Abstract
The analysis of inflammatory cytokines and chemokines produced during Hepatitis C virus (HCV) infection has advanced our understanding of viral-host interactions and identified predictors of treatment response. Administration of interferons (IFN) made it difficult to interpret biomarkers of immune activation during treatment. Direct acting antiviral (DAA) regimens without IFN are now being used to treat HCV with excellent efficacy. To gain insight into HCV-host interactions occurring before, during and after HCV treatment we performed a case-control study that measured serial plasma levels of IP-10, MCP-1, MIP-1β, and IL-18 in 131 patients with chronic HCV treated with sofosbuvir (SOF) plus ribavirin (RBV). A linear regression analysis using baseline factors identified strong positive associations between elevated ALT and pre-treatment IP-10 and between the presence of cirrhosis and elevated pre-treatment IL-18. Mean IP-10, MCP-1, MIP-1β and IL-18 levels all decline on therapy but display different dynamics late in treatment and following cessation of therapy. On treatment IP-10 and MIP-1β levels were significantly higher in individuals who achieved sustained virologic response (SVR). Logistic regression analyses examining treatment response in all patients demonstrated significant associations between higher baseline MIP-1β levels and smaller decreases in MIP-1β early in treatment and SVR. Higher early MIP-1β levels were also significantly associated with SVR in subsets of patients with cirrhosis and individuals with GT3 infection, two factors associated with decreased responsiveness to treatment. Conclusion: Our study shows that changes in IP-10 levels mirror HCV RNA suggesting IP-10 is an indicator of innate immune viral recognition. MIP-1β levels remained elevated in GT2/3 patients who achieved SVR suggesting differential immune activation in those who respond to SOF/RBV therapy and a potential role in predicting treatment responses.
Keywords: Hepatitis C virus, MIP-1 beta, IP-10, IL-18, direct acting antiviral
INTRODUCTION
HCV causes significant human morbidity and mortality infecting approximately 3% of the world's population (1, 2). HCV infection is a leading cause of liver disease, cirrhosis, and hepatocellular carcinoma and consequently it is a principal indication for liver transplantation in the U.S. and Europe (3, 4). HCV is a single stranded RNA virus that replicates at very high rates using an error prone RNA-dependent RNA polymerase generating 10 fold higher circulating viral diversity than Human Immunodeficiency Virus (HIV) (5-7). Extrapolating from HIV, where durable viral suppression requires the concurrent use of three or more potent anti-virals with varying mechanisms of action to overcome high HIV mutation rates, one may hypothesize that HCV therapy would similarly require 3 or more potent anti-viral drugs from several classes to achieve viral eradication (8, 9). Surprisingly, HCV treatment with DAA regimens containing one (with RBV) or two direct antivirals have shown remarkable efficacy achieving up to 90-95% SVR rates in certain populations (10-14). One possible explanation for this unanticipated efficacy is that suppression of HCV by DAAs releases host immune responses from active HCV suppression augmenting the effectiveness of HCV therapies (15).
SOF, a potent nucleotide NS5B polymerase inhibitor, in combination with RBV is approved for the treatment of GT2 and GT3 chronic HCV infection based on the results of multiple phase 3 clinical trials (12-14). Despite the improved efficacy of this regimen over pegylated-IFN (peg-IFN) plus RBV, a subset of patients does not achieve SVR. Multivariable logistic regression analysis of demographic and clinical data in the POSITRON, FISSION and FUSION studies identified infection with HCV GT3 and cirrhosis as independent predictors of relapse after treatment (12, 13).
In the pre-DAA treatment era, analysis of circulating chemokines or cytokines improved our understanding of immune responses to HCV infection and identified predictors of treatment response (16-19). Higher baseline levels of IFNγ-inducible protein 10 (IP-10), a CXC chemokine produced in response to IFNγ, are inversely correlated with rates of SVR during therapy with IFN and RBV (16-18, 20, 21). Interleukin 18 (IL-18) levels are elevated during HIV and HCV disease and decrease after viral suppression (22-27) and human gene polymorphisms in IL-18 have been associated with differences in hepatitis B or C viral clearance (28-33). Macrophage inflammatory protein 1β (MIP-1β), a CC chemokine expressed in the portal vein endothelium involved in macrophage and lymphocyte recruitment, was significantly higher after 24 hours of peg-IFN treatment in patients who achieved SVR compared to those who relapsed (19, 34, 35). Monocyte chemoattractant protein 1 (MCP-1) is a CC chemokine that recruits monocytes and T-cells to areas of inflammation and elevated MCP-1 levels have been associated with rapid progression to liver failure in patients with HCV (36). The first line treatment for HCV has transitioned from IFN based therapy to DAA containing regimens, and it is not known if cytokines or chemokines can act as predictors of treatment response. Additionally, due to direct immune stimulation exerted by peg-IFN, a cornerstone of previous treatment regimens, it has been difficult to interpret the relationship of circulating cytokines and chemokines during therapy with the coincident immune responses. Since DAAs do not directly stimulate cellular immunity, analysis of immune molecules during treatment with IFN-free regimens has the potential to provide new insight into immune responses during HCV therapy.
In order to gain new insight into host-HCV interactions and potentially identify predictors of therapeutic response we measured serum chemokines and cytokines (IP-10, MCP-1, MIP-1β, IL-18) in 131 patients treated with SOF plus RBV for chronic HCV infection.
EXPERIMENTAL PROCEDURES
Study Population and Study Design
We conducted a case-control study of 131 patients with GT2 or GT3 HCV treated with SOF (Gilead Sciences) 400 mg orally once daily plus RBV (Ribasphere, Kadmon) orally in divided dose according to body weight (1000 mg daily <75 kg and 1200 mg daily ≥75 kg). Enrolled patients from three previous published studies (FISSION, POSITRON, or FUSION) (12, 13) had plasma samples collected at multiple time points including a pre-treatment baseline, on drug at weeks 1 (w1), 2 (w2), and 12 (w12) and at 4 weeks post-treatment (4wPT). The FISSION trial was a multicenter, randomized, open-label, active-control study comparing SOF plus RBV versus peg-IFN alfa-2a plus RBV in previously untreated patients (13). The POSITRON trial was a multicenter, blinded, placebo controlled study comparing SOF and RBV with placebo in patients for whom treatment with peg-IFN was not an option. The FUSION trial was a multicenter, blinded, active-control study involving patients who did not respond to treatment with peg-IFN (12). Full study designs and details are available online (at http://www.clinicaltrials.gov). For the current study, cases and controls were selected based on treatment response (Relapse or SVR). Cases and controls were further stratified based on HCV genotype (GT2 or GT3) and cirrhosis status (cirrhosis or no cirrhosis). Subjects were randomly selected from SOF plus RBV treated patients enrolled in the FISSION, FUSION, and POSITRON studies in an attempt to obtain 20 individuals in each of the stratified groups shown at the top of Table 1. However, due to the high frequency of SVR in individuals with GT2 infection treated with SOF + RBV we were unable to analyze 20 individuals for all groups (Table 1). SVR was defined as absence of detectable plasma HCV RNA 12 weeks or more after completion of therapy. Relapse was defined as presence of plasma HCV RNA 12 weeks or more after completion of therapy. Baseline data collected include standard clinical laboratory testing (baseline Alanine Aminotransferase (ALT) treated as a binary categorical variable (<1.5 upper limit of normal (ULN) or ≥1.5 ULN), creatinine clearance, serum HCV RNA levels, IL28B genotype (CC vs. CT or TT), HCV genotype (GT2 or GT3), body mass index ((BMI) <30 or ≥ 30) and cirrhosis status (cirrhosis or no cirrhosis); and demographic variables age, sex, race (White, Black, American Indian/Alaskan native, Asian) and ethnicity (Hispanic vs. non-Hispanic). Individuals in this analysis were pooled from three prospective clinical trials. To determine if the study origin of the patients were associated with treatment outcome, study (FISSION, POSITRON, FUSION) was included as a categorical independent variable in the logistic regression analysis. In the FUSION study, some individuals were randomized to 12 versus 16 weeks of SOF plus RBV therapy. Thus, treatment duration (12 or 16 weeks) was included as an independent binary variable in the logistic regression analyses. Treatment-experienced (n = 65) individuals are defined as those who received any duration of IFN therapy for treatment of HCV. This group includes IFN treatment failures (n = 41) and individuals who were intolerant of IFN therapy (n = 24). Banked plasma samples were then analyzed retrospectively to determine IP-10, MCP-1, MIP-1β, and IL-18 levels by immunoassay as described below.
