Abstract
Methods for rapidly assessing sequence-structure-function landscapes and developing conditional gene-regulatory devices are critical to our ability to manipulate and interface with biology. We describe a framework for engineering RNA devices from preexisting aptamers that exhibit ligand-responsive ribozyme tertiary interactions. Our methodology utilizes cell sorting, high-throughput sequencing, and statistical data analyses to enable parallel measurements of the activities of hundreds of thousands of sequences from RNA device libraries in the absence and presence of ligands. Our tertiary interaction RNA devices exhibit improved performance in terms of gene silencing, activation ratio, and ligand sensitivity as compared to optimized RNA devices that rely on secondary structure changes. We apply our method to building biosensors for diverse ligands and determine consensus sequences that enable ligand-responsive tertiary interactions. These methods advance our ability to develop broadly applicable genetic tools and to elucidate understanding of the underlying sequence-structure-function relationships that empower rational design of complex biomolecules.
Introduction
Engineered biological systems hold potential in programming cell behavior to advance sustainable technologies, materials synthesis, and human health. However, incomplete understanding of the sequence-structure-function relationships that govern the design space limits our capacity to access, process, and act on information in living systems. Methods for assessing sequence-structure-function landscapes and developing conditional gene-regulatory devices are thus critical to advancing our ability to manipulate and interface with biology.
Programmable RNA-based gene-regulatory devices comprise parts that encode sensing, information transmitting, and actuating functions1. RNA device architectures connect sensor and actuator components, such that sensor-detected information is transmitted into controlled activity of the actuator. One class of RNA devices utilizes a hammerhead ribozyme (HHRz) actuator to modulate the stability of a target transcript through conditional control of cleavage activity via binding of the cognate ligand1. The ribozyme-based device framework supports genetic controllers in different organisms2–6, responsive to diverse ligands1,3,7–9, exhibiting complex computation10, and applied to regulate complex phenotypes11,12. Sensor and actuator components are linked through a rationally designed1 or screened13 transmitter that guides secondary structure changes in the components. As RNA folding is largely hierarchical and dictated by localized hydrogen bonding and base stacking14, secondary structure changes are tractable. While this approach enables sequence-level modular device design1, it limits regulatory potential. The relatively slow kinetics associated with the transmitter-induced secondary structure rearrangement15 places a limit on self-cleavage kinetics13,16, over which a trade-off between gene-silencing activity and ligand sensitivity is observed17.
To address performance limitations inherent with secondary structure switching RNA devices, a new device architecture that achieves faster switching is needed. The natural diversity of HHRz tertiary interactions18, inspires a tertiary structure switching architecture that removes the transmitter and encoded secondary structure rearrangement. A platform modulating HHRz tertiary interactions16 (Fig. 1a) may achieve improved performance by eliminating the slow secondary structure conformational change14, thereby supporting ribozymes with faster cleavage kinetics. Since ribozyme tertiary interactions are only functionally conserved18, a library framework that supports the creation of RNA devices with ligand-responsive tertiary interactions can be screened for functional sequences.
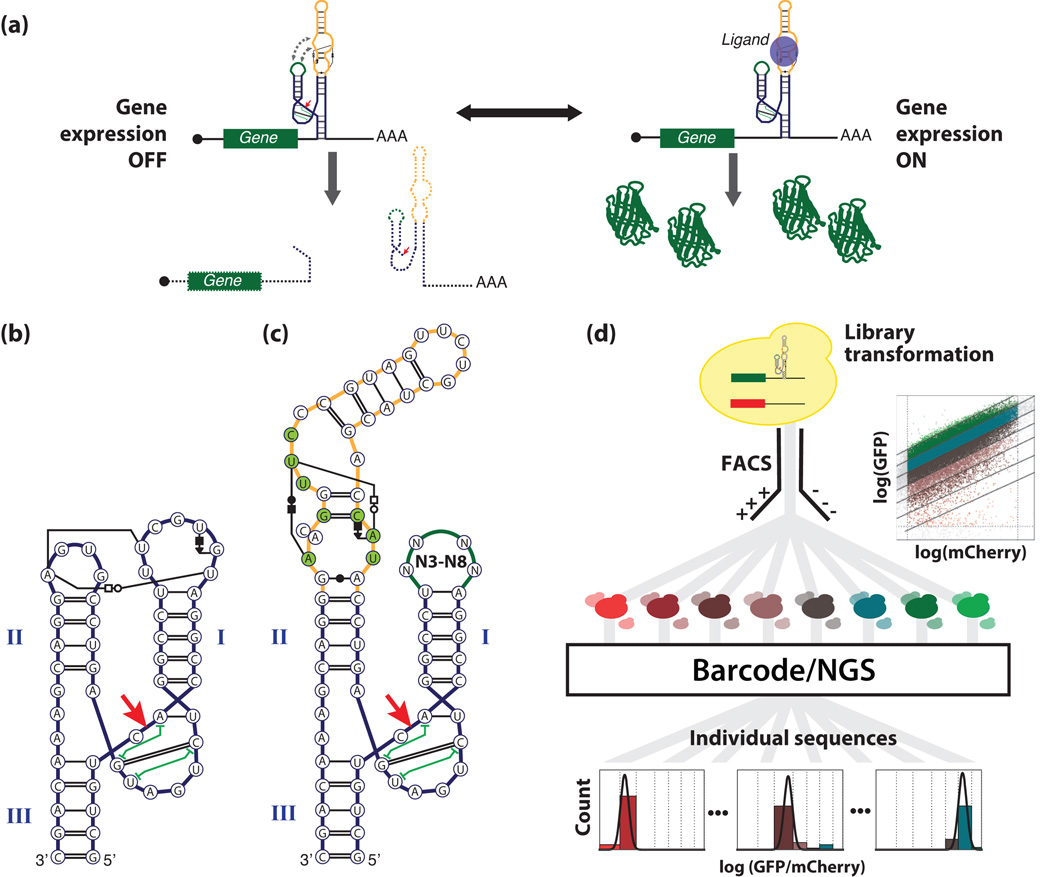
Fig. 1.
High-throughput RNA device engineering method. (a) RNA device gene-regulatory mechanism. The RNA device is encoded into the 3’ UTR of a gene, such that device cleavage results in transcript destabilization and reduced expression levels16. Binding of ligand (blue circle) to the RNA device disrupts tertiary interactions required for self-cleavage, thereby stabilizing the transcript and upregulating gene expression1. (b) HHRz (sTRSV) interactions. Interactions are indicated following Leontis-Westhof notation39 with the addition of green “I-beams” showing non-adjacent base-stacking interactions (c) Library design for theophylline-responsive tertiary interaction switches. Loop libraries (N3-N8; green) are grafted onto stem II of the sTRSV HHRz (blue), with the theophylline aptamer (gold) on the opposing stem. The aptamer and loop library sequences replace tertiary-interacting regions of the ribozyme, constituted naturally by loops I and II. Red arrow indicates the ribozyme cleavage site. Nucleotides in contact with the theophylline ligand are indicated in green. The single nucleotide difference between the CAG and AAG aptamer variants is shown with a joint cytosine/adenine nucleotide. The library also contains the corresponding structure with the aptamer grafted onto stem I (not shown). (d) Overview of the FACS-Seq method for high-throughput RNA device engineering. Device libraries are gap-repaired into a two-color reporter construct in yeast. Cells harboring the libraries are grown under selected conditions and separately sorted 8-ways using gates uniformly log-spaced over the GFP/mCherry ratio (upper-right inset). For each sorted bin, plasmid DNA is extracted and uniquely barcoded before the entire set is mixed and sequenced. The activities (µ) of each sequence for each particular condition are computed from the NGS bin counts.
High-throughput in vitro and in vivo selection and screening strategies for creating RNA devices have been described. In vitro selections7,19 have largely been supplanted by cell-based (in vivo) strategies to avoid any change in activities when transitioning from in vitro to in vivo environments19. In vivo strategies link device activity to a readily measureable expression output, such as fluorescence13,20–22, motility23, or viability24. These strategies only reveal sequence-activity information on a small number of individually-tested sequences. Strategies that provide sequence-activity information for all members in large libraries are needed to rapidly identify all high-functioning RNA devices and gain a complete understanding of the sequence-structure-function landscape to enable more robust design strategies. Methods that integrate fluorescence activated cell sorting (FACS) and high-throughput next generation sequencing (NGS) have been applied to investigate and/or develop gene-regulatory elements such as translation initiation sites25, N-terminal codons26, and various cis-regulatory elements27–33.
