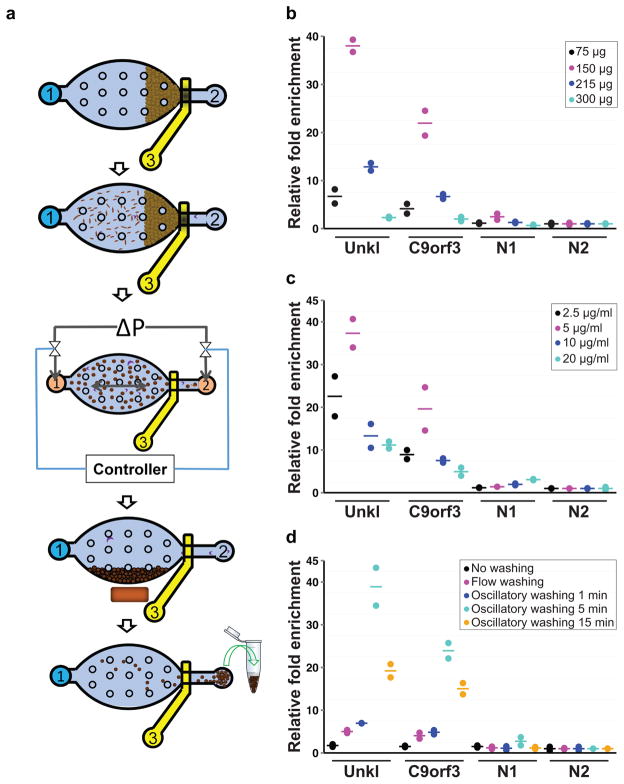
Figure 1. Overview of the MOWChIP-Seq protocol and its optimization.
(a) Schematic illustration for the five major steps of the protocol: step 1: Formation of a packed bed of IP beads; step 2: ChIP by flowing the chromatin fragments through the packed bed; step 3: Oscillatory washing; step 4: Removal of the unbound chromatin fragments and debris by flushing the chamber; step 5: Collection of the IP beads. The microfluidic chamber contains supporting pillars (shown as small circles) that prevent collapsing. (b–d) Optimization of the MOWChIP-Seq protocol. Major parameters of the protocol were optimized by checking for IP fold enrichment of known positive (UNKL and C9orf3) and negative loci (N1 and N2). IP was done against H3K4me3 in GM12878 cells. All experiments were conducted in duplicate and the horizontal lines represent the mean. Parameters optimized include: amount of beads in device chamber (b); concentration of antibody used for coating IP beads (c); washing duration in each of the two washing buffers (d). The relative fold enrichment was normalized against that of N2. 1000-cell samples were used in (b–d). 150 μg IP beads were used in (c, d). The antibody concentration for coating was 5 μg/ml for (b) and (d). The duration of oscillatory washing was 5 min for (b, c). Flow washing in (d) was implemented by flowing each washing buffer unidirectionally for 3 min under 1.5 μl/min.
We found that the quality and amount of ChIP DNA were affected by several parameters of the protocol, including amount of IP beads in the device, antibody concentration used for coating IP beads, duration of the oscillatory washing, and cell sample size. We optimized these parameters by using MOWChIP-qPCR to examine fold enrichment at known positive and negative loci for H3K4me3 (with the primer sequences listed in Supplementary Table 1) in a human lymphoblastoid cell line, GM12878 (Fig. 1b–d). The fold enrichment reached a peak value at an intermediate bead amount, likely due to increased nonspecific adsorption and trapping when too many beads were used (Fig. 1b). Similarly, for antibody used for coating IP beads, we obtained the highest fold enrichment at an intermediate antibody concentration (Fig. 1c). This is likely due to insufficient antibody coverage on the beads at low concentration, which decreases the amount of binding for chromatin targets. On the other hand, excessive amount of antibody on the beads may promote binding to low-affinity or nonspecific chromatin. The high-efficiency adsorption by the packed bed of IP beads also led to increased nonspecific adsorption and physical trapping. We found that oscillatory washing was essential for the high quality of ChIP DNA (Fig. 1d). At the same time, excessive washing needs to be avoided in order to reduce DNA loss. Using optimized conditions that balanced both DNA yield and quality, we were able to obtain ~1.3 ng ChIP DNA from 10000 cells (5.3% of the total chromatin) and ~180 pg from 1000 cells (6.2% of the total chromatin) for H3K4me3 after DNA purification (Supplementary Fig. 3). This yield was almost 2 orders of magnitude higher than that reported in previous work2 and within the range of the theoretical limit (2.2–7.8% of the genome is marked by H3K4me3 based on ENCODE data10). To assess the amount of background reads in our data, we computed the fraction of reads in peaks (FRiP)11. The values were 35.6% and 21.6% for our 10000- and 1000-cell data, respectively. These were substantially higher than 1% guideline recommended by ENCODE, suggesting low background in our recovered chromatin. As a result, we used ChIP DNA directly for sequencing library construction without pre-amplification.