Table 1.
Baseline demographic and clinical characteristics with virologic outcomes.
| Relapse | SVR | Total | ||
|---|---|---|---|---|
| Genotype 2 | Cirrhosis | 8 | 19 | 27 |
| No Cirrhosis | 6 | 20 | 26 | |
| Genotype 3 | Cirrhosis | 18 | 20 | 38 |
| No Cirrhosis | 21 | 19 | 40 | |
| Total | 53 | 78 | 131 | |
| Study | FISSION | 2 (2%) | ||
| POSITRON | 88 (67%) | |||
| FUSION | 41 (31%) | |||
| Treatment Duration | 12 weeks | 112 (85%) | ||
| 16 weeks | 19 (15%) | |||
| Race | AA | 10 (8%) | ||
| AI/AN | 2 (2%) | |||
| Asian | 5 (4%) | |||
| White | 114 (87%) | |||
| Ethnicity | Hispanic | 11 (8%) | ||
| Non-Hispanic | 120 (92%) | |||
| BMI | < 30 | 79 (60%) | ||
| ≤ 30 | 52 (40%) | |||
| IL28b Genotype | CC | 71 (54%) | ||
| CT | 43 (33%) | |||
| TT | 17 (13%) | |||
| ALT | < 1.5×ULN | 54 (41%) | ||
| ≥ 1.5×ULN | 77 (59%) | |||
| Age in years | 52.4 (10.1) | |||
| Plasma Log10HCV RNA (IU/ml) | 6.29 (0.82) | |||
| Creatinine Clearance in ml/min | 117.9 (33.5) | |||
Number and (Percentage) noted for all categorical variables
Mean and (SD) noted for all continuous variables
AA, African American; AI/AN, American Indian/Alaskan Native; ULN, upper limit of normal; HCV, Hepatitis C Virus; ALT, Alanine Aminotransferase; BMI, Body Mass Index; SVR, Sustained Virologic Response; GT2, genotype 2; GT3, genotype 3
Cytokine Measurements
The Meso Scale Discovery multiplexed immunoassay was used to measure IP-10, MCP-1, MIP-1β, and IL-18 (Meso Scale Discovery, Rockville, MD). The assay was performed per the manufacturer's recommendations using plasma from the patients taken from each of the study time points. Each sample was tested in duplicate and the average was calculated.
HCV Genotype and Quantification
HCV RNA level were measured using the COBAS TaqMan HCV Test version 2.0 (Roche Molecular Systems), with the lower limit of quantification equal to 25 IU per milliliter. For all analyses involving HCV viral load, an undetectable viral load was given a numeric quantity of 24 IU/ml that is just below the limit of detection for this assay. HCV genotype was determined using the Siemens Versant HCV Genotype 2.0 Assay.
IL28b Genotyping
IL-28b genotype was determined using PCR amplification and sequencing of the rs12979860 single-nucleotide polymorphism (37).
STATISTICAL ANALYSIS
We compared demographic, clinical, and treatment differences (Sex, Race, Ethnicity, BMI, cirrhosis status, IL28B genotype, baseline ALT, Study origin, Treatment duration) between patients with and without SVR using Fisher's exact test. Levene's test was applied to all continuous demographic, clinical, and cytokine variables to ensure equality of variances between patients who did or did not achieve SVR. Continuous variables were analyzed for differences using independent samples t tests with appropriate correction for those variables that did not display equality of variances. The untransformed cytokine data (IP-10, MCP-1, MIP-1β, or IL-18) demonstrated non-normal distributions. Log10-based transformation of cytokine levels improved normality. The goal was to identify potential predictors of outcome that could be measured early during treatment prior to failure or success of treatment, thus only cytokine data from baseline, week 1 and week 2 were used in the logistic regression analyses. Demographic and clinical characteristics included as independent variables in the logistic regression analyses were patient sex, age, BMI, race, ethnicity, baseline creatinine clearance, baseline ALT, baseline log10HCV plasma RNA, cirrhosis status and IL28b genotype. Differences in response to therapy (SVR v. Relapsers) were evaluated first in a univariable logistic regression model to determine unadjusted odds of achieving SVR with 95% confidence intervals (95% CI). Variables that achieved a p-value ≤ 0.20 in an unadjusted analysis were included in an additive multivariable model. A stepwise (backward/forward) logistic regression was used to determine the final model that best predicted SVR. Variables were then tested for interactions and if significant were included in the final model. Variables that achieved a p-value of ≤ 0.05 are specifically denoted. To determine if baseline demographic variables and HCV viral load levels were significantly associated with baseline log10 transformed IP-10, MCP-1, MIP-1β, or IL-18 we used a univariable linear regression analysis. Variables that achieved a p-value ≤ 0.20 in an unadjusted analysis were included in an additive multivariable model. A stepwise (backward/forward) linear regression was used to determine the final model that best predicted individual cytokine levels. The models that provided the fit best for each cytokine on the basis of Akiake's Information Criterion are displayed as the adjusted regression coefficient. Individual factors that achieved a p-value of ≤ 0.05 in the multivariable model are specifically denoted. Publicly available packages in R (v. 3.1.1; R Foundation for Statistical Computing, Vienna, Austria) were used for all analyses and figure creation.
RESULTS
Characteristics of the study participants
Cases and controls for this observational analysis were selected based on treatment response (SVR vs Relapse) with stratification by viral genotype (GT2 vs GT3) and pre-treatment clinical status (Cirrhotic vs Non-Cirrhotic) (Table 1). Accordingly, patient groups are similar with respect to cirrhosis status (Table 1). However, due to the efficacy of SOF plus RBV therapy in patients with GT2, fewer patients with GT2 (40%) and treatment failure (40%) were included in this analysis. The study participants were selected predominantly from the POSITRON and FUSION clinical trials. The majority received 12 weeks of SOF plus RBV therapy (85%) while nineteen individuals from FUSION received 16 weeks of therapy (15%). The study participants were mostly middle-aged (mean age 52.4, SD 10.1)) white (87%) non-Hispanic (92%) men (66%). The IL28b genotypes of the participants were CC (54%), CT (33%), and TT (13%). Slightly more patients (59%) had baseline ALT levels ≥ 1.5 × upper limit of normal ULN, were not obese (BMI<30, 60%), and had high baseline HCV viral loads (mean 6.29 million IU/ml).