We establish a framework for developing RNA devices that exhibit ligand-responsive ribozyme tertiary interactions. Our new device architecture forgoes strict sequence modularity and displays design-level modularity, where the sequence of the actuator changes with the sensor. We describe a reliable closed-end method for building high dynamic range RNA devices starting from preexisting aptamers based on a FACS/NGS approach (FACS-Seq) and statistical data analyses that enables parallel measurements of the activities of hundreds of thousands of sequences from device libraries. Our tertiary interaction RNA devices show improved performance in terms of basal level, activation ratio, and ligand sensitivity as compared to the highest activity secondary-structure switching RNA devices described to date. Through our massively parallel characterization method we determine consensus sequences that enable ligand-responsive tertiary interactions for each aptamer-integrated device. This method greatly increases our capacity to rapidly and reliably build genetic tools and provides insight into the sequence-structure-function relationships needed to guide rational design.
Results
Simultaneously assaying all members of an RNA device library
Our hypothesis that it is possible to build RNA devices that function based on interference with the tertiary interactions between the two loops of a HHRz (Fig. 1b) relies on (i) the ability to obtain catalytic activity in a ribozyme with an arbitrary sequence on one loop by varying the opposite loop sequence, and (ii) target molecule binding to an aptamer on one loop interfering with that activity. The first property allows replacing one of the ribozyme loops with an aptamer for an arbitrary target and identifying a corresponding sequence on the opposite loop that restores cleavage activity in the absence of target (Fig. 1c). The second property allows this structure to behave as a switch through ligand binding to the aptamer interfering with the tertiary interactions.
We designed tertiary interaction switch libraries based on the theophylline aptamer and assayed the activities of all library members using a massively parallel FACS-Seq method (Fig. 1d, Supplementary fig. 1). The libraries were designed based on modifying the loop sequences of the tobacco ringspot virus (sTRSV) HHRz. One of two theophylline aptamer variants34 was grafted onto the ribozyme to replace either loop I or II, while the opposite loop was substituted with a library of all possible sequences ranging in length from three to eight nucleotides (Fig. 1c), requiring a library size of 349,440 sequences not including controls. The in vivo gene-regulatory activity of every library member was simultaneously measured through a FACS-Seq assay (Fig. 1d). The RNA device libraries were cloned by gap-repair into the 3’ untranslated region (UTR) of a reporter construct (encoding GFP), where cleavage of the reporter transcript (or high ribozyme activity) results in low GFP expression13. The reporter construct was placed within a low-copy plasmid that harbored a second reporter construct (encoding mCherry) that served as a control to normalize for cell-to-cell variability in gene expression13.
Following transformation and cell growth, populations harboring the RNA device library were FACS-sorted to enrich for cells exhibiting a reduced GFP/mCherry expression ratio (µ), indicative of ribozyme catalytic activity (see Online Methods). This initial sort served to enrich the population of cells for those harboring sequences with self-cleavage activity, which are more likely to exhibit expression levels modulated by the presence of the target. The prescreened cells were grown separately in the presence and absence of ligand, and individual cells from these populations were sorted based on the measured GFP/mCherry ratio (µ) into eight different bins. Library members in each bin were recovered through plasmid extraction and separately barcoded. An NGS analysis determined the frequency of occurrence of each library member in the different activity bins as a function of ligand condition (Fig. 1d). Biological replicates were carried forward at every step of the process, starting from parallel library-scale transformations.
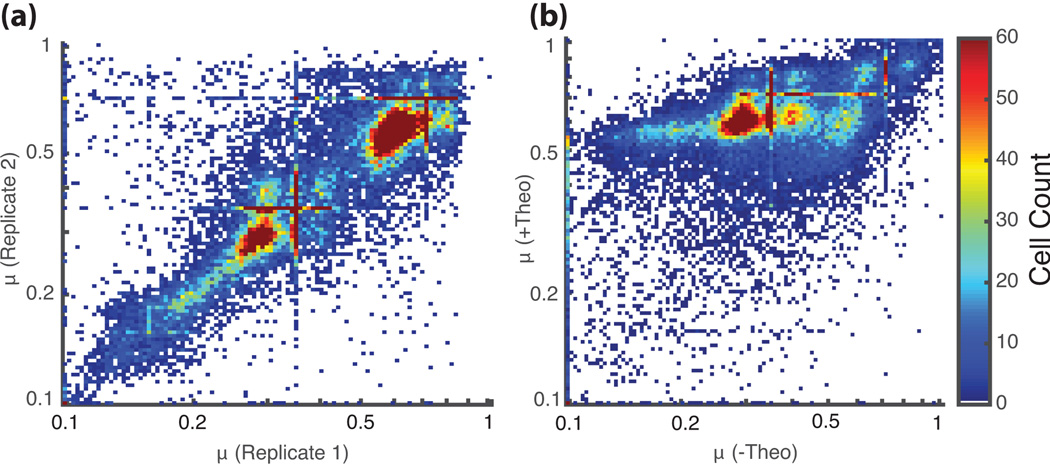
Data were analyzed to reduce the bin counts into a point estimate for µ for each library sequence (see Online Methods). Under the no-theophylline condition, most sequences in the prescreened library showed low µ with a median value of 0.30 for both replicates (Fig. 2a). These results indicated that the prescreen selection was effective at enriching for cells that exhibit low GFP/mCherry ratio in the absence of ligand. In the presence of theophylline, both replicates exhibited higher µ with median values of 0.62 and 0.61 for the replicates (Fig. 2b). The majority of the sequences exhibited switching (73% have a fold change of at least 1.3), with the activation ratio of the switch predominantly determined by the basal GFP level in the absence of theophylline. Trends observed in the data are consistent with our hypothesis of competition between binding of the target to the aptamer loop and tertiary interactions resulting in self-cleavage.
Fig. 2.
GFP/mCherry activity ratios (µ) for all members of the theophylline aptamer libraries based on FACS-Seq assays. Each point on the plots represents a unique library sequence that has the indicated GFP/mCherry values under the specified conditions. (a) GFP/mCherry values (µ) for theophylline aptamer library members from the two replicate runs without target present (N=5,389). (b) GFP/mCherry values (µ) for theophylline aptamer library members in the presence (5 mM) and absence of theophylline (N=16,024; combined replicate data). Only data for which at least 50 cells were counted is used in each analysis. The striations at particular values result from sequences for which all NGS reads for that sequence were from the same FACS bin, resulting in an estimate at the midpoint of the bin.