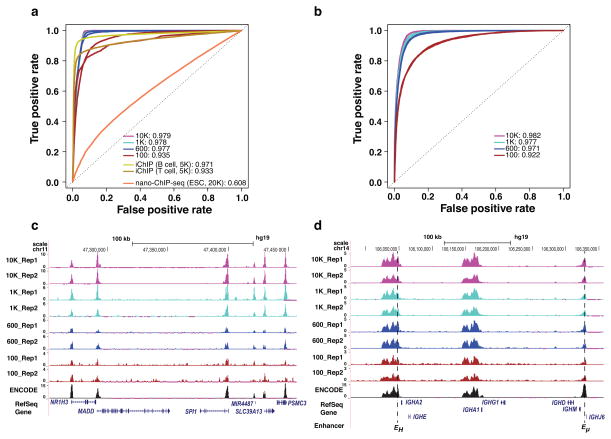
To evaluate the performance of MOWChIP-Seq, we used it to profile H3K4me3 and histone 3 lysine 27 acetylation (H3K27Ac) marks with various amounts of chromatin from GM12878 cells. We prepared sonicated chromatin using 10000 cells and aliquoted chromatin samples equivalent to 10000, 1000, 600 and 100 cells. For all four cell sample sizes, replicate experiments were highly correlated (average r = 0.933 and 0.894 for H3K4me3 and H3K27Ac, respectively, Supplementary Fig. 4). Using published ChIP-Seq data generated by conventional protocol with millions of cells per sample as the gold standard, we compared the performance of MOWChIP-Seq to two other methods, nano-ChIP-Seq2 and iChIP5 (detailed in Online Methods and Supplementary Table 2). We used the Receiver Operating Characteristic (ROC) curve to quantify the data quality in terms of the agreement with a gold standard. The Area Under the ROC Curve (AUC) is a standard metric for quantifying balanced sensitivity and specificity. For H3K4me3, MOWChIP-Seq with 100–600 cells showed performance that was comparable to iChIP with 5000 cells and superior to nano-ChIP-Seq with 20000 cells (Fig. 2a). For H3K27Ac, MOWChIP-Seq also produced data with excellent AUC values when using as few as 100 cells (Fig. 2b). Normalized MOWChIP-Seq signals (see Online Methods) for H3K4me3 at the Spi1 gene locus show consistency among samples of various sizes (Fig. 2c). This gene encodes an important transcription factor for B cell development. The Spi1 promoter region was highly enriched for H3K4me3 signal in MOWChIP-Seq data generated using 100–10000 cells. H3K27Ac is a mark for active transcriptional enhancers12. We show normalized MOWChIP-Seq signals for H3K27Ac across the immunoglobin heavy chain locus (Fig. 2d). As expected, the AUC values decreased with decreasing number of cells. Nevertheless, all data had good quality and reproducibility that enabled analysis of important genome-wide features (Supplementary Table 3).
Figure 2. MOWChIP-Seq generates high quality data using as few as 100 cells.
The performance of MOWChIP-Seq was compared to those of two other methods: nano-ChIP-seq and iChIP. (a) Receiver Operating Characteristic (ROC) curves for H3K4me3 data. ROC curves were constructed by comparing the ChIP-Seq data generated by various methods to published gold-standard data generated using conventional protocols with millions of cells. Nano-ChIP-Seq data was from Adli et al. 2; iChIP data was from Lara-Astiaso et al. 5. Values shown are average Area Under the ROC curve (AUC) of two replicate experiments. (b) ROC curves for H3K27Ac data generated by MOWChIP-Seq. (c) Normalized H3K4me3 MOWChIP-Seq signals at the SPI1 gene locus using data generated with various sample sizes. ENCODE data were generated using millions of cells and shown for comparison. (d) Normalized H3K27Ac MOWChIP-Seq signals at the immunoglobin heavy chain locus. Known B-cell enhancers are indicated at the bottom of the figure.
We applied MOWChIP-Seq to study the epigenome of hematopoietic stem and progenitor cells (HSPCs) isolated from mouse fetal liver. Little is known about the dynamics of the epigenome during embryonic hematopoiesis, largely due to the difficulty in isolating sufficient quantities of highly purified HSCs from developing embryos. This challenge makes mouse HSPC study an ideal test case for our technology. Definitive HSCs first appear in embryonic day 10.5 aorta-gonads-mesonephros and thereafter migrate to the fetal liver (FL) where they proliferate before they eventually colonize bone marrow (BM). Previously, histone modifications had been mapped in adult BM HSCs but not stem and progenitor cells at any earlier stage. We mapped H3K4me3 and H3K27Ac using chromatin equivalent to 10000, 1000, 600 and 100 purified FL HSPCs (Supplementary Fig. 5). For all four cell sample sizes, replicate experiments were highly correlated (average r = 0.864 and 0.881 for H3K4me3 and H3K27Ac, respectively, Supplementary Fig. 6). We also computed the correlation of our data with three published datasets on BM HSPC data. Since the HSPCs being compared are highly related but not identical, our data showed lower but reasonable correlation with published data, further supporting the quality of our data (average r = 0.677 and 0.745 for H3K4me3 and H3K27Ac, respectively, Supplementary Fig. 6). Histone modification signals of the promoter regions were also correlated with gene expression levels (r ranged from 0.47 to 0.60, Supplementary Fig. 7). Taken together, these results suggest that our FL HSPC data is of high quality.