The effects of baseline demographics and clinical characteristics on baseline cytokine levels
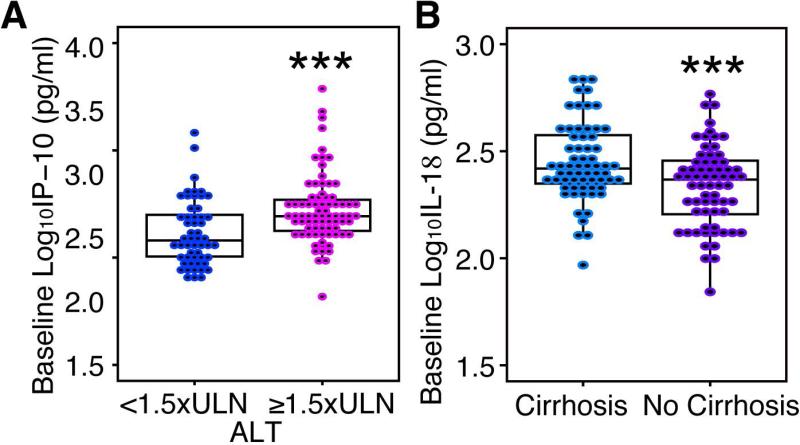
To determine if demographic and clinical characteristics are correlated with baseline IP-10, MCP-1, MIP-1β, and IL-18 levels we performed a univariable and multivariable linear regression analysis; results of which are detailed in Table 2. In a univariable analysis elevated levels of baseline log10IP-10 were associated with elevated ALT (Figure 1A). Additionally, elevated baseline log10IP-10 showed a weak inverse correlation with log10 plasma HCV RNA level (Supplementary Figure 1A). After stepwise multivariable regression elevated baseline ALT and lower HCV viral load were significant independent predictors of higher baseline IP-10 levels (Table 2). The positive correlation between higher IP-10 and elevated ALT has been previously described in treatment naïve individuals with chronic HCV infection (15). Using a similar approach, a univariable analysis demonstrated that increasing age and decreasing creatinine clearance were significantly associated with elevated baseline MCP-1 and MIP-1β (Table 2 and Supplementary Figure 1B). Increasing age remained a significant independent predictor of both increasing baseline MCP-1 and MIP-1β in our best-fit multivariable model (Table 2). In both univariable and multivariable analyses of IL-18, the presence of cirrhosis and the CC IL28b genotype were significantly associated with elevated baseline log10IL-18 (Figure 1D and Table 2). This strong positive association between cirrhosis and elevated IL-18 is similar to findings from previous studies that demonstrated associations between cirrhosis, the degree of liver disease, or the activity and severity of HCV infection and elevated IL-18 levels (25, 38, 39).
Table 2.
Association of demographic and clinical characteristics with baseline cytokine/chemokine levels.
| IP-10 baseline | MCP-1 Baseline | MIP-1β Baseline | IL-18 Baseline | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β (SE) | Aβ (SE) | β (SE) | Aβ (SE) | β (SE) | Aβ (SE) | β (SE) | Aβ (SE) | ||
| Sex | Male | −0.03 (0.06) | −0.01 (0.03) | −0.06 (0.04) | --- | 0.06 (0.04) | -- | ||
| Female | REF | REF | REF | -- | REF | -- | |||
| Age | CONTINUOUS (1yr) | 0.003 (0.003) | 0.004(0.001)** | 0.004 (0.001)* | 0.005 (0.002)** | 0.005 (0.002)* | −0.0002(0.002) | ||
| BMI | <30 | REF | REF | REF | REF | ||||
| >30 | 0.02 (0.06) | −0.14 (0.13) | −0.11 (0.17) | 0.01 (0.03) | |||||
| Race | Asian | 0.22 (0.28) | −0.13 (0.13) | 0.11 (0.17) | −0.06(0.17) | ||||
| AA | 0.14 (0.26) | 0.04 (0.12) | 0.07 (0.16) | 0.14(0.15) | |||||
| AI/AN | REF | REF | REF | REF | |||||
| White | 0.09 (0.24) | 0.02 (0.11) | 0.10 (0.15) | 0.02(0.14) | |||||
| Ethnicity | Hispanic | REF | REF | REF | -- | REF | |||
| Non-Hispanic | 0.05 (0.11) | 0.01 (0.05) | 0.11 (0.07) | -- | −0.06(0.06) | ||||
| Creat clearance | CONTINUOUS | −0.001 (0.0009) | −0.0009 (0.0004)* | -- | −0.001 (0.0005)* | -- | 0.0002(0.0005) | ||
| ALT | <1.5×ULN | REF | REF | REF | REF | REF | REF | REF | |
| ≥ 1.5×ULN | 0.22 (0.06)*** | 0.20 (0.06)*** | 0.05 (0.03) | 0.04 (0.03) | 0.07 (0.04)* | 0.06 (0.04) | |||
| Genotype | GT2 | 0.09 (0.06) | -- | −0.01 (0.03) | −0.01 (0.04) | 0.02(0.03) | |||
| GT3 | REF | -- | REF | REF | REF | ||||
| Log10HCV RNA baseline | CONTINUOUS | −0.10 (0.03)** | −0.08 (0.03)* | −0.01 (0.02) | −0.002 (0.02) | −0.002(0.02) | |||
| Cirrhosis | Yes | REF | REF | REF | REF | REF | |||
| No | −0.02 (0.06) | −0.03 (0.03) | 0.03 (0.04) | −0.11(0.03)*** | −0.11(0.03)*** | ||||
| IL28b genotype | CC | REF | REF | REF | REF | REF | |||
| CT or TT | −0.02 (0.06) | 0.002 (0.03) | −0.02 (0.04) | −0.07 (0.03)* | −0.06 (0.03)* | ||||
β = regression coefficient
Aβ = adjusted regression coefficient
SE = Standard Error
AA, African American; AI/AN, American Indian/Alaskan Native; ULN, upper limit of normal; HCV, Hepatitis C Virus; ALT, Alanine Aminotransferase; BMI, Body Mass Index; SVR, Sustained Virologic Response; GT2, genotype 2; GT3, genotype 3
p <0.05
p<0.01
p <0.001; calculated using the Students T distribution
“--” indicates variables that were dropped from the exploratory multivariable model
Figure 1.
Correlates of baseline cytokine/chemokine levels. (A) Comparison of baseline Log10IP-10 and baseline ALT. (B) Comparison of Log10IL-18 at baseline by cirrhosis status. t test p-value <0.001 ***. ULN = Upper limit of normal; ALT, Alanine Aminotransferase.