Identifying highly functional tertiary interaction switches
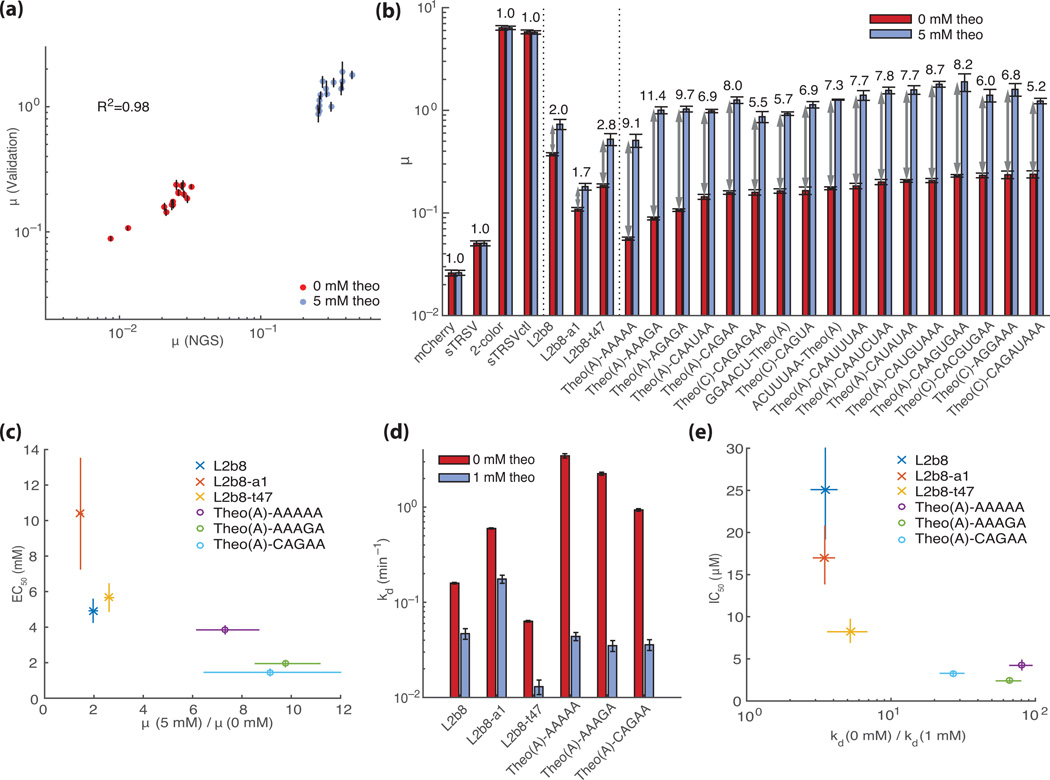
The FACS-Seq method can rapidly assess in vivo activities of large libraries of RNA devices. These data can be mined to identify sequences that result in highly functional gene-regulatory switches. We identified seventeen sequences from our theophylline aptamer library, five with the CAG aptamer variant and twelve with the AAG variant34, that exhibit the largest activation ratios (Supplementary table 1) to validate through additional characterization assays. The RNA devices were individually synthesized, integrated into the two-color characterization plasmid, and assayed in yeast via flow cytometry (Supplementary fig. 2). The µ values obtained from flow cytometry analysis of the reconstructed sequences are tightly correlated with those obtained through the FACS-Seq analysis (Fig. 3a; R2=0.98). We compared the validated activation ratios (µ-target/µ + target) for several of the best performing switches from the tertiary interaction switch libraries with those from previously optimized RNA devices that function through secondary structure rearrangements13 (Fig. 3b, Supplementary table 1). The data indicated that the switches identified from the tertiary interaction switch libraries exhibit higher activation ratios (11.4±0.8 fold change for Theo(A)-AAAGA, 2.8±0.3 for L2b8-t47, where L2b8 and its variants refer to secondary-structure switching devices) and stringencies (basal level of 0.056±0.002 for Theo(A)-AAAAA, 0.109±0.004 for L2b8-a1) than those that function through secondary-structure switching mechanisms. These values compare favorably with the basal level attainable by the wild-type ribozyme (sTRSV; µ=0.051±0.003), whereas the inactive control ribozyme (sTRSVctl; Supplementary table 2) exhibits a µ of 5.8±0.3.
Fig. 3.
Validation of theophylline-responsive tertiary interaction switches identified through the FACS-Seq method. (a) Comparison of NGS- and flow cytometry-based GFP/mCherry activity measurements (µ) for individual library members. Each point (N=30) is a single library sequence identified from the NGS analysis. Values are reported at 0 mM (red) and 5 mM (blue) theophylline. Error bars for the flow cytometry validation represent the standard error of the mean over at least three biological replicates from independent transformants. Error bars for measurements from the NGS analysis represent the range covered by two biological replicates. (b) Flow cytometry-based activity measurements for selected theophylline-responsive switches identified from the NGS analysis. Median GFP/mCherry ratios are reported for cells harboring the indicated switches and controls grown in 0 mM (red) and 5 mM (blue) theophylline. Activation ratios are reported above each construct. Error bars represent standard error of each mean over at least three biological replicates from independent transformants. (c) EC50 values versus activity ratios for theophylline-responsive switches (cross, secondary structure device; circle, tertiary interaction device). Flow cytometry data (Supplementary fig. 3) were fit to a 4-parameter logistic model with the Hill slope fixed at 1.0, from which the EC50 and 80% confidence interval were determined. Activity ratio is reported as the ratio of µ for switches grown in 0 and 5 mM theophylline; error bars indicate standard deviation over four biological replicate experiments for each condition. (d) Ribozyme dissociation rate constants as measured through a SPR cleavage assay for devices measured at 0 mM (red) and 1 mM (blue) theophylline. Error bars represent the standard error of the mean from at least triplicate assays. (e) IC50 values versus dissociation rate ratios for theophylline-responsive switches (cross, secondary structure device; circle, tertiary interaction device). For each condition, the measurements from at least three separate sets of assays were fit to a 4-parameter logistic model with the Hill slope fixed at 1.0, to compute an IC50 value for each set (Supplementary fig. 4). Dissociation rate ratio is reported as the ratio of kd at 0 and 1 mM theophylline; error bars represent the 80% confidence interval for the fitted values.
For a subset of the theophylline-responsive switches, we measured the activity as a function of target concentration by performing dose-response assays on reconstructed sequences (Fig. 3c, Supplementary fig. 3, Supplementary table 3). The data shows that compared to the secondary-structure switching devices, the identified tertiary interaction switch devices exhibit greater maximal activation ratios (fold change at 5 mM theophylline: ≥7.3 for tertiary interaction devices, ≤2.6 for secondary-structure switching device) and ligand sensitivities (mean EC50 of 7.0 µM for secondary-structure switching devices and 2.4 µM for tertiary interaction devices).
We further investigated device activities and ligand sensitivities using an in vitro SPR-based cleavage assay35. We observed that the highest in vitro cleavage activities of the tertiary interaction devices are ~6-fold higher than that of the highest previously-designed secondary-structure switching devices in the absence of ligand (Fig. 3d, Supplementary fig. 4; kd: 3.5 min−1 for Theo(A)-AAAAA, 0.6 min−1 for L2b8-a1) and ~4-fold lower in the presence of 1 mM theophylline (Fig. 3d; kd: 0.044 min−1 for Theo(A)-AAAAA, 0.17 min−1 for L2b8-a1). The ligand concentration at which the cleavage kinetics are half-maximal is 5-fold lower, comparing the average over the tertiary interaction devices with the average over the secondary-structure switching devices (Fig. 3e; IC50: 3.3 µM for tertiary interaction devices, 17 µM for secondary-structure switching devices). These data support the in vivo findings and indicate that improved cleavage activity and ligand sensitivity can be achieved with the tertiary interaction architecture.
Design-level modularity is extendable to other aptamers
The widespread applicability of our tertiary interaction switching architecture relies on the ability to restore activity to a ribozyme that has one loop sequence modified by integration of an aptamer. Restoration of cleavage activity is accomplished through the selection of an appropriate opposite loop sequence that restores tertiary interactions and geometries conducive to self-cleavage. We investigated the generality of this strategy, by characterizing the activities of all members of a HHRz library with loops I and II randomized. We verified HHRz library members activities span a wide range of activities, with consistent coverage from the activity level of the wildtype (sTRSV) HHRz to the inactive control (Supplementary note 1 and Supplementary fig. 5–Supplementary fig. 8). This graded ribozyme library also provides a new genetic tool for modulating gene expression levels over a 77-fold range through choice of the particular ribozyme sequence. A subset of validated sequences, which uniformly span the range, are provided in Supplementary table 4.
We next explored extension of the tertiary interaction RNA device architecture and FACS-Seq strategy as a general method for generating highly functioning gene-regulatory switches. Utilizing the same general architecture, we designed libraries for aptamers to neomycin36 and tetracycline37, where the aptamer sequences were placed on either loop I or II of the ribozyme and a library of all possible sequences ranging in length from three to eight nucleotides was placed on the opposite loop (Supplementary fig. 9), requiring a library size of 174,720 sequences for each aptamer. To analyze the in vivo gene-regulatory activities of every library member, the FACS-Seq method was performed on these libraries as previously described.