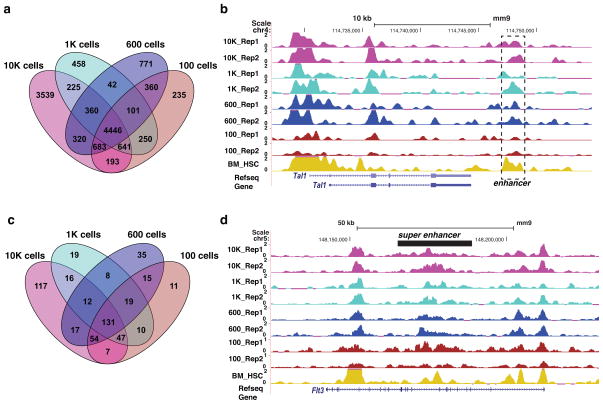
Little is known about the enhancer repertoire of FL HSPCs. Using our MOWChIP-Seq data, we predicted active enhancers in FL HSPCs using the signature of H3K4me3lo + H3K27Achi. In total, we predicted 10407, 6523, 7083, and 6909 enhancers (FDR < 0.5%) using the 10000, 1000, 600, and 100-cell data sets, respectively. The average pairwise overlap of the four sets of enhancers was 81.8% (Fig. 3a). In total, 4446 enhancers were shared among all four data sets, which we used as the final set of enhancers in this study (Fig. 3a). We identified many known transcriptional enhancers that are active in FL HSPCs, such as the Tal1 +19 enhancer (Fig. 3b) 13, Erg +85 enhancer and Runx1 +24 enhancer (Supplementary Fig. 8) 14,15. DNA motif analysis revealed that the set of enhancers were enriched for binding motifs of 45 transcription factors, including many well-known hematopoietic TFs such as ERG, ETV6, FLI1, PU.1 and RUNX1 (Supplementary Table 4). To identify the unique enhancers in FL HSPCs, we compared our enhancer set to the enhancer catalog covering 16 blood cell types5. We found that 58% (2,561) of enhancers identified in this study were unique to FL HSPCs (Supplementary Table 5), suggesting enhancer activity is highly dynamic during early hematopoiesis.
Figure 3. Epigenomics-aided discovery of novel enhancers and super enhancers in fetal liver HSPCs.
(a) Venn diagram of sets of enhancers predicted using epigenomic data generated using various sample sizes. (b) Normalized H3K27Ac ChIP-Seq signals at the known Tal +19 enhancer. BM_HSC denotes data on bone marrow HSC generated by Lara-Astiaso et al. 5. (c) Venn diagram of sets of super enhancers predicted using epigenomic data generated using various sample sizes. (d) Normalized H3K27Ac ChIP-Seq signals at the super enhancer of the Flt3 gene that plays a critical role in hematopoiesis.
Super enhancer (SE) is a newly discovered class of enhancers that are typically much longer than single enhancers 16. They play a critical role in regulating genes that determine lineage identity. Almost nothing is known about super enhancers in HSPCs. Using our epigenomic data, we discovered 131 SEs in FL HSPCs (Fig. 3c and Supplementary Table 6). Consistent with the notion that SEs often regulate lineage-conferring genes, target genes of our predicted SEs were enriched for genes involved in hematopoiesis (p-value = 6.5E-3, hypergeometric test, Supplementary Table 7). Example target genes included many known key regulators of hematopoiesis such as Erg, Etv6, Fli1, Flt3, Runx1, and Spi1. The super enhancer controlling the Flt3 gene plays an important role in hematopoiesis, especially for FL HSPCs (Fig. 3d)17.
The chromatin used for generating 1000-, 600-, and 100-cell data above (Fig. 2 and 3) was aliquoted from a stock chromatin sample prepared from 10000 cells. In order to use MOWChIP-Seq directly on samples with 100–600 cells, we replaced washing of cross-linked cells with dilution by the sonication buffer in order to minimize chromatin loss due to centrifugation and resuspension (see Online Methods). Our modified cross-linking and sonication procedures generated desired chromatin size distribution for sequencing after library preparation (Supplementary Fig. 9). We generated additional MOWChIP-Seq data using the modified protocol with starting cell numbers of 100 and 600 and assessed the data quality by correlation and ROC curve analysis. Data generated using the two protocols (i.e. using the stock chromatin vs. starting directly with 100 or 600 cells) were highly correlated (average r = 0.843 over two histone marks, Supplementary Fig. 10). Data generated using the two protocols also had similar AUC values (Fig. 2 and Supplementary Fig. 11).