IP-10, MCP-1, MIP-1β, IL-18 levels during SOF plus RBV therapy
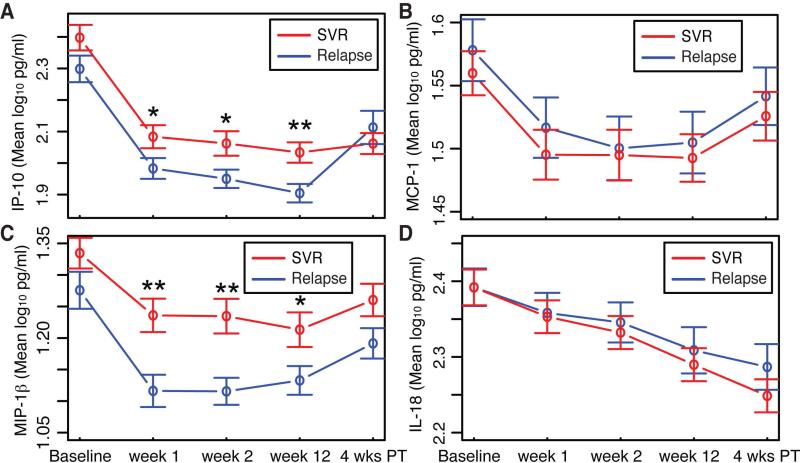
To gain insight into the immune response during DAA therapy for chronic HCV we measured IP-10, MCP-1, MIP-1β, and IL-18 plasma levels at baseline, w1, w2, and w12 on SOF plus RBV therapy, and 4wPT. Following initiation of therapy mean IP-10, MCP-1, MIP-1β, and IL-18 levels all decreased (Figure 2A-D). On average patients achieving SVR (red line) had significantly increased levels of IP-10 and MIP-1β at weeks one, two, and twelve weeks on therapy compared to patients who relapsed (blue line) (Figure 2A-C). At 4wPT mean IP-10 significantly increased only in individuals with relapse. IL-18 levels fell at a relatively constant rate throughout the study in both outcome groups and did not increase 4wPT (Figure 2D).
Figure 2.
Temporal dynamics of cytokines and chemokines during HCV infection and treatment. (A) Mean IP-10 (B) MCP-1 (C) MIP-1β (D) IL-18 levels (pg/ml) at baseline, weeks 1, 2, and 12 on SOF and RBV, and 4 weeks post-treatment by treatment outcome. t test p-value <0.01 **, <0.05 *. 4 wks PT, 4 weeks post treatment; SVR, sustained virologic response.
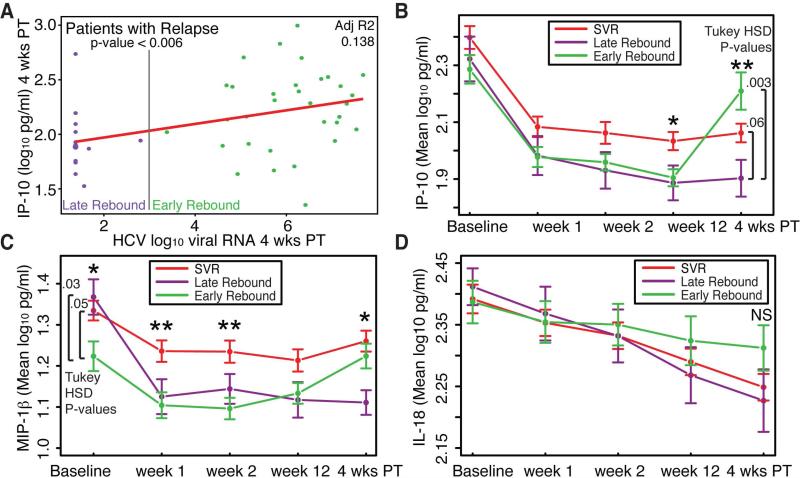
Our analysis revealed no significant differences in these four cytokines at 4wPT between patients that achieved SVR or relapse. We wondered whether the lack of a clear separation in some of the cytokines at 4wPT was because some individuals who relapsed late had cytokine profiles similar to those who achieved SVR at this early post-treatment time point. To determine if the timing of viral relapse was associated with differences in circulating cytokines we compared patients with early viral rebound (plasma HCV RNA level ≥1000 IU/ml at 4wPT) and late viral rebound (HCV Plasma HCV RNA <1000 IU/ml at 4wPT). In individuals who experienced relapse there was a weak but significant positive linear association between log10HCV plasma HCV RNA level and log10IP-10 at 4wPT (p-value <0.006) (Figure 3A). Additionally, individuals with early viral rebound had significantly higher mean log10IP-10 levels at 4wPT compared to individuals with late viral rebound (Tukey HSD p-value = 0.003) (Figure 3B). In addition, to discover differences in post-treatment cytokine levels in early versus late rebounders we found that individuals with early viral rebound had significantly lower baseline MIP-1β levels compared to those with late viral rebound or SVR (Figure 3C). By w1 MIP-1β levels in both groups of relapsers were similar and remained so throughout treatment. No statistically significant differences in circulating IL-18 were found, however, we did detect a trend towards increasing IL-18 in relapsers with early viral rebound compared to those with late viral rebound or SVR. This trend appeared to develop during treatment and thus preceded the development of detectable plasma HCV RNA (Figure 3D).
Figure 3.
Changes in cytokines and chemokines based magnitude of viral rebound at 4 weeks post-treatment. (A) Association of Log10IP-10 and HCV Log10HCV viral RNA at 4 weeks post treatment. Mean (B) IP-10 (C) MIP-1β (D) IL-18 at baseline, weeks 1, 2, and 12 on SOF and RBV, and 4 weeks post-treatment in individuals with SVR, early and late viral rebound. ANOVA p-value <0.01 **, <0.05 *. 4 wks PT, 4 weeks post treatment; SVR, sustained virologic response. Early rebound is defined as the presence of ≥ 1000 IU/ml HCV RNA at 4 weeks post treatment.
Analysis of treatment outcome based on demographic and clinical variables and cytokine levels
Our analysis of cytokines early in treatment demonstrated significant differences in IP-10 and MIP-1β based on treatment outcome. To test the hypothesis that specific demographic variables, clinical characteristics or cytokine (IP-10, MCP-1, MIP-1β, or IL-18) levels early in treatment could potentially serve as predictors of treatment outcome (SVR v. Relapse) in patients with GT2 and GT3 treated with SOF and RBV, we performed a univariable and multivariable logistic regression analysis. Compared to individuals who failed treatment, those that achieved SVR were more likely to have significantly higher w1 log10IP-10 (p-value 0.05, t-test) and log10MIP-1β (p-value 0.002, t-test) levels as well as higher w2 log10IP-10 (p-value 0.02, t-test) and log10MIP-1β (p-value <0.001, t-test) (Table 3). After mixed (backwards and forward) stepwise regression, in a multivariable model elevated baseline log10MIP-1β (OR 1.32 (1.08, 1.66) 95%CI, p-value 0.01) and smaller changes in log10MIP-1β during the first two weeks of therapy (OR 0.74 (0.58, 0.91) 95%CI, p-value 0.008) were independent predictors of SVR (Table 3).
Table 3.