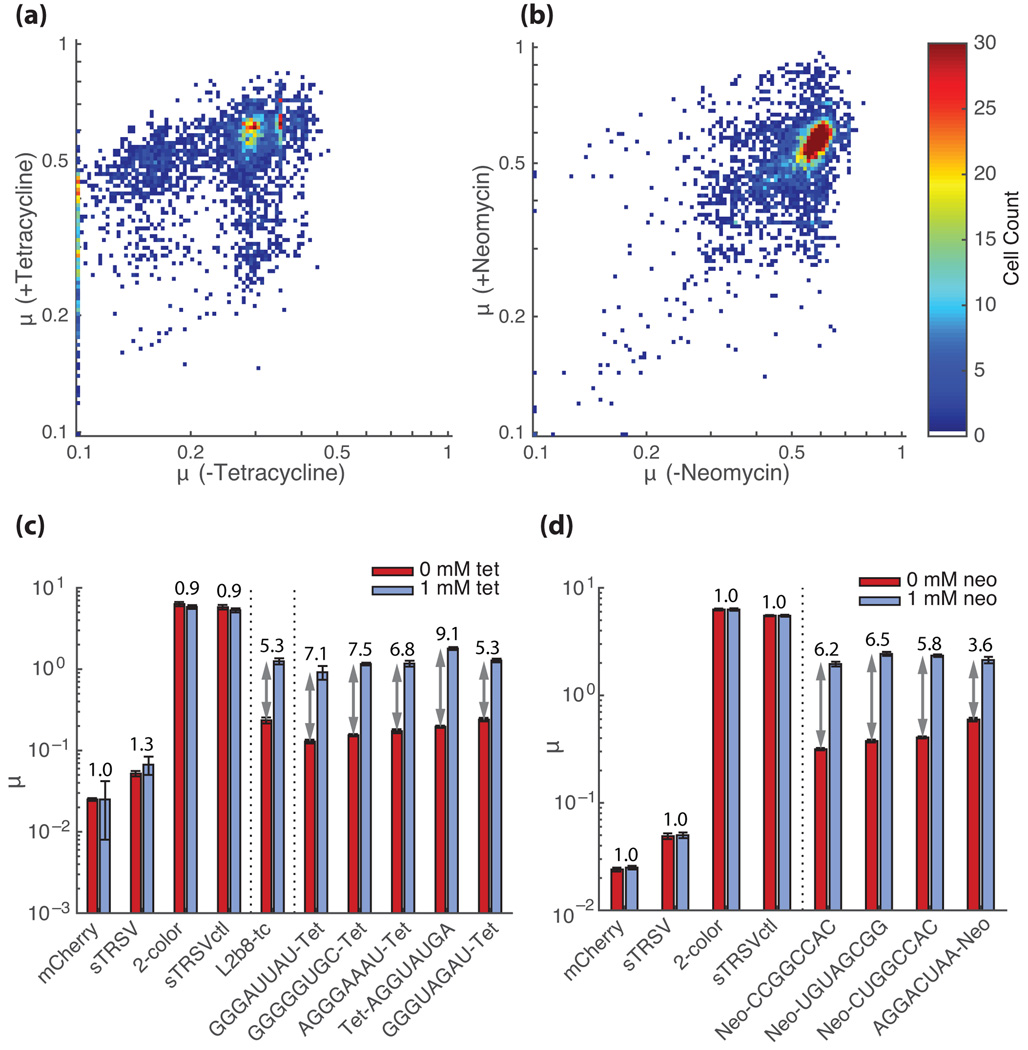
The resulting NGS data were analyzed as previously described. The activity trends of the neomycin and tetracycline libraries exhibited notable differences from those observed for the theophylline libraries. Members of the tetracycline aptamer library displayed similar activity distributions; however, a smaller fraction of the sequences exhibited low µ values in the absence of ligand (Fig. 4a). The median µ was 0.28 in the absence of tetracycline for both replicates and 0.46 and 0.53 in the presence of tetracycline for the replicates. In contrast, the sequences from the neomycin library showed negligible reduction in the median µ in the absence of ligand (Fig. 4b). The neomycin library exhibited a median µ of 0.58 and 0.55 in the absence of neomycin and 0.57 and 0.55 in the presence of neomycin for the replicates. The data indicated that few of the sequences in the neomycin library exhibit a reduction in GFP levels, and thus self-cleaving activity. The low number of sequences exhibiting cleavage activity may be due to the design of the neomycin library (Supplementary fig. 9), which incorporated one extra base pair in the stem harboring the aptamer than the theophylline library.
Fig. 4.
Extension of the FACS-Seq method to identifying tertiary interaction switches for other aptamer-target pairs. (a, b) GFP/mCherry ratios (µ) for tetracycline (a) and neomycin (b) aptamer library members in the presence and absence of target based on FACS-Seq assays. Each point on the plots represents a unique library sequence (tetracycline: N=3,873; neomycin: N=5,286; combined replicate data) that has the indicated (by color) µ under the specified conditions. Only sequences for which at least 50 cells were counted over the two combined replicates are used in each analysis. (c, d) Flow cytometry-based activity measurements for selected tetracycline-responsive (c) and neomycin-responsive (d) switches identified from the NGS analysis. Median GFP/mCherry ratios are reported for cells harboring the indicated switches and controls grown in the absence (red) and presence (blue; tetracycline: 1 mM, neomycin: 0.1 mM) of target. Error bars represent the standard error of the mean over at least three biological replicates. Activation ratios are reported above each construct. Vertical dotted lines separate ribozyme/controls, secondary structure switches, and tertiary structure switches. Additional compensation was performed for samples containing tetracycline (see Online Methods) due to fluorescence properties of this molecule.
We mined the NGS data from these libraries to identify highly functional gene-regulatory switches responsive to tetracycline and neomycin. We identified five sequences from the tetracycline aptamer library and four sequences from the neomycin aptamer library, that exhibit the largest switching ratios (Supplementary table 1) to validate. While the vast majority of sequences in the neomycin library, reduction in GFP levels or response to ligand was not observed (Fig. 4b), we were able to identify rare sequences that exhibit switching activity. These selected RNA devices were individually reconstructed as previously described and assayed via flow cytometry (Fig. 4c, d). The µ values obtained through the flow cytometry analysis were compared with the µ values obtained from the FACS-Seq analysis. The best switches exhibited activation ratios of 9.1 for tetracycline and 6.5 for neomycin (Fig. 4c, d).
Identifying aptamer-loop consensus sequences
The datasets obtained through the FACS-Seq analysis of the HHRz aptamer libraries (Supplementary note 1) were analyzed to identify consensus loop sequences that pair with different aptamers on the opposing loop and result in functional switches. Such consensus loop sequences provide additional support for particular interactions occurring between the modified loops. Starting with the theophylline AAG-variant on loop I and an eight-nt random loop II, we successively fixed one of the nucleotides on loop II and computed the 10th-percentile µ over the measured sequences with that particular nucleotide identity. The computed 10th-percentile µ ranged from 0.17 (loop II=CNNNNNNN) to 0.33 (loop II=NNNNNNNC) (Supplementary fig. 10a). Similarly, we examined the effect of nucleotide identity pairwise by computing the µ for each of the 896 possible combinations and found 10th-percentile µ values ranging from 0.08 (CANNNNNN) to 0.37 (NCNNNNGN) (Supplementary fig. 10a). We used these results to select the “best” consensus (lowest 10th-percentile µ; CNNNNNNN) and repeated the analysis on the remaining nucleotides (Supplementary fig. 10b) to determine a consensus of CANNNNNN. Continuing this process, we arrived at an overall consensus sequence of CANNNNAN for loop II with a 10th-percentile µ of 0.06; 4-fold lower than the 10th-percentile µ for the entire library of 0.24. Similarly, for the CAG-variant theophylline aptamer on loop I we identified a consensus sequence of NANNNNAA for loop II (Supplementary fig. 11; 10th-percentile µ of 0.04, 5-fold lower than the 10th-percentile of 0.22 for the library). The consensus sequences for the other aptamers also exhibit an improvement over the full library, ranging between 1.2- and 3.0-fold (Supplementary fig. 12–Supplementary fig. 17; Supplementary table 5). The results provide support for particular interactions occurring between the aptamer sequence and modified loop sequence restoring ribozyme cleavage activity.