In summary, we demonstrated that MOWChIP-Seq with as few as 100 cells generated high quality genome-wide profiles of histone modifications. Our microfluidic technology is fundamentally different from other high-sensitivity ChIP technologies which rely on superior amplification2,3,18 and indexing-pooling5 schemes, thus may potentially complement other methods. The microfluidic device has a simple structure and is easy to operate. The platform allows running of multiple assays in parallel. Our technology paves the way for epigenomic studies involving extremely low number of cells from animals and patients.
Online Methods
Fabrication of the microfluidic ChIP device
The microfluidic chip consisted of a microfluidic chamber, connecting channels, and a micromechanical valve (Fig. 1 and Supplementary Fig. 2). The microfluidic chamber had an elliptic shape with a major axis of 6 mm, a minor axis of 3 mm and a depth of 40 μm. Micropillars were positioned inside the microfluidic chamber to prevent collapsing. The on-chip micromechanical valve, which allowed partial closure, was employed to stop magnetic IP beads while allowing liquid flow.
The microfluidic device was fabricated out of polydimethylsiloxane (PDMS) using multilayer soft lithography with minor modifications7. Briefly, two photomasks were generated with the microscale patterns designed using FreeHand MX (Macromedia) and printed on high-resolution (5080 dpi) transparencies. The patterns in the photomasks were replicated onto two masters (i.e. silicon wafers with photoresist patterns) for the control layer (~50 μm thick, SU-8 2025, Microchem) and the fluidic layer (~40 μm thick, SU-8 2025) with the photoresist spun on a 3-inch silicon wafer (978, University Wafer). Prepolymer PDMS (General Electric silicone RTV 615, MG chemicals) with a mass ratio of A:B = 5:1 was poured onto the fluidic layer master in a Petri dish to generate ~5 mm thick fluidic layer. PDMS at a mass ratio of A:B = 20:1 was spun onto the control layer master at 1100 rpm for 35 s, resulting in the thin PDMS control layer (~108 μm thick). Both layers of PDMS were partially cured at 80°C for 30 min. The fluidic layer was then peeled off the master. The fluidic layer feature was aligned with and bonded to that of the control layer from the top. The two-layer PDMS structure was baked at 80°C for 60 min, peeled off from the control layer master, and punched to produce the inlet and the outlet. The two-layer PDMS and a pre-cleaned glass slide were treated with oxygen plasma cleaner (PDC-32G, Harrick Plasma) and immediately brought into contact against each other to form closed channels and chamber. Finally, the assembled chip was baked at 80°C for 1 h to strengthen the bonding between PDMS and glass. Glass slides were cleaned in a basic solution (H2O: 27% NH4OH: 30% H2O2= 5:1:1, volumetric ratio) at 75°C for 2 h and then rinsed with ultra-pure water and thoroughly blown dry.
Setup of the microfluidic device
The microfluidic chip was mounted on an inverted microscope (IX 71, Olympus) and the operation was monitored by a CCD camera (ORCA-285, Hamamatsu) attached to the port of the microscope. Prior to experiments, the control channel was pre-filled with water to prevent bubble formation in the fluidic channel. The reagents were introduced into the inlet via perfluoroalkoxyalkane (PFA) high purity tubing (1622L, ID: 0.02 in. and OD: 0.0625 in., IDEX Health & Science) with the flow driven by a syringe pump (Fusion 400, Chemyx). The on-chip micromechanical valve was actuated by a solenoid valve (18801003-12V, ASCO Scientific) and a pressure source (either a gas cylinder or a compressed air outlet). A data acquisition card (NI SCB-68, National Instruments) and a LabVIEW (LabVIEW 2012, National Instruments) program were employed to control the switching of the solenoid valve. The applied pressure (35–40 psi) in the PDMS control channel deformed the thin PDMS membrane between the fluidic and control channels and closed the fluidic channel partially to stop beads while allowing liquid to flow. During oscillatory washing, the inlet and outlet of the microfluidic chamber were attached to two solenoid valves via PFA tubing and the pressure pulses were applied via the two solenoid valves under the automation by the data acquisition card and the LabVIEW program.