Correlates of sustained virologic response (SVR) in all patients treated with Sofosbuvir and Ribavirin.
| Relapse | SVR | P-value | OR (95% CI)a |
|||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | |||||
| Sex | Female | 14 (10.7%) | 31 (23.7%) | 0.14b | Reference | -- |
| Male | 39 (29.8%) | 47 (35.9%) | 0.54 (0.25, 1.15) | |||
| Log10IP10 baselined | 2.299 (0.299) | 2.398 (0.351) | 0.10c | 1.10 (0.99, 1.24) | -- | |
| Log10MIP1β baselined | 1.276 (0.208) | 1.334 (0.211) | 0.12c | 1.15 (0.97, 1.37) | 1.32 (1.08, 1.66)* | |
| Log10IP10 week 1d | 1.983 (0.233) | 2.084 (0.317) | 0.05c | 1.14 (1.00, 1.32) | -- | |
| Log10MIP1β week 1d | 1.116 (0.181) | 1.236 (0.230) | 0.002c | 1.32 (1.11, 1.61) | -- | |
| Log10IP10 week 2d | 1.950 (0.205) | 2.062 (0.336) | 0.02c | 1.16 (1.02, 1.35) | -- | |
| Log10MIP1β week 2d | 1.115 (0.151) | 1.235 (0.238) | <0.001c | 1.34 (1.12, 1.64) | -- | |
| Δ Log10MIP1β w0-1d | 0.160 (0.191) | 0.103 (0.184) | 0.10c | 0.85 (0.69, 1.03) | -- | |
| Δ Log10MIP1β w0-2d | 0.160 (0.184) | 0.099 (0.207) | 0.09c | 0.85 (0.70, 1.02) | 0.74 (0.58, 0.91)** | |
Variables meeting p-value <0.2 threshold for inclusion in initial multivariable analysis are reported. Percentages have been rounded and might not total 100.
Univariable (unadjusted) and multivariable (adjusted) ORs are presented.
P-values calculated by Fisher Exact Test
P-values calculated by independent sample t tests
Odds ratios were calculated per 0.1 unit increase
p <0.05
p<0.01
*** p <0.001; calculated in final multivariable model
“--” indicates variables that were dropped from the exploratory multivariable model
Mean and (SD) noted under Relapse and SVR for all continuous variables
Number and (Percentage) noted for all categorical variables
Analysis of treatment outcomes in patients stratified by cirrhosis status
Cirrhosis is associated with worse treatment outcomes in patients treated with SOF plus RBV (12, 13). To determine if cirrhosis changes the predictive capacity of demographic, clinical, and cytokine (IP-10, MCP-1, MIP-1β, or IL-18) data on treatment outcomes, we analyzed subsets of patients with or without cirrhosis. Patients with cirrhosis (n=65) (Table 1) who achieved SVR were less likely to be Hispanic (p-value 0.03, Fisher exact test) and more likely to have higher plasma Log10MIP-1β at w1 (p-value 0.001, t-test) and w2 (p-value 0.01, t-test) (Table 4). After mixed stepwise regression higher Log10MIP-1β at baseline (OR 1.67 (1.11, 2.79)) and a smaller decline in Log10MIP-1β levels between baseline and w1 (OR 0.59 (0.37, 0.88)) were independent predictors of SVR in patients with cirrhosis (Table 4).
Table 4.
Correlates of sustained virologic response (SVR) in patients with cirrhosis.
| Relapse | SVR | P-value | OR (95% CI)a |
|||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | |||||
| Sex | F | 4 (6.2%) | 14 (21.5%) | 0.09b | 1 [Reference] | |
| M | 22 (33.8%) | 25 (38.5%) | 0.32 (0.082, 1.06) | -- | ||
| Ethnicity | Hispanic | 5 (7.7%) | 1 (1.5%) | 0.03b | 1 [Reference] | |
| Non-Hispanic | 21 (32.3%) | 38 (58.5%) | 9.05 (1.34, 179.54) | 7.64 (0.98, 161.3) | ||
| Log10MIP1β baselined | 1.240 (0.209) | 1.332 (0.208) | 0.08c | 1.25 (0.98, 1.65) | 1.67 (1.11, 2.79)* | |
| Log10MIP1β week 1d | 1.114 (0.191) | 1.280 (0.197) | 0.001c | 1.65 (1.21, 2.45) | -- | |
| Log10MIP1β week 2d | 1.119 (0.162) | 1.241 (0.225) | 0.01c | 1.38 (1.06, 1.87) | -- | |
| Δ Log10MIP1β w0-1d | 0.126 (0.158) | 0.053 (0.185) | 0.10c | 0.77 (0.55, 1.04) | 0.59 (0.37, 0.88)* | |
| Δ Log10MIP1β w0-2d | −0.005 (0.105) | 0.039 (0.146) | 0.16c | 1.32 (0.89, 2.03) | -- | |
Variables meeting p-value <0.2 threshold for inclusion in initial multivariable analysis are reported. Percentages have been rounded and might not total 100.
Univariable (unadjusted) and multivariable (adjusted) ORs are presented.
P-values calculated by Fisher Exact Test
P-values calculated by independent sample t tests
Odds ratios were calculated per 0.1 unit increase
“--” indicates variables that were dropped from the exploratory multivariable model
Mean and (SD) noted under Relapse and SVR for all continuous variables
Number and (Percentage) noted for all categorical variables
In patients without cirrhosis (n=66) (Table 1), individuals who achieved SVR were significantly more likely to have higher w1 and w2 IP-10 levels as well as higher plasma w2 MIP-1β compared to individuals who failed therapy (Supplementary Table 1). A mixed stepwise regression identified higher baseline Log10IP-10 (OR 1.28 (1.06, 1.62)) levels and a smaller decrease in log10MIP-1β during the first two weeks (OR 0.72 (0.53, 0.94)) of therapy as independent predictors of SVR in patients without cirrhosis.
Analysis of treatment outcomes in patients stratified by HCV genotype
Although SOF plus RBV therapy has excellent overall efficacy, patients with GT3 infection are more likely to fail therapy than patients with GT2 infection (12, 13). To determine significant predictors of treatment efficacy in GT3 infection we performed a univariable and multivariable analysis of patients infected with GT3 (n=78) (Table 1). Individuals who attained SVR were significantly more likely to have higher baseline Log10IP-10 levels (p-value 0.05, t-test), w1 Log10MIP-1β levels (p-value 0.05 t-test) and have received 16 weeks of therapy (p-value 0.01 Fisher exact test) compared to those who relapsed in an unadjusted analysis (Table 5). After mixed stepwise regression, longer duration therapy (16 weeks) (OR 1.80 (1.59, 31.0)), higher w1 Log10MIP-1β (OR 1.35 (1.04, 1.80)) and a larger decrease in Log10IP-10 (OR 1.32 (1.04, 1.71)) between baseline and w1 on therapy were independent predictors of SVR in patients with GT3 infection (Table 5). To determine if these results were restricted to GT3 infection we conducted a similar analysis in individuals infected with GT2 HCV (n = 53). Similar to our analysis of GT3 infection, in individuals with GT2 infection higher early levels of MIP-1β were significantly associated with SVR in univariable and multivariable analyses (Supplementary table 2).