Discussion
RNA folding is largely hierarchical and an ensemble of tertiary structures are formed for each secondary structure14. We postulate that secondary structure switching mechanisms can exhibit significant misfoldings and/or conformation interconversion timescales15 that restrict switching activity and thus gene silencing efficacy13. In contrast, the tertiary interaction switches adopt one secondary structure conformation, with aptamer and ribozyme secondary structures preformed, enabling the interactions involved in ligand-binding and ribozyme cleavage to directly compete to determine the ON and OFF states. In support of this hypothesis, in vitro cleavage assays indicated that the cleavage kinetics of the tertiary interaction switches, unlike the secondary-structure switching devices, are completely inhibited at high ligand concentrations (Supplementary fig. 4). In addition, the ligand sensitivities of the tertiary interaction switches (IC50 2.4–4.2 µM) unlike the secondary-structure switching devices are near the equilibrium dissociation constant of the initial theophylline aptamers measured under the assay conditions (Supplementary fig. 18; KD 2.4–4.4 µM), suggesting that ligand binding is directly competing with cleavage activity.
Tertiary interaction switches cannot be developed through rational design strategies, as existing RNA folding software does not accurately predict tertiary interactions and ligand binding. Our methodology, comprising a novel device framework and FACS-Seq strategy, provides a framework for efficiently generating tertiary interaction devices with design-level modularity rather than sequence-level modularity. The broader application of our approach to diverse aptamer-ligand pairs is dependent on the ability to restore activity of a ribozyme that has one loop modified with an arbitrary sequence by generating an appropriate opposing loop sequence that restores tertiary interactions. The feasibility of this approach is supported by the loop sequence flexibility observed in our analysis of active sequences within a ribozyme library. The data generated through the FACS-Seq assay can be used to define consensus loop sequence requirements for activity with different aptamer sequences, thereby increasing our understanding of sequence-structure-function relationships.
A method for modulating HHRz tertiary interactions in response to binding of protein ligands at a ribozyme loop was recently described9. The method searches databases of wild-type HHRzs for variants with stem loop sequences similar to the selected aptamer. The stem loop is replaced with the aptamer on the most similar ribozyme variant, and point mutations and/or FACS-based screening on small libraries is performed to identify mutations that restore in vivo cleavage activity. The methodology is less generalizable in that it is limited to a small subset of protein-binding aptamer sequences that closely resemble HHRz loop sequences. Our tertiary switch framework is robust to aptamers of varying length and complexity and identifies solutions that current structure-guided design methods are unable to obtain. Our massively parallel assay characterizes each member of large libraries under identical conditions providing extensive data for understanding sequence-structure-function relationships and a resource for improving computational models that attempt to predict these relationships.
We applied a combination of binned FACS and NGS on libraries larger than any that have previously used these methods. Our data analysis extends these methods by combining information about the distribution statistics of the measurements to produce maximum likelihood estimates of the activity of individual library members at a resolution better than the binning widths. Thus, the number of cells captured and sequenced rather than the bin widths, determine the resolution of the measurements. Our data indicate that these measurements are highly reproducible and are tightly predictive of subsequent single-sequence cytometry validation.
We have described an efficient pipeline for engineering ligand-responsive ribozyme tertiary interactions to generate RNA devices. We also developed a graded ribozyme library with gene-regulatory activities spanning a 77-fold range in vivo, thereby expanding the tools available for precisely controlling expression across diverse biological systems38. Our FACS-Seq approach supports parallel measurements of the activities of large RNA regulator libraries under chosen conditions. By assaying every member of these libraries in parallel within a single culture, this method enables elucidation of consensus sequences for genetic devices. The non-iterative method of combining existing aptamers, including those derived from naturally occurring riboswitches, with a ribozyme to build genetic sensors that outperform those currently available will advance our ability to develop sophisticated genetic tools and our understanding of the underlying sequence-structure-function relationships that empower rational design of complex biomolecules.
Online Methods
Tertiary interaction RNA switch library design
Tertiary interaction switch libraries were constructed based on the sequence of the tobacco ringspot virus (sTRSV) HHRz by replacing either the wild-type loop I or II sequences with previously identified minimal aptamer sequences and the other loop with a randomized sequence between three and eight nucleotides (Supplementary table 2). This library design resulted in 174,720 distinct sequences for each aptamer. The aptamer sequences have a structurally conserved terminal helix that is reconstituted by a ribozyme stem in our design architecture (Fig. 1c). The TCT8-4 theophylline aptamer sequence40 and also a variant sequence with a single base change (C28A, in the postulated binding pocket of the aptamer34) were used with the terminal stem removed. The first base pair of the terminal stems of the tetracycline37 and neomycin aptamer36 were retained in our device design as it has been shown to be important to ligand binding (Supplementary fig. 9b). Each of the switches was flanked by a spacer sequence designed to minimize interactions between the surrounding sequences and the switch sequence1. Example predicted three-dimensional structures for sequences from these libraries are presented in Supplementary fig. 19.
Library construction and high-efficiency yeast transformation
All RNA device libraries were assembled from two oligonucleotide fragments through overlap-extension PCR using PFU Ultra II HS DNA polymerase (Agilent Technologies). The fragments were designed to overlap in the region between the two stems, allowing the random loop regions to be modularly coupled with the four aptamer sequences (Supplementary table 2). The resulting sequences were combined into three distinct libraries based on the target ligand. In preparation for yeast-mediated gap-repair cloning, each DNA library was amplified by PCR (PFU Ultra II HS; Agilent) with primers (Supplementary table 2) with overhangs homologous to portions of a previously described two-color screening plasmid (pCS1748)13. The low-copy plasmid backbone is designed to place the switches in the 3’ UTR of a GFP reporter gene, and also harbors a separate mCherry expression cassette13 (Supplementary fig. 20).
Briefly, as previously described41, for each of the three libraries, 50 ml yeast culture (OD600 1.3–1.5) was incubated with Tris-DTT buffer (2.5 M DTT, 1 M Tris, pH 8.0) for 15 min at 30°C, pelleted, washed, and resuspended in Buffer E (10 mM Tris, pH 7.5, 2 mM MgCl2) to 300 µl. To 50 µl of the yeast cell suspension, 2 µg of linearized plasmid and 1 µg of library insert DNA was added and the DNA-cell suspension was electroporated (2 mm gap cuvette, 540 V, 25 µF, 1000 Ω). Transformed cells were diluted to 1 ml volume in yeast peptone dextrose (YPD) media, incubated for 1 hr, then further diluted in selective media (synthetic complete media with a uracil dropout solution containing 2% dextrose; SC-URA) and propagated for FACS screening13. Each of the libraries was independently transformed into yeast twice providing two biological replicates (or six library samples in total), which were handled separately through all subsequent steps of the FACS-Seq method. The budding yeast strain W303α (MATα leu2–3,112 trp1-1 can1–100 ura3-1 ade2-1 his3–11,15) was used in all experiments. All fungal growth and propagation steps were carried out in a 30°C incubator, shaking at 230 rpm, unless otherwise stated.
Library prescreening for active ribozyme sequences
Following high-efficiency transformation and subsequent cell growth, the six samples were prescreened through FACS to enrich for cells that exhibit reduced GFP expression or by extension ribozyme cleavage activity in the absence of ligand. Cells harboring the libraries were back-diluted 20:1 to an approximate OD600 of 0.07 in SC-URA media and grown for 6 hrs to OD600 ~0.8. Cells were washed, resuspended in PBS (Life Technologies) with 1% BSA (Sigma-Aldrich), stained with DAPI viability dye (Life Technologies), then filtered through a 40 µm cell strainer (BD Biosciences) prior to analysis on a FACSAria II cell sorter (BD Biosciences).