Preparation of sonicated chromatin
10000-cell samples
10000-cell samples were centrifuged at 1,600xg for 5 min at room temperature in a swing bucket centrifuge with soft deceleration. Cells were then washed twice with 1.0 ml 1x PBS (14190-144, Sigma-Aldrich) at room temperature by centrifugation and resuspension. Cells were cross-linked for 5 min with 1ml 1% freshly-prepared formaldehyde (28906, Thermo Scientific). Cross-linking was terminated by adding 0.05 ml 2.5 M glycine (R000333, Covaris) and shaking for 5 min at room temperature. Cross-linked cells were pelleted and washed with pre-cooled PBS buffer and resuspended in 130 μl of the sonication buffer (Covaris, 10mM Tris-HCl, pH8.1, 1mM EDTA, 0.1% SDS, and 1× protease inhibitor cocktail (R000306, Covaris)). Cross-linked cells were sonicated with a Covaris E220 sonicator for 14 min with 5% duty cycle, 105 peak incident power and 200 cycles per burst. The sonicated lysate was centrifuged at 14000×g for 10 min at 4°C. Sonicated chromatin in the supernatant was transferred to a new 1.5 ml LoBind Eppendorf tube (17014013, Denville) for MOWChIP-Seq. From this stock chromatin preparation, samples equivalent to 1000, 600 and 100 cells were aliquoted and diluted to give a final volume of 50 μl for MOWChIP-Seq. 10% of the sample was used as the input. After this procedure, we typically obtained ~2.7 pg DNA per cell from the pre-ChIP chromatin samples. DNA was extracted using the IPure kit from Diagenode (C03010012). DNA concentration was measured using a Qubit 2.0 fluorometer with dsDNA HS Assay kit (Q32851, Life Technologies).
100- or 600-cell samples
The procedure was different for preparing sonicated chromatin from 100 or 600 cells directly. Cells were counted with a hematocytometer and then 100 or 600 cells were transferred to a 1.5 ml LoBind Eppendorf tube containing 10 μl 10% FBS in PBS. Cells were then cross-linked for 5 min at room temperature by adding 0.625 μl 16% formaldehyde to yield a final concentration of 1%. Cross-linking was quenched by adding 1.25 μl 2.5 M glycine for 5 min at room temperature. The cross-linked sample was then diluted using 120 μl Covaris sonication buffer (to give a total volume of 130 μl) and sonicated with a Covaris E220 sonicator for 8 min with 5% duty, 105 peak incident power and 200 cycles per burst in a Covaris microtube (520045, Covaris). The sonicated lysate was centrifuged at 14000×g for 10 min at 4°C. Sonicated chromatin in the supernatant was transferred to a new 1.5 ml LoBind Eppendorf tube for MOWChIP-Seq. After this procedure, we typically obtained ~3.8 pg DNA per cell from the pre-ChIP chromatin samples. This per-cell yield was substantially higher than that obtained using the above procedure because we replaced washing of cross-linked cells (involving centrifugation and resuspension) with dilution by the sonication buffer to minimize chromatin loss.
Preparation of immunoprecipitation (IP) beads
Superparamagnetic Dynabeads® Protein A (2.8 μm, 30 mg/ml, 10001D, Invitrogen) were used for immunoprecipitation. 150 μg (5 μl of the original suspension) beads were washed twice with freshly-prepared IP buffer (20 mM Tris-HCl, pH8.0, 140 mM NaCl, 1mM EDTA, 0.5mM EGTA, 0.1%(w/v) sodium doxycholate, 0.1% SDS, 1%(v/v) Triton-100X) and resuspended in 150 μl IP buffer which contained antibody. Beads were gently mixed with the antibody at 4°C on a rotator mixer at 24 rpm for 2 h. Antibody-coated beads were washed twice with the IP buffer, and resuspended in 5 μl IP buffer. We optimized the antibody concentration for the bead coating step based on our ChIP-qPCR results. The optimal antibody concentration for MOWChIP-Seq with anti H3K4me3 antibody (07-473, Millipore) and anti H3K27Ac antibody (ab4729, Abcam) was 3.3 μg/ml for 100~600 cells, 5 μg/ml for 1000 cells, and 6.6 μg/ml for 10000 cells. These conditions were equivalent to using 495, 750, and 990 ng antibody in the preparation of 150 μg IP beads.
MOWChIP
The MOWChIP process involved several steps (Fig. 1a and Supplementary Fig. 2). The microfluidic device was first rinsed with the IP buffer for conditioning. The antibody-coated magnetic IP beads were then loaded into the microfluidic chamber via the combined effects of pressure-driven flow (provided by the syringe pump) and magnetic force generated by a cylindrical permanent magnet (NdFeB, D48-N52, 0.25 in. dia. and 0.5 in. thick, K&J Magnetics). The on-chip micromechanical valve was partially closed and the IP beads were packed against the valve to form a packed bed. After the loading of the IP beads (~150 μg under optimal condition), the IP buffer (with freshly added 1 mM PMSF (78830-1G, Sigma-Aldrich) and 1% protease inhibitor cocktail (P8340, Sigma-Aldrich)) containing sonicated chromatin fragments (with a total volume of either 50 or 130 μl) was flowed through the packed bed of IP beads at a flow rate of 1.5 or 3.5 μl/min, respectively. Under these flow rates, the immunoprecipitation step was finished around 40 min.