Table 5.
Correlates of SVR in patients infected with Genotype 3 HCV
| Relapse | SVR | P-value | OR (95% CI)a |
|||
|---|---|---|---|---|---|---|
| Unadjusted | Adjusted | |||||
| Treatment Duration (wks) | 12 | 36 (46.2%) | 26 (33.3%) | 0.01b | 1 [Reference] 6.0 (1.73, 28.1) |
1 [Reference] 1.80 (1.59, 31.0)* |
| 16 | 3 (3.8%) | 13 (16.7%) | ||||
| Clinical Trial | FUSION | 9 (11.5%) | 18 (23.1%) | 0.06b | 1 [Reference] 0.35 (0.13, 0.91) |
-- |
| POSITRON | 30 (38.5%) | 21 (26.9%) | ||||
| Log10HCV RNA baselinee | 6.064 (0.804) | 6.334 (0.750) | 0.13c | 1.58 (0.885, 2.966) | -- | |
| Log10IP-10 baselined | 2.331 (0.262) | 2.459 (0.298) | 0.05c | 1.19 (1.01, 1.43) | -- | |
| Log10MIP1β week 1d | 1.124 (0.166) | 1.215 (0.234) | 0.05c | 1.26 (1.00, 1.61) | 1.40 (1.08, 1.89)* | |
| Log10IP-10 week 2d | 1.953 (0.173) | 2.062 (0.345) | 0.09c | 1.17 (0.99, 1.43) | -- | |
| Log10MIP1β week 2d | 1.111 (0.132) | 1.202 (0.258) | 0.06c | 1.25 (1.00, 1.61) | -- | |
| A Log10IP-10 w0-1d | 0.328 (0.227) | 0.402 (0.235) | 0.16c | 1.15 (0.95, 1.42) | 1.34 (1.06, 1.75)* | |
Variables meeting p-value <0.2 threshold for inclusion in initial multivariable analysis are reported. Percentages have been rounded and might not total 100.
Univariable (unadjusted) and multivariable (adjusted) ORs are presented.
P-values calculated by Fisher Exact Test
P-values calculated by independent sample t tests
Odds ratios were calculated per 0.1 unit increase
Odds ratio was calculated per 1 log10 increase in viral load
“--” indicates variables that were dropped from the exploratory multivariable model
Mean and (SD) noted under Relapse and SVR for all continuous variables
Analysis of treatment outcomes in patients stratified based on prior treatment with IFN
We performed univariable and multivariable analysis on the subsets of individuals who were treatment-experienced (n = 65) or treatment-naïve (n = 66). In a univariable analysis, treatment-experienced individuals who attained SVR showed a non-statistically significant trend towards higher w1 MIP-1β levels (p-value 0.06 t-test) and longer treatment duration (16 weeks) (p-value 0.06 Fisher exact test) (Supplementary table 3). A mixed stepwise regression identified 16 weeks of treatment (OR 6.53 (1.52, 36.7)), lower baseline Log10MCP1 (OR 0.51 (0.26, 0.88) and higher week 1 Log10MIP-1β (OR 1.62 (1.06, 2.68) as significant independent predictors of SVR in treatment experienced patients. In treatment-naïve individuals, SVR was significantly associated with higher baseline, w1 and w2 IP-10 as well as higher w1 and w2 MIP-1β (Supplementary table 4). After mixed stepwise logistic regression, both increased baseline Log10IP-10 (OR 1.37 (1.08, 1.83)) and w2 Log10MIP-1β (OR 1.42 (1.07, 2.00)) were significant independent predictors of SVR in treatment-naïve individuals.
Previous IFN treatment failure, especially in those individuals with cirrhosis and GT3 infection, have been associated with lower SVR rates in individuals treated with SOF plus RBV (12, 14). In the subset of individuals with cirrhosis and genotype 3 infection who had previously failed IFN therapy (n = 27), a univariable analysis demonstrated a significant association between higher baseline HCV levels and SVR (p-value 0.01 t-test) (Supplementary table 5). The mixed stepwise logistic regression analysis identified longer treatment duration, higher baseline HCV levels, lower MCP1 at baseline, and higher w1 MIP-1β as statistically significant independent predictors of SVR (Supplementary table 5). In the larger subset of all individuals who failed IFN therapy regardless of HCV genotype and liver histology (n = 41) only w1 MIP-1β was statistically associated with SVR in a univariable (p-value 0.03 t-test) and multivariable (OR 1.49 (1.05, 2.40)) analysis (Supplementary table 6).
DISCUSSION
In this case-control study we measured plasma IP-10, MCP-1, MIP-1β, and IL-18 in a cohort of 131 patients pooled from the FISSION, POSITRON, and FUSION trials who were infected with HCV GT2 or GT3 and treated with SOF plus RBV. Of the cytokines tested MIP-1β and IP-10 showed significant differences based on treatment outcome. After logistic regression analysis, a best-fit multivariable model included higher baseline MIP-1β levels and smaller decreases in MIP-1β levels early in treatment as predictive of SVR. Previously, the FISSION, POSITRON, and FUSION trials identified patients infected with HCV GT3 and/or cirrhosis as more likely to relapse after treatment. When we analyzed patients with cirrhosis, baseline MIP-1β and early changes in MIP-1β remained significant independent predictors of treatment outcome with elevated levels at baseline and smaller changes during w1 of treatment predicting SVR. In patients infected with HCV GT3 higher w1 MIP-1β and larger declines in IP-10 during the first week of treatment were significant independent predictors of SVR in a multivariable model. To our knowledge only one previous study has reported a significant association of MIP-1β with HCV treatment outcome (19). In that study, Florholmen et al. analyzed individuals treated with peg-IFN and found that MIP-1β was significantly higher at 24 hours in patients who achieved SVR compared to those who relapsed. Additional studies will be needed to confirm the predictive capacity of early MIP-1β levels on treatment response and determine whether MIP-1β levels will be predictive in treatment with other DAA regimens and in other viral genotypes.
After initiation of SOF plus RBV, plasma HCV RNA decreases rapidly and mean circulating IP-10, MCP-1, MIP-1β, and IL-18 levels decline at varying rates. IP-10, MCP-1, and MIP-1β display similar temporal response curves during treatment characterized by rapidly decreasing levels during w1 leading to a nadir between w1 and w12 on therapy. Following completion of therapy IP-10 levels rise in non-responders in a manner correlated with HCV viremia at 4wPT. This suggests that the sharp increase in w4PT IP-10 in individuals with relapse is at least in part driven by increasing viral replication with subsequent IFN stimulated IP-10 production.
Higher baseline, w1, or w2 MIP-1β levels or smaller decreases in MIP-1β early in treatment were found to be important predictors of treatment outcome in all patient groups and subgroups analyzed in our cohort. Our analysis of patients based on timing of viral relapse found that individuals with early viral rebound had significantly lower baseline MIP-1β levels. If we consider individuals who demonstrate later viral rebound after cessation of therapy (late rebound) to have had a better response than individuals who develop high viral loads very soon after cessation of therapy (early rebound) this data is consistent with the cumulative data presented here that lower MIP-1β levels are associated with poorer response to SOF plus RBV therapy.