GFP was excited at 488 nm and measured with a splitter of 505 nm and bandpass filter of 525/50 nm. mCherry was excited at 532 nm and measured with a splitter of 600 nm and bandpass filter of 610/20 nm. Fluorescence levels of cells harboring a negative-control plasmid (pCS4) were used to determine background, autofluorescence levels of both colors13. Initial gates based on the forward scatter area, side scatter area, side scatter height, and side scatter width were used to gate out cell debris and non-viable cells. Next, a gate which removed cells with mCherry levels comparable to the no-color control was applied (~15% of cells removed), followed by a gate which removed any cells with GFP levels that saturated the instrument measurement (~2% of cells removed). Finally, a gate based on the ratio of GFP to mCherry expression (µ) established to collect cells with a µ below a threshold value. This threshold was set such that ~10% cells that passed the parent gates were collected (Supplementary fig. 21). The final sort gate was applied to enough cells to ensure at least 15 cells per library sequence were considered. The actual counts of cells sorted and collected are reported in Supplementary table 6.
Sorting of RNA device libraries into activity bins
The prescreened cell populations were grown for 14.5 hrs at 30°C in SC-URA, after which cell counts were measured using a MACSQuant VYB flow cytometer (Miltenyi Biotec GmbH). The six samples were then normalized to 3.1×106 cells/ml by addition of media, and growth was continued for 12 hrs under the same conditions, after which they were back-diluted 100:1 to OD600 ~0.05 and grown an additional 7 hrs to OD600 ~1.3 keeping them in the exponential growth phase throughout. In parallel to the above, a separate culture of cells, which contained a set of four graded ribozymes in approximately equal ratio, was similarly transformed and grown. This reference culture was kept separate for use in setting the final gating, as described below. Each of these six cultures was back-diluted 20:1 into two separate 50 ml samples of fresh media to an OD600 ~0.07, with the target molecule added to one of the two samples. The target molecules were added to the following final concentrations: theophylline 5 mM, neomycin 0.1 mM, and tetracycline 1 mM. The cultures were grown for 6 hrs at 30°C to OD600 ~0.8 to ~1.0.
The yeast cultures were spun down and resuspended in PBS (Life Technologies) to a final concentration of 2×107 cells/ml. The twelve samples were then combined into four mixtures prior to sorting; 1− (replicate 1, no target), 1 + (replicate 1, with target), 2− (replicate 2, no target), 2 + (replicate 2, with target). The sequence differences between the aptamers would allow for the three combined samples to be resolved during subsequent NGS processing, while reducing the number of samples to sort.
Sorting of the samples into activity bins was performed on a FACSAria II Cell Sorter (BD Biosciences). Excitation and emission filters for GFP and mCherry and scatter gating were as described above. In addition, a viability gate based on DAPI and side-scatter area was applied to exclude the DAPI-positive dead cells from subsequent analysis. DAPI was excited at 355 nm and measured with a bandpass filter of 450/50 nm. The cells that passed these gates were then divided into one of eight gates based on the GFP/mCherry ratio to allow binned sorting of the cells. The gates were set using the reference culture of four graded ribozymes. These ribozymes were chosen to have GFP/mCherry levels that uniformly span the range of interest. Gate edges between bins 1&2, 3&4, 5&6, and 7&8 were set on the log(GFP) vs. log(mCherry) display to equally split the populations for each of these graded ribozymes as shown in Supplementary fig. 22. The remaining three bin edges (i.e., between bins 2&3, 4&5, 6&7) were then set to approximately halfway between each of these. Since the sorter has a maximum capability of four-way sorting, the samples were each sorted twice based on the defined gates. Cells falling into bins 1–4 were collected in the first sort and all other cells were discarded. In the second sort, cells falling into bins 5–8 were collected. SC-URA at a volume (3 ml) of at least 3:1 was added to each collection tube immediately after sorting. Sorting of each sample, except 2 +, was continued until at least ~6 million cells were collected. The following number of cells were collected over the eight bins for each sample over a 2.5 hr period: 1− 7.6 million, 1 + 7.1 million, 2− 7.1 million, and 2 + 5.9 million. Details of cells sorted per bin are included in Supplementary table 7.
NGS sample preparation
Sorted samples were grown in SC-URA at 30°C for up to 32 hrs, with samples stored at 4°C once they reached OD600 ~0.7. The volumes for each culture were chosen such that each sample contained at least 50x the number of cells that were initially sorted into that bin. In addition to these 32 cultures, seven additional cultures were processed in parallel. These were cultures taken prior to the prescreening (the three target libraries pooled in each of the two replicates), just prior to the main sort (four samples of pooled target libraries), and a culture of cells containing an unmodified plasmid (no switch inserted) as a negative control. Cells from each of these 39 samples were collected, lysed, and the DNA from each sample was extracted using the ZR Fungal/Bacterial DNA MiniPrep™ (Zymo Research), according to the manufacturer’s instructions. A diversity control was then added to each sample of prepared DNA (Supplementary note 2). This control consisted of a 17-nt random region of DNA (synthesized using a machine mix of the four nucleotides) flanked by the spacer sequence used with the switch sequences. Since almost every molecule of this control has a unique sequence, subsequent occurrence counting of each distinct sequence within the control was used to compute the mean number of reads due to any single molecule that existed in the sample at this point. This method was used to verify that all bins had less than 1.25 reads/molecule with most less than 1.05 read/molecules (Supplementary table 8).
The DNA encoding the RNA devices was amplified from the bulk DNA in each sample through 14 cycles of PCR using PFU Ultra II HS (Agilent Technologies) and 400 nM primers based on the spacer sequences (T7_W_Primer, X_Primer-RC; Supplementary table 2). Each reaction was sized such that the number of molecules in the template was at least 10x the number of NGS reads planned for that sample, while keeping the template volume at or below 25% of the total PCR volume. The PCR products for each bin were used as the template for a second PCR, which used primers with overhang regions corresponding to the standard Illumina adapter sequences. DNA barcodes were also added to allow identification of the particular sample from the NGS reads. These barcodes are a sequence of up to seven nucleotides that were added to each end of the sequence of interest. The variable length also increased base diversity at each read position, which can improve read quality during Illumina sequencing. In addition to the 39 samples from the DNA extractions (Supplementary fig. 1), an individually barcoded sample containing an equimolar mix of the original DNA libraries used for the transformations was also included as a control to verify the pre-transformation library distribution.
Samples were quantified on a Bioanalyzer 2100 (Agilent Technologies) and sequenced on an Illumina HiSeq 2500 by Elim Biopharmaceuticals, Inc. using 2×100 paired-end reads. The sample was run using Illumina standard procedures, with PhiX (Illumina) added (to 15% by molarity) to further increase diversity at nucleotide positions which would, otherwise, have a significant fraction of the sample sharing the same base call and result in lower read quality42.
NGS data processing
The paired-end reads were first joined using PEAR43. The joined sequences were then split using the concatenated barcodes on each end into 40 separate files corresponding to the 32 bins (2 conditions x 8 bins x 2 replicates) plus 8 control samples consisting of the DNA library, post-transformation plasmid prep (2 replicates, each pooling the three libraries), pre-sort plasmid prep (2 replicates x 2 conditions), and a blank plasmid prep (cells with the parent plasmid, no switch integrated; controls for cross-contamination). Sequences without an exact match to expected barcodes, spacer, and library entry sequences were ignored during the main analyses, although the full set of sequences was used for assessing controls. The matching data (46.7M reads) were then collapsed into tables that gave the count of occurrences of each designed sequence for each bin or control sample.
Prior to beginning the main FACS-Seq experiment, we collected flow cytometry data on cells harboring the two-color expression constructs that incorporate four graded ribozymes that span the expression range of interest (Supplementary fig. 21b). Analysis of these data and prior cytometry on cells harboring a single switch sequence incorporated into the expression construct indicate that the GFP/mCherry ratio follows a log-normal distribution with a uniform variance over a wide range of ratios as is often the case for biological quantities44. The observed coefficient of variation for these samples was measured to be 0.31. Based on this observation, a method was developed for estimating the underlying mean GFP/mCherry ratio of a population of cells from the binned cell counts with a resolution better than the bin width, limited only by the model mismatch and the number of cells counted.