After ChIP, a low-salt washing buffer (20 mM Tris-HCl, pH 8.0,150 mM NaCl, 2 mM EDTA, 0.1%SDS, 1%(v/v) Triton-100X) was flowed into the microfluidic chamber. Oscillatory washing was conducted (for 5 min unless otherwise noted) to remove nonspecifically adsorbed or physically trapped materials from the bead surface. We prefilled the tubing with 10 μl washing buffer at each end of the microfluidic chamber and kept the on-chip valve open. Pressure pulses (each at 3 psi, with a pulse width of 0.5 s and an interval of 0.5 s between two pulses) were applied alternatingly at either end of the microfluidic chamber. The duration and frequency of the pressure pulses were set in a LabVIEW program and implemented via the regulation of the two solenoid valves by the data acquisition card (Supplementary Fig. 1b). After the oscillatory movement, the IP beads were retained by the NdFeB magnet on one side of the chamber while the unbound chromatin fragments and other debris/waste were flushed out of the microfluidic chamber by a clean washing buffer flow at 2 μl/min. The process of oscillatory washing was repeated once using a high salt washing buffer (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1%(v/v) Triton-100X). Finally the IP beads were flowed out of the microfluidic chamber under a flow rate of 50 μl/min and collected into a 1.5 ml LoBind Eppendorf tube containing 100 μl IP buffer. The optimal duration for washing was 5 min for both washing buffers.
Extraction of ChIP DNA and input DNA
Chromatin samples (either ChIP or input chromatin) were processed by IPure kit (C03010012, Diagenode) to extract DNA. Purified DNA was dissolved in 10 μl DNase-free water and used directly for ChIP-qPCR or for sequencing library construction. DNA concentrations were measured using a Qubit 2.0 fluorometer with dsDNA HS Assay kit (Q32851, Life Technologies).
Construction of sequencing libraries
Sequencing libraries were prepared using ThruPLEX-FD kit (Rubicon Genomics). This kit reduces the assay time and the risk of contamination by using a single tube and eliminating intermediate purification steps. The process involved template preparation, library synthesis, and library amplification. Adaptor-based PCR amplification (98 °C for 20 s, 72 °C for 50 s for each cycle) was used during library amplification. We used 11 cycles for input DNA, 12~13 cycles for ChIP DNA from 10000 cells, and 14~17 cycles for ChIP DNA from 1000 or fewer cells. The libraries were purified using Ampure XP beads (A63880, Beckman Coulter). Library fragment size was determined using high sensitivity DNA analysis kit (5067-4626, Agilent) on an Agilent 2200 TapeStation. KAPA library quantification kit (KK4809, Kapa Biosystems) was used to determine effective library concentrations. The final concentrations of libraries submitted for sequencing were ~2 nM. The libraries were sequenced on an Illumina HiSeq 2500 with single-end 50 nt read. Typically 15–20 million reads were generated per library.
Cell culture
GM12878 cells were obtained from Coriel Institute for Medical Research. Species of origin of the cell line was confirmed by PCR targeting the glucose-6-phosphate dehydrogenase gene. Donor subject has a single bp (G-to-A) transition at nucleotide 681 in exon 5 of the CYP2C19 gene (CYP2C19*2) which creates an aberrant splice site. Donor origin of the cell line was confirmed using PCR against the point mutation. The cell line was tested for mycoplasma contamination using ABI MycoSEQ mycoplasma detection assay (Applied Biosystems). Cells were propagated in RPMI 1640 (11875-093, Gibco) plus 15% fetal bovine serum (26140-079, Gibco), 100 U penicillin (15140-122, Gibco), and 100mg/ml streptomycin (15140-122, Gibco) at 37°C in a humidified incubator containing 5% CO2. Cells were sub-cultured every two days to maintain them in exponential growth phase.
Mouse strain, embryo dissection and cell sorting by fluorescence-activated cell sorting (FACS)
The University of Iowa Office of the Institutional Animal Care and Use Committee review board approved these studies. Wild type C57BL/6 (Stock No. 000664) and B6129S6F1 (Stock No. 101043) mice were purchased from the Jackson Laboratory. To obtain embryonic day 14.5 (E14.5) fetal liver (FL), B6129SF1 females were mated with C57BL/6 males (6–9 weeks old) late in the afternoon and females were checked the following morning for the presence of a vaginal plug, which was designated as E0.5. FLs were dissected from E14.5 embryos. Single cell suspensions were prepared by dissociating mechanically and expelling the cells through 40 μm nylon filter (352340, Falcon), followed by red blood cell lysis (ACK Lysing Buffer, 10-548E, Lonza). Cells were resuspended in 1 ml staining buffer (2% FBS in PBS) per 1×108 cells. FACS was performed as previously described19,20 with a few modifications. To remove nonspecific binding, anti-mouse CD16/CD32 (Fc Block, 101302, Biolegend) were added to single cell suspension and incubated for 10 min at 4°C. Next, cells were stained with a cocktail of antibodies against lineage markers (Ly-6G/Ly-6C (108417), CD45R/B220 (103225), CD3ε (100321), TER-119b (116215), CD4 (100529), CD8a (100723), CD19 (115521)), Kit (17-1171-83), and Sca-1 (12-5981-83). Lineage antibodies were purchased from Biolegend. Kit and Sca-1 antibodies were purchased from eBiosciences. Stained samples were first subjected to yield sort for Lineage−Sca-1+ Kit+ (LSK) cells and collected into a 12×75-mm polystyrene tube containing 500μl 1× IMDM (12440-053, Gibco)+20% FBS. Collected cells were then subjected to purity sort using the same gating strategy and sorted into a 1.5 ml DNA LoBind tube containing 0.8 ml 1× IMDM+50% FBS. On average, ~10000 FL LSK cells can be obtained per mouse embryo.