Regarding IL-18 levels, analysis of patients with early versus late viral rebound demonstrated that individuals with early viral rebound showed a non-statistically significant trend towards higher IL-18 levels. This difference is interesting given previous data identifying elevated plasma IL-18 as an indicator of sustained HCV viremia in patients with acute infection (22). In this study, IL-18 levels diverged in patients with early viral rebound and the divergence began prior to cessation of therapy and detectable viral rebound. Although not statistically significant this data provides some suggestion that higher IL-18 levels late in therapy may precede eventual viral relapse. It will be interesting to analyze IL-18 levels at later time points following therapy to see if IL-18 levels in those with early rebound reach statistically higher levels and if individuals with late viral rebound begin to show increasing IL-18 levels over those with SVR.
Higher baseline or early on-treatment IP-10 levels were significantly associated with SVR in our overall analysis or in subgroups of patients without cirrhosis, GT3 infection, or in individuals who are treatment naïve. Previous studies from the IFN treatment era found the opposite correlation that lower baseline IP-10 levels were associated with SVR (16-18, 20, 21). It had been proposed that individuals with lower baseline IP-10 levels had better responses to IFN therapy because the therapeutic IFN was able to stimulate a larger net increase in IFN stimulated genes (ISGs) that collectively induce an antiviral state and eradicate HCV. This paradoxical difference in IP-10 and prediction of treatment response with DAA therapy requires additional investigation to both corroborate these findings and begin to develop a mechanistic understanding.
The subgroup analysis of individuals with cirrhosis and genotype 3 infection who failed IFN therapy identified higher early MIP-1β (week 1), longer treatment duration (16 weeks), lower baseline MCP and higher baseline HCV as independent predictors of SVR in a multivariable model. It is unclear why a higher baseline HCV RNA should be associated with SVR based on our current understanding of HCV treatment. The number of individuals in this subgroup is small (n = 27) compared to the number of predictor variables and therefore the results of this subgroup analysis should be cautiously interpreted. When the subgroup population was expanded to include all individuals who previously failed IFN therapy (n = 41), baseline HCV viral load was no longer significantly associated with SVR. Additional studies will be needed to further define important predictors in this difficult to treat population.
A major limitation of this study is its case-control design that is exploratory and hypothesis generating in nature. Additional, prospective studies will be needed to solidify the associations described herein. The patient population is primarily male, white, and non-Hispanic and this may limit the ability of our results to be generalized to all populations. However, it should be noted that these patients were selected from Phase 3 studies with multicenter international designs and this enrollment strategy increases the potential for generalizability of our results. Our logistic regression analysis of treatment outcomes in patients with GT3 infection, included serum samples taken from patients treated with SOF and RBV for 12 (FISSION, POSITRON) or 16 weeks (FUSION). A subsequent clinical trial (VALENCE(14)) has demonstrated higher rates of SVR in patients with GT3 infection after 24 weeks of treatment. It is unclear if higher Log10MIP-1β levels will be predictive of treatment outcome in longer duration treatment. Lastly, the described analysis was limited to studying the levels of only four cytokine/chemokines that were potentially important in HCV infection. Measurements of additional cytokines or serum biomarkers could significantly improve the overall predictive capacity of our multivariable models.
Supplementary Material
Acknowledgements
We thank Scott Letendre and Michael Potter for advice and technical support related to Meso Scale cytokine determinations, and Florin Vaida for critical comments regarding statistical analyses.
Financial Support:
Gilead Sciences, AI097061 (DLW), AI076558 (RTS, DLW), T32 AI007036-35, 2P30CA023100-28S2 (PA)
List of Abbreviations
- HCV
hepatitis c virus
- IFN
interferon
- DAA
direct acting antiviral
- SOF
sofosbuvir
- RBV
ribavirin
- SVR
sustained virologic response
- HIV
Human Immunodeficiency Virus
- peg-IFN
peg-interferon
- IP-10
IFNγ-inducible protein 10
- IL-18
interleukin 18
- MIP-1β
Macrophage inflammatory protein 1β
- MCP-1
Monocyte chemoattractant protein 1
- GT2
genotype 2
- GT3
genotype 3
- w1
week 1 on treatment
- w2
week 2 on treatment
- w12
week 12 on treatment
- 4wPT
4 weeks post treatment
- ALT
Alanine Aminotransferase
- ULN
upper limit of normal
- BMI
body mass index
- PRR
pattern recognition receptor
Footnotes
Disclosures: DLW is a paid consultant of and receives research support from Gilead Sciences paid to UC Regents for involvement in clinical trials. RTS is a member of the Gilead Sciences Scientific Advisory Board and receives research support from Gilead Sciences. Support for both activities is paid to the Regents of the University of California. QS, HW, MSP and LMS are employees of Gilead Sciences.
REFERENCES
- 1.NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1–46. [PubMed] [Google Scholar]
- 2.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Adam R, McMaster P, O'Grady JG, Castaing D, Klempnauer JL, Jamieson N, Neuhaus P, et al. Evolution of liver transplantation in Europe: report of the European Liver Transplant Registry. Liver Transpl. 2003;9:1231–1243. doi: 10.1016/j.lts.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Seeff LB, Hoofnagle JH. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology. 2002;36:S1–2. doi: 10.1053/jhep.2002.36992. [DOI] [PubMed] [Google Scholar]
- 5.Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, et al. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee A, Guedj J, Perelson AS. Mathematical modelling of HCV infection: what can it teach us in the era of direct-acting antiviral agents? Antivir Ther. 2012;17:1171–1182. doi: 10.3851/IMP2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci P. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis. 2011;31:356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 8.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 9.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 10.Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, et al. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med. 2014;370:1483–1493. doi: 10.1056/NEJMoa1316366. [DOI] [PubMed] [Google Scholar]
- 11.Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, et al. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med. 2014;370:211–221. doi: 10.1056/NEJMoa1306218. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson IM, Gordon SC, Kowdley KV, Yoshida EM, Rodriguez-Torres M, Sulkowski MS, Shiffman ML, et al. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med. 2013;368:1867–1877. doi: 10.1056/NEJMoa1214854. [DOI] [PubMed] [Google Scholar]