Sequencing results were separated by barcode and sequence identity to produce a histogram of read counts, ri,b, per sequence, i, in each of the eight FACS bins, b. The read counts were then normalized by a factor Cb/Rb, where Cb is the total number of cells sorted in bin b and Rb is the total number of NGS reads with barcode corresponding to bin b, to give an estimate of cells per bin, ci,b. This accounts for the differences between the bins in post-sort growth, plasmid preparation, or NGS mixing. The average number of cells per read for each bin over each of the samples is reported in Supplementary table 9. With the GFP/mCherry fluorescence ratios, Ab,b + 1, used to set the FACS gates between bins b and b + 1, the ci,b were fit to a model that assumes that these ratios are random variables that follow a log-normal distribution with a constant variance of 0.3. That is, we assumed:
where N(x,µ,σ) is the normal probability density function with mean µ, variance σ2, evaluated at x, Ci=∑ci,b, b=0..8 and σ=0.30 (CV=0.31).
The fits were performed using custom MATLAB (MathWorks) code available at http://github.com/btownshend/TwoColor. These fits resulted in an estimate for each sequence, ai, of the GFP/mCherry ratio for that sequence. The method can also produce confidence intervals for µ based on the bin statistics, but this captures only the variability due to counting statistics of the reads, ri,b, and does not model systematic variability in σ or µ such as post-sort growth bias or model mismatch. We also determined error bounds on each of these calculated values based on the difference between the two biological replicates and found that these were in agreement with the model confidence intervals with approximately 80% of the replicate µ values falling within the 80% confidence intervals.
Identification of switches
Potential switches sensitive to each of the target molecules were identified by analysis of the µ values in the −target and + target conditions. For the theophylline aptamers, sequences were considered that satisfied the following constraints, with the two replicates combined: at least 20 cells measured, µ− target <0.10, µ+ target >0.50 (Fig. 2b). These values were chosen to identify switches with activation ratios of at least 5 fold and gene expression levels in the absence of ligand close to that of the wild-type sTRSV ribozyme. Of the 205 sequences that satisfied these constraints, seventeen representative sequences were selected for validation. Similarly, for the tetracycline aptamer, sequences were considered that had at least 40 cells measured, µ− target <0.025, µ+ target >0.25. These criteria were satisfied by seventeen sequences of which five were selected for further validation. For the neomycin aptamer, fewer sequences exhibited strong switching so the criteria were relaxed: µ− target <0.15, µ+ target >0.25 over at least 40 cell measurements, resulting in seven hits with four selected for further validation.
Flow cytometry validation of reconstructed sequences
Specific switch sequences were synthesized from overlapping oligonucleotides using overlap-extension PCR as described for the device library constructions. These were gap-repair transformed into the yeast two-color screening plasmid along with control plasmids by the lithium acetate/single-stranded carrier DNA/polyethylene glycol method45, with each switch sequence verified using Sanger sequencing. At least three individual colonies were picked and inoculated in SC-URA media. Cultures were grown overnight, back-diluted 20:1 to an OD600 ~0.07 and then grown 6 hrs in the absence and presence of a ligand target, at the same ligand concentrations as used for the FACS-Seq assays. The cells were then spun down and resuspended in an equal volume of 1×PBS buffer (Life Technologies) with 1% BSA (Fraction V, EMD Millipore) and a DAPI viability dye (Life Technologies). GFP was excited at 488 nm and measured with a bandpass filter of 525/50 nm. mCherry was excited at 561 nm and measured with a bandpass filter of 615/20 nm. DAPI was excited at 405 nm and measured with a bandpass filter of 450/50 nm. Prior to each use, voltages of fluorescence PMT detectors were calibrated with MACSQuant calibration beads to fix GFP and mCherry levels. For each culture, 10 µl of sample was analyzed, which captured 50,000–150,000 events while also providing cell density measurements. The data was analyzed using a custom MATLAB program to gate for mCherry expression above the no-color controls and non-saturating values for GFP and mCherry, and then extract µ, the median GFP/mCherry ratio. Since cultures that contain tetracycline produce non-specific fluorescence in the GFP emission region, the µ values for this condition were corrected by subtracting a fixed offset. This offset, 0.17, was determined from the mean difference in the plus and minus-tetracycline conditions for control samples with an “mCherry-only” plasmid that did not contain a GFP gene.
Note that the NGS data is based on cells sorted through a FACSAria II Cell Sorter. An in-house flow cytometer (Miltenyi Biotec MACSQuant VYB) was used for validation measurements. The GFP and mCherry levels are given in arbitrary fluorescence units that differ between the two instruments, but in all cases are treated as a linear function of the actual protein levels in order to compute µ. Thus, the µ values from the validation and the NGS data each incorporate a different linear scale factor.
Surface plasmon resonance validation of reconstructed sequences
Representative FACS-Seq sort identified theophylline-responsive RNA device cleavage kinetics and ligand sensitivity were determined by surface plasmon resonance (SPR; Supplementary fig. 4), using previously described protocols3,35. Briefly, the RNA device DNA templates were amplified by PCR (PFU UltraII HS; Agilent) with primers containing overhangs corresponding to the T7 RNAP promoter and cis-blocking sequences that prevent device cleavage during in vitro T7 transcription35 (Supplementary table 2; SPR templates). A second PCR (KAPA HiFi PCR Kit; Kapa Biosystems) with short primers was performed to enrich the product for full-length sequences (Supplementary table 2; SPR_fwd_primer, SPR_rev_primer). A total of 100–200 ng of PCR product was transcribed in a 50 µl reaction, consisting of the following components: 1×RNA Pol Reaction Buffer (New England Biolabs), 2.5 mM of each rNTP, 2 µl Superase•In (Life Technologies), an additional 4 mM MgCl2 (Ambion), 2 µl T7 RNA Polymerase (New England Biolabs). After incubation at 37°C for 2 hrs, the transcription reaction was purified with the RNA Clean and Concentrator™-25 kit (Zymo Research) according to the manufacturer’s instructions and estimated by Nanodrop.
The Biacore X100 sensor chip (GE Healthcare) surface immobilized with DNA activator was generated as previously described35. The Biacore X100 instrument (GE Healthcare) was equilibrated with the physiologically-relevant reaction buffer at 25°C prior to all ribozyme cleavage assays. The SPR baseline was stabilized by performing 2–5 startup cycles, where each cycle includes a capture and a regeneration step. The capture step was performed by an injection of a total of 10–25 ng transcribed cis-blocked RNA diluted in HBS-N (GE Healthcare) buffer over the reaction flow cell (FC2) for 1 min at a flow rate of 10 µl/min. The capture step typically yielded ~50–700 RU of the SPR signal for the described constructs. The regeneration step was performed by an injection of 25 mM NaOH over both flow cells for 30 s at a flow rate of 30 µl/min. Following the startup cycles, assay cycles were performed. Each assay cycle includes a capture, a reaction, and a regeneration step. The capture and regeneration steps in an assay cycle were performed as described for those in the startup cycle. The reaction step was performed by an injection of the running buffer containing 500 µM MgCl2 with or without theophylline over both FCs for 300–500 s at a flow rate of 10 µl/min. Biacore sensorgram processing and analysis were performed using custom Matlab software. Due to the slight time delay at which injected analyte reaches the respective flow cells, the resultant sharp spikes at the beginning and the end of injection were excluded from the analysis46. The processed sensorgram (R) was fit to a simple exponential equation R = R0 [fc e− kdt + 1 − fc ], where R0 (fit locally for each replicate) is the initial SPR signal before the cleavage reaction, fc (fit globally for a given RNA sample) is the extrapolated residual response at the end of the cleavage reaction as a fraction of the captured RNA signal, and kd is the first-order RNA cleavage (dissociation) rate constant. Reported values are the mean of at least three independent experiments.
SPR-based cleavage assays were performed at various theophylline concentrations to generate dose-response curves. The RNA dissociation rate constant (kd) at each theophylline concentration ([theo]) was fit to the sigmoidal equation kd = kd,min + (kd,max – kd,min)/(1 + [theo]/IC50) using MATLAB, where kd,max and kd,min are the maximum and minimum RNA dissociation rate constants, evaluated in absence of and with the highest theophylline concentration assayed, respectively. The IC50 here is defined as the theophylline concentration at which kd is halfway between the minimum and maximum values. Replicate dose response measurements were fit to the three parameter logistic equation, with a shared kd,max, and kd,min, and IC50 for all replicate assays for a given device. For each device the fitted parameters are reported in Supplementary fig. 4.