MOWChIP-qPCR data analysis
Real-time PCR was done using iQ SYBR Green Supermix (1708882, Bio-Rad) on an CFX96 real-time PCR machine (Bio-Rad) with C1000Tm thermal cycler base. All PCR assays were performed using the following thermal cycling profile: 95°C for 10 min followed by 40 cycles of (95°C for 15 s, 58°C for 40 s, 72°C for 30s). Primer concentrations were 400 nM. All primers were ordered from Integrated DNA Technologies. The results were represented as relative fold enrichment, which is the ratio of percent input between a positive locus and a negative locus. Percent input was computed using the following equation:
where Ctinput and CtIP are the Ct values of input and ChIP DNA, respectively; dilution factor (DF) is defined as (sample volume of input + sample volume of IP)/(sample volume of input).
MOWChIP-Seq reads mapping and normalization
Sequencing reads were mapped to the mouse genome (mm9) and human genome (hg19) using Bowtie2 (v2.2.2) 21 with default parameter settings. Uniquely mapped reads from both ChIP and input samples were used to compute a normalized signal for each 100 nt bin across the genome. Normalized signal is defined as following:
Peak calling of MOWChIP-Seq data
Only uniquely mapped reads were used for peak calling. Two peak callers were used with the following parameter settings: MACS (p-value < 10−5) 22 and SPP (z-score > 4) 23 with other parameters set at default values. The final set of high-confidence peaks was those that were called by both methods.
Construction of Receiver Operating Characteristic (ROC) curves
Using ROC curves, we compared the performance of MOWChIP-Seq to that of two state-of-the-art methods nano-ChIP-seq2 and iChip5. We focused on promoter regions (defined as 2000 bp upstream and 500 bp downstream of a transcription start site (TSS)). We obtained published ChIP-Seq data generated using conventional protocol with a large sample size (typically 10 million cells per sample) as the gold standard. The gold-standard true positives were defined as the set of high-confidence promoter peaks identified as described in the peak calling section. The set of promoter regions that did not overlap with any peaks were defined as the gold-standard negative set. Using the gold-standard sets, the following quantities were defined to compute the ROC curve: True Positives (TPs), peaks that were supported by the gold-standard positive set; False Positives (FPs), peaks that were not supported by the gold-standard positive set; False Negatives (FNs), gold-standard positives that were not called peaks in an experiment; True Negatives (TN), peaks that were not called in an experiment and were in the gold-standard negative set. True positive rate (TPR) was defined as TP/(TP+FN) and false positive rate (FPR) was defined as FP/(FP+TN). ROC curves were generated by computing TPR and FPR values on prediction sets obtained by varying the peak calling threshold.
The gold-standard datasets used for constructing the ROC curves for MOWChIP-Seq, nano-ChIP-seq and iChIP were summarized in Supplementary Table 2. Briefly, we generated H3K4me3 and H3K27ac data using GM12878 cells. The corresponding gold-standard data were generated by the ENCODE consortium. The authors of Nano-ChIP-Seq generated H3K4me3 data using mouse ESCs. The corresponding gold-standard data were from Marson et al.24 and Goren et al.25. The authors of iChIP generated H3K4me3 data using mouse CD4 T and B cells. The corresponding gold-standard data were from Wei et al.26 and Heinz et al.27.
Correlation analysis of MOWChIP-Seq data with other published ChIP-Seq data sets
To evaluate the quality of our FL HSPC data, we selected four published datasets of H3K4me3 and H3K27ac using BM LSK2, BM LT-HSC5, B cell5 and macrophage5. For a given histone modification, normalized ChIP-Seq signals in all promoter regions in the genome were extracted. Promoter regions were defined as +/− 2kb around transcription start sites (TSS). TSS annotation was based on RefSeq. Averaged signals across the promoter region was used. Promoter regions with zero signals in both data sets were excluded for computing Pearson correlation coefficient.