- 13.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 14.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, et al. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med. 2014;370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 15.Lin JC, Habersetzer F, Rodriguez-Torres M, Afdhal N, Lawitz EJ, Paulson MS, Zhu Y, et al. Interferon gamma-induced protein 10 kinetics in treatment-naive versus treatment-experienced patients receiving interferon-free therapy for hepatitis C virus infection: implications for the innate immune response. J Infect Dis. 2014;210:1881–1885. doi: 10.1093/infdis/jiu325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butera D, Marukian S, Iwamaye AE, Hembrador E, Chambers TJ, Di Bisceglie AM, Charles ED, et al. Plasma chemokine levels correlate with the outcome of antiviral therapy in patients with hepatitis C. Blood. 2005;106:1175–1182. doi: 10.1182/blood-2005-01-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darling JM, Aerssens J, Fanning G, McHutchison JG, Goldstein DB, Thompson AJ, Shianna KV, et al. Quantitation of pretreatment serum interferon-gamma-inducible protein-10 improves the predictive value of an IL28B gene polymorphism for hepatitis C treatment response. Hepatology. 2011;53:14–22. doi: 10.1002/hep.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeremski M, Markatou M, Brown QB, Dorante G, Cunningham-Rundles S, Talal AH. Interferon gamma-inducible protein 10: a predictive marker of successful treatment response in hepatitis C virus/HIV-coinfected patients. J Acquir Immune Defic Syndr. 2007;45:262–268. doi: 10.1097/QAI.0b013e3180559219. [DOI] [PubMed] [Google Scholar]
- 19.Florholmen J, Kristiansen MG, Steigen SE, Sorbye SW, Paulssen EJ, Kvamme JM, Konopski Z, et al. A rapid chemokine response of macrophage inflammatory protein (MIP)-1alpha, MIP-1beta and the regulated on activation, normal T expressed and secreted chemokine is associated with a sustained virological response in the treatment of chronic hepatitis C. Clin Microbiol Infect. 2011;17:204–209. doi: 10.1111/j.1469-0691.2010.03206.x. [DOI] [PubMed] [Google Scholar]
- 20.Askarieh G, Alsio A, Pugnale P, Negro F, Ferrari C, Neumann AU, Pawlotsky JM, et al. Systemic and intrahepatic interferon-gamma-inducible protein 10 kDa predicts the first-phase decline in hepatitis C virus RNA and overall viral response to therapy in chronic hepatitis C. Hepatology. 2010;51:1523–1530. doi: 10.1002/hep.23509. [DOI] [PubMed] [Google Scholar]
- 21.Lagging M, Askarieh G, Negro F, Bibert S, Soderholm J, Westin J, Lindh M, et al. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One. 2011;6:e17232. doi: 10.1371/journal.pone.0017232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chattergoon MA, Levine JS, Latanich R, Osburn WO, Thomas DL, Cox AL. High plasma interleukin-18 levels mark the acute phase of hepatitis C virus infection. J Infect Dis. 2011;204:1730–1740. doi: 10.1093/infdis/jir642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iannello A, Boulassel MR, Samarani S, Tremblay C, Toma E, Routy JP, Ahmad A. HIV-1 causes an imbalance in the production of interleukin-18 and its natural antagonist in HIV-infected individuals: implications for enhanced viral replication. J Infect Dis. 2010;201:608–617. doi: 10.1086/650314. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe D, Uehira T, Yonemoto H, Bando H, Ogawa Y, Yajima K, Taniguchi T, et al. Sustained high levels of serum interferon-gamma during HIV-1 infection: a specific trend different from other cytokines. Viral Immunol. 2010;23:619–625. doi: 10.1089/vim.2010.0065. [DOI] [PubMed] [Google Scholar]
- 25.Sharma A, Chakraborti A, Das A, Dhiman RK, Chawla Y. Elevation of interleukin-18 in chronic hepatitis C: implications for hepatitis C virus pathogenesis. Immunology. 2009;128:e514–522. doi: 10.1111/j.1365-2567.2008.03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vecchiet J, Falasca K, Cacciatore P, Zingariello P, Dalessandro M, Marinopiccoli M, D'Amico E, et al. Association between plasma interleukin-18 levels and liver injury in chronic hepatitis C virus infection and non-alcoholic fatty liver disease. Ann Clin Lab Sci. 2005;35:415–422. [PubMed] [Google Scholar]
- 27.Yoneda S, Umemura T, Katsuyama Y, Kamijo A, Joshita S, Komatsu M, Ichijo T, et al. Association of serum cytokine levels with treatment response to pegylated interferon and ribavirin therapy in genotype 1 chronic hepatitis C patients. J Infect Dis. 2011;203:1087–1095. doi: 10.1093/infdis/jiq165. [DOI] [PubMed] [Google Scholar]
- 28.An P, Thio CL, Kirk GD, Donfield S, Goedert JJ, Winkler CA. Regulatory polymorphisms in the interleukin-18 promoter are associated with hepatitis C virus clearance. J Infect Dis. 2008;198:1159–1165. doi: 10.1086/592047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong JY, Cho SW, Oh B, Kimm K, Lee KM, Shin SJ, Lee JA, et al. Association of interleukin-18 gene polymorphisms with hepatitis B virus clearance. Dig Dis Sci. 2010;55:1113–1119. doi: 10.1007/s10620-009-0819-z. [DOI] [PubMed] [Google Scholar]
- 30.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20–38. doi: 10.1111/j.1600-065X.2008.00624.x. [DOI] [PubMed] [Google Scholar]
- 31.Barr DP, Belz GT, Reading PC, Wojtasiak M, Whitney PG, Heath WR, Carbone FR, et al. A role for plasmacytoid dendritic cells in the rapid IL-18-dependent activation of NK cells following HSV-1 infection. Eur J Immunol. 2007;37:1334–1342. doi: 10.1002/eji.200636362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reading PC, Smith GL. Vaccinia virus interleukin-18-binding protein promotes virulence by reducing gamma interferon production and natural killer and T-cell activity. J Virol. 2003;77:9960–9968. doi: 10.1128/JVI.77.18.9960-9968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reading PC, Whitney PG, Barr DP, Wojtasiak M, Mintern JD, Waithman J, Brooks AG. IL-18, but not IL-12, regulates NK cell activity following intranasal herpes simplex virus type 1 infection. J Immunol. 2007;179:3214–3221. doi: 10.4049/jimmunol.179.5.3214. [DOI] [PubMed] [Google Scholar]
- 34.Ahlenstiel G, Woitas RP, Rockstroh J, Spengler U. CC-chemokine receptor 5 (CCR5) in hepatitis C--at the crossroads of the antiviral immune response? J Antimicrob Chemother. 2004;53:895–898. doi: 10.1093/jac/dkh239. [DOI] [PubMed] [Google Scholar]
- 35.Shields PL, Morland CM, Salmon M, Qin S, Hubscher SG, Adams DH. Chemokine and chemokine receptor interactions provide a mechanism for selective T cell recruitment to specific liver compartments within hepatitis C-infected liver. J Immunol. 1999;163:6236–6243. [PubMed] [Google Scholar]
- 36.Farci P, Wollenberg K, Diaz G, Engle RE, Lai ME, Klenerman P, Purcell RH, et al. Profibrogenic chemokines and viral evolution predict rapid progression of hepatitis C to cirrhosis. Proc Natl Acad Sci U S A. 2012;109:14562–14567. doi: 10.1073/pnas.1210592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludwiczek O, Kaser A, Novick D, Dinarello CA, Rubinstein M, Vogel W, Tilg H. Plasma levels of interleukin-18 and interleukin-18 binding protein are elevated in patients with chronic liver disease. J Clin Immunol. 2002;22:331–337. doi: 10.1023/a:1020600230977. [DOI] [PubMed] [Google Scholar]
- 39.Yamano T, Higashi T, Nouso K, Nakatsukasa H, Kariyama K, Yumoto E, Kobayashi Y, et al. Serum interferon-gamma-inducing factor/IL-18 levels in primary biliary cirrhosis. Clin Exp Immunol. 2000;122:227–231. doi: 10.1046/j.1365-2249.2000.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.