The binding affinities of the CAG- and AAG-variant theophylline aptamers were determined at the same conditions as the SPR-based cleavage assay (500 µM MgCl2, 150 mM NaCl and 10 mM HEPES (pH 7.4), at 25°C) using a previously described SPR-based binding assay47. Aptamer equilibrium dissociation constants (KD) were determined from fit of binding responses to theophylline, measured at concentrations spanning four orders of magnitude, to a steady-state affinity model using MATLAB. The binding curves are presented in Supplementary fig. 18.
Consensus Analyses
Analyses of NGS data for consensus sequences were performed using custom MATLAB software. For each possible identity of one nucleotide, or pair of identities for two nucleotides, the 10th percentile of µ was formed over all sequences that match that nucleotide or nucleotides. In this way, sequence positions that can result in low µ values are found without being overly sensitive to the sequences, which may, due to the effects of other sequence positions, have a much higher µ. Raw NGS data was pooled from the two biological replicates and only sequences for which we have at least 20 cells sorted were used in the computations. Initially all degenerate loop nucleotides were allowed to vary. After computing each stage, the nucleotide position with the greatest effect on the average was fixed at the value that gave the lowest average µ, and the process was repeated four times. The reported consensus sequence is the last of these with at least 100 sequences used in the averaging.
Structural Modeling
Leontis-Westhof interactions between nucleotides in the HHRz (Fig. 1) are based on 3D structures from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Databank (PDB)48 entry 2QUS49. Interactions between nucleotides in the theophylline RNA device are based on 3D structures from the RCSB PDB entries 2QUS49 (ribozyme) and 1O1550 (theophylline aptamer). Base pair interactions were extracted from the PDB entries using FR3D51.
Supplementary Material
Acknowledgements
We thank M. McKeague, C. Schmidt for valuable feedback in preparation of this manuscript; C. Crumpton, M. Bigos, and B. Gomez of the Stanford Shared FACS facility. This work was supported by funds from the US National Institutes of Health [grant to C.D.S. and Shared Instrumentation Grant S10RR025518-01], Defense Advanced Research Projects Agency [grant to C.D.S.], Natural Sciences and Engineering Research Council of Canada [fellowship to A.B.K.], Agency for Science, Technology, and Research [fellowship to J.S.X.].
Footnotes
Author contributions
A.B.K. and B.T. have contributed equally to this work. The author order was chosen at random. A.B.K., B.T., and C.D.S. conceived the project and wrote the manuscript. A.B.K., B.T., and J.S.X. conducted the experiments. B.T. developed the software to analyze the cytometry and NGS data. B.T., A.B.K., J.S.X., and C.D.S. designed the experiments and analyzed the results.
Competing interests statement
The authors declare competing financial interests in the form of a pending patent application.
References
- 1.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei KY, Chen YY, Smolke CD. A yeast-based rapid prototype platform for gene control elements in mammalian cells. Biotechnol. Bioeng. 2013;110:1201–1210. doi: 10.1002/bit.24792. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy AB, Vowles JV, d’Espaux L, Smolke CD. Protein-responsive ribozyme switches in eukaryotic cells. Nucleic Acids Res. 2014:1–16. doi: 10.1093/nar/gku875. at < http://www.ncbi.nlm.nih.gov/pubmed/25274734>. [DOI] [PMC free article] [PubMed]
- 4.Ausländer S, Ketzer P, Hartig JS. A ligand-dependent hammerhead ribozyme switch for controlling mammalian gene expression. Mol. Biosyst. 2010;6:807–814. doi: 10.1039/b923076a. [DOI] [PubMed] [Google Scholar]
- 5.Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- 6.Nomura Y, Zhou L, Miu A, Yokobayashi Y. Controlling mammalian gene expression by allosteric hepatitis delta virus ribozymes. ACS Synth. Biol. 2013;2:684–689. doi: 10.1021/sb400037a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittmann A, Suess B. Selection of tetracycline inducible self-cleaving ribozymes as synthetic devices for gene regulation in yeast. Mol. Biosyst. 2011;7:2419–2427. doi: 10.1039/c1mb05070b. [DOI] [PubMed] [Google Scholar]
- 8.Klauser B, Atanasov J, Siewert LK, Hartig JS. Ribozyme-Based Aminoglycoside Switches of Gene Expression Engineered by Genetic Selection in S. cerevisiae. ACS Synth. Biol. 2014 doi: 10.1021/sb500062p. [DOI] [PubMed] [Google Scholar]
- 9.Ausländer S, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat. Methods. 2014 doi: 10.1038/nmeth.3136. at < http://www.ncbi.nlm.nih.gov/pubmed/25282610>. [DOI] [PubMed]
- 10.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galloway KE, Franco E, Smolke CD. Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science. 2013;341:1235005. doi: 10.1126/science.1235005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. U. S. A. 2010:1–6. doi: 10.1073/pnas.1001721107. at < http://www.ncbi.nlm.nih.gov/pubmed/20421500>. [DOI] [PMC free article] [PubMed]
- 13.Liang JC, Chang AL, Kennedy AB, Smolke CD. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012;40:e154. doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tinoco I, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 15.Mustoe AM, Brooks CL, Al-Hashimi HM. Hierarchy of RNA functional dynamics. Annu. Rev. Biochem. 2014;83:441–466. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat. Struct. Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 17.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput. Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perreault J, et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link KH, et al. Engineering high-speed allosteric hammerhead ribozymes. Biol. Chem. 2007;388:779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- 20.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 21.Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. Chembiochem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- 22.Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomura Y, Yokobayashi Y. Dual selection of a genetic switch by a single selection marker. Biosystems. 2007;90:115–120. doi: 10.1016/j.biosystems.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Topp S, Gallivan JP. Random walks to synthetic riboswitches--a high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 25.Noderer WL, et al. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014;10:748. doi: 10.15252/msb.20145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- 27.Kinney JB, Murugan A, Callan CG, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9158–9163. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raveh-Sadka T, et al. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nat. Genet. 2012;44:743–750. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 30.Shalem O, et al. Measurements of the impact of 3’ end sequences on gene expression reveal wide range and sequence dependent effects. PLoS Comput. Biol. 2013;9:e1002934. doi: 10.1371/journal.pcbi.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patwardhan RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosuri S, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann GR, Shields TP, Jenison RD, Wick CL, Pardi A. A semiconserved residue inhibits complex formation by stabilizing interactions in the free state of a theophylline-binding RNA. Biochemistry. 1998;37:9186–9192. doi: 10.1021/bi980082s. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy AB, Liang JC, Smolke CD. A versatile cis-blocking and trans-activation strategy for ribozyme characterization. Nucleic Acids Res. 2013;41:e41. doi: 10.1093/nar/gks1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigand JE, et al. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 38.Redden H, Morse N, Alper HS. The synthetic biology toolbox for tuning gene expression in yeast. FEMS Yeast Res. 2014 doi: 10.1111/1567-1364.12188. [DOI] [PubMed] [Google Scholar]
- 39.Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 40.Jenison RD, Gill SC, Pardi a, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 41.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 42.Krueger F, Andrews SR, Osborne CS. Large scale loss of data in low-diversity illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One. 2011;6:e16607. doi: 10.1371/journal.pone.0016607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ron Milo Phillips R. Cell Biology by the Numbers. 2014;368 at < http://book.bionumbers.org/how-much-cell-to-cell-variability-exists-in-protein-expression/>. [Google Scholar]
- 45.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 46.Myszka D. Improving biosensor analysis. J. Mol. Recognit. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 47.Chang AL, McKeague M, Liang JC, Smolke CD. Kinetic and equilibrium binding characterization of aptamers to small molecules using a label-free, sensitive, and scalable platform. Anal. Chem. 2014;86:3273–3278. doi: 10.1021/ac5001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi YI, et al. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:2060–2068. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clore GM, Kuszewski J. Improving the accuracy of NMR structures of RNA by means of conformational database potentials of mean force as assessed by complete dipolar coupling cross-validation. J. Am. Chem. Soc. 2003;125:1518–1525. doi: 10.1021/ja028383j. [DOI] [PubMed] [Google Scholar]
- 51.Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB. FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 2008;56:215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.