Prediction of enhancers and super enhancers using epigenomic data
We used “H3K4me3lo + H3K27Achi” to define enhancers. Specifically, enhancers were predicted using the CSI-ANN algorithm28 and normalized H3K4me3 and H3K27ac MOWChIP-Seq signals across the genome. The coordinates of the predicted enhancers and H3K27ac MOWChIP-Seq data were then used as the input to predict super enhancers using the ROSE software by the Young lab (http://bitbucket.org/young_computation/rose). We set the parameters to allow enhancers within 15000 bp to be stitched together. In addition, we excluded the constituent enhancers located within +/− 2000 bp from annotated TSSs.
Transcription factor motif enrichment analysis
We compiled a set of 1207 TF binding motifs from three major public databases, JASPAR 29, UniPROBE 30, Transfac 31 and motifs of ten hematopoietic transcription factors32. We used the program CentriMo 33 to identify over-represented motifs in a given set of enhancer sequences. Default parameters of CentriMo were used.
Gene ontology (GO) term enrichment analysis of super enhancer targets
Genes closest to the super enhancers were used as their targets. Database for Annotation, Visualization, and Integrated Discovery (DAVID) 34 was used for GO analysis of the target genes. Nominal p-values were corrected for multiple testing using the method by Benjamini and Hochberg 35. GO terms with a corrected p-value of 0.05 were regarded as significant.
Assumptions of statistical tests
All statistical tests were performed using large sample sizes and underlying distribution assumptions were met. Sample sizes were reported in figure legends. All reported p-values were corrected for multiple testing.
Supplementary Material
Acknowledgments
We thank Genomics Research Laboratory of Virginia Bioinformatics Institute and Genomics Division of Iowa Institute of Human Genetics for providing sequencing service; L. Van Tol and University of Iowa Institute for Clinical and Translational Science for providing computing support. This work was supported by US National Institutes of Health grants EB017855 (K.T. and C.L.), CA174577 (C.L.), EB017235 (C.L.), HG006130 (K.T.), GM104369 (K.T.), and a seed grant from Virginia Tech Institute for Critical Technology and Applied Science (C.L.).
Footnotes
Accession codes. Gene Expression Omnibus: MOWChIP-Seq data were deposited under accession number GSE65516.
Author contributions: C.L. designed the microfluidic device. C.L., K.T. and Z.C. developed the MOWChIP-Seq technology. Z.C. generated the MOWChIP-qPCR and MOWChIP-Seq data. K.T., C.C. and B.H. designed the biological experiments, isolated the primary cells from mice, and conducted data analysis. All authors wrote the manuscript together.
Competing financial interests: The authors declare competing financial interests. Virginia Polytechnic Institute and State University (on behalf of C.L. and Z.C.) filed an US utility patent (US letters patent serial no. 14/511,422) on the MOWChIP-Seq technology on Oct 10, 2014.
References
- 1.Park PJ. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adli M, Zhu J, Bernstein BE. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shankaranarayanan P, et al. Nat Methods. 2011;8:565–U565. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 4.Zwart W, et al. BMC Genomics. 2013;14:232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lara-Astiaso D, et al. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu AR, et al. Lab Chip. 2009;9:1365–1370. doi: 10.1039/b819648f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng T, et al. Lab Chip. 2011;11:2842–2848. doi: 10.1039/c1lc20253g. [DOI] [PubMed] [Google Scholar]
- 8.Wu AR, et al. Lab Chip. 2012;12:2190–2198. doi: 10.1039/c2lc21290k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong JW, Studer V, Hang G, Anderson WF, Quake SR. Nat Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- 10.Kundaje A, et al. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landt SG, et al. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rada-Iglesias A, et al. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottgens B, et al. Embo J. 2002;21:3039–3050. doi: 10.1093/emboj/cdf286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoms JA, et al. Blood. 2011;117:7079–7089. doi: 10.1182/blood-2010-12-317990. [DOI] [PubMed] [Google Scholar]
- 15.Nottingham WT, et al. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hnisz D, et al. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buza-Vidas N, et al. Blood. 2009;113:3453–3460. doi: 10.1182/blood-2008-08-174060. [DOI] [PubMed] [Google Scholar]
- 18.Jakobsen JS, et al. BMC Genomics. 2015;16:46. doi: 10.1186/s12864-014-1195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinney-Freeman S, et al. Cell Stem Cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinney-Freeman SL, et al. Blood. 2009;114:268–278. doi: 10.1182/blood-2008-12-193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharchenko PV, Tolstorukov MY, Park PJ. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marson A, et al. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goren A, et al. Nat Methods. 2010;7:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei G, et al. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, et al. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firpi HA, Ucar D, Tan K. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Portales-Casamar E, et al. Nucleic Acids Res. 2010;38:D105–110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newburger DE, Bulyk ML. Nucleic Acids Res. 2009;37:D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matys V, et al. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson NK, et al. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 33.Whitington T, Frith MC, Johnson J, Bailey TL. Nucleic Acids Res. 2011;39:e98. doi: 10.1093/nar/gkr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Lempicki RA. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.