Abstract
Peroxisomes are highly metabolic, autonomously replicating organelles that generate ROS as a by product of fatty acid β-oxidation. Consequently, cells must maintain peroxisome homeostasis, or risk pathologies associated with too few peroxisomes, such as peroxisome biogenesis disorders, or too many peroxisomes, inducing oxidative damage and promoting diseases such as cancer. We report that the PEX5 peroxisome import receptor binds ataxia-telangiectasia mutated (ATM) and localizes this kinase to the peroxisome. In response to reactive oxygen species (ROS), ATM signaling activates ULK1 and inhibits mTORC1 to induce autophagy. Specificity for autophagy of peroxisomes (pexophagy) is provided by ATM phosphorylation of PEX5 at Ser141, which promotes PEX5 mono-ubiquitination at K209, and recognition of ubiquitinated PEX5 by the autophagy adapter protein p62, directing the autophagosome to peroxisomes to induce pexophagy. These data reveal an important new role for ATM in metabolism as a sensor of ROS that regulates pexophagy.
Peroxisomes participate in β-oxidation of branched and very long chain fatty acids (VLCFAs), which results in the production of reactive oxygen species (ROS)1, 2. When in excess, ROS can cause cellular damage, and trigger catabolic functions such as autophagy3-6. As autonomously replicating organelles, maintaining the balance between peroxisome biogenesis and degradation is critical for normal cellular homeostasis7-11, and if dysregulated, can give rise to diseases such as peroxisome biogenesis disorders (PBDs) 7, 11, 12, white matter disease9, 13 and Alzheimer's disease8, 13. While the importance of maintaining peroxisome homeostasis is clear, mechanisms for recognition and removal of excessive or aberrant peroxisomes to prevent pathologies associated with too few or too many peroxisomes, are not well understood.
Selective autophagy of peroxisomes (pexophagy) is a major pathway by which excess peroxisomes are eliminated14-18. During selective autophagy, adaptor proteins mediate target recognition, such as the ubiquitin-binding protein p62, which contains both an LC3-interacting region (LIR) that binds to LC3-associated with the nascent autophagosome, and a ubiquitin-associated (UBA) domain that binds to monoubiquitinated lysine residues in the target19. p62 is known to be involved in pexophagy20, however, the peroxisomal targets recognized by p62, and mechanisms responsible for regulation of pexophagy have not been elucidated.
Recently, we reported that ataxia-telangiectasia mutated (ATM) signals to the tuberous sclerosis complex (TSC) in the cytoplasm to regulate autophagy in response to ROS3. ATM is activated by ROS via formation of a disulfide-cross-linked dimer21, and this kinase has been localized previously to the peroxisome22, 23. Importantly, we recently found that the TSC signaling node that regulates mTORC1 (a suppressor of autophagy) is also resident at the peroxisome in liver cells, the predominant cell type in the body for β-oxidation of fatty acids24, 25. These data led us to hypothesize that ROS may serve as a rheostat for peroxisomal homeostasis, activating signaling molecules at the peroxisome to regulate pexophagy.
RESULTS
ATM is a peroxisome-localized kinase activated by ROS
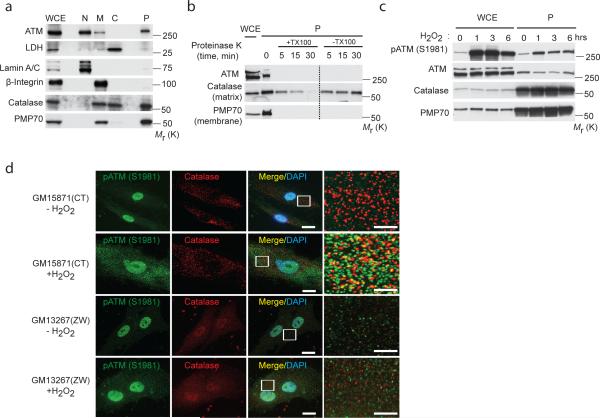
Endogenous ATM was detected in the nuclear fraction of cells (Fig. 1a), consistent with what is known about the function of this kinase as DNA damage response sensor26, 27. ATM was also found in the membrane and peroxisome compartments (Fig. 1a), consistent with previous reports that ATM was localized to this organelle22, 23. To determine whether peroxisomal ATM localized to the exterior (membrane) or interior (matrix) of this organelle, isolated peroxisomes were treated with proteinase K in the absence or presence of the membrane disrupting detergent Triton X-100. Like the peroxisome membrane protein PMP70, but not peroxisome matrix protein catalase which is resistant to degradation when peroxisome membranes are intact, ATM was rapidly degraded in both absence and presence of Triton X-100, indicating that ATM was associated with the outer (proteinase K accessible) surface of peroxisomes (Fig. 1b).
Figure 1. ATM kinase is localized at peroxisome and activated in response to ROS.
(a) Subcellular fractionation of HEK293 cells. Catalase and PMP70 were used as subcellular markers of the peroxisome (P). LDH, Lamin A/C and β-integrin were used as markers for cytosolic (C), nuclear (N) and membrane (M) fractions, respectively. WCE, whole cell extract. (b) Proteinase K assay in the presence or absence of Triton-X 100 performed with peroxisomal fractions obtained from HEK293 cells. Immunoblotting was performed with ATM, catalase and PMP70 antibodies. WCE, whole cell extract; P, peroxisome fraction. (c) HepG2 cells treated with H2O2 (0.4 mM) at 1, 3, 6 h. Whole cell extracts (WCE) and peroxisomal fractions (P) were probed with the indicated antibodies. (d) Representative image of wild type (GM15871) and Zellweger (GM13267) fibroblasts treated with or without H2O2 for 1 h and immunostained for active p-ATM (S1981) (green) and catalase (red). Scale bar, 15 μm. High-magnification images of boxed areas are indicated to the right (Scale bar, 5 μm). Uncropped images of western blots are shown in Supplementary Fig. S9.
We also observed an increase in activated ATM in the peroxisome fraction (increased immunoreactivity with a phospho-specific ATM (S1981) antibody) in response to H2O2 (Fig. 1c), which was confirmed by deconvolution microscopy, showing co-localization of pATM with the peroxisomal protein catalase in peroxisomes (Fig. 1d). Co-localization was not observed in peroxisome-deficient human fibroblasts from the well-characterized Zellweger peroxisome biogenesis disorder (mutated in PEX6 gene) (Fig. 1d) while nuclear localization and activation (phosphorylation) of ATM (pATM) was observed in control and Zellweger fibroblasts (Fig. 1d and Supplementary Fig. S1a). Together, these data identify the peroxisome as a site for activation of ATM in response to ROS.
ATM is localized to the peroxisome by PEX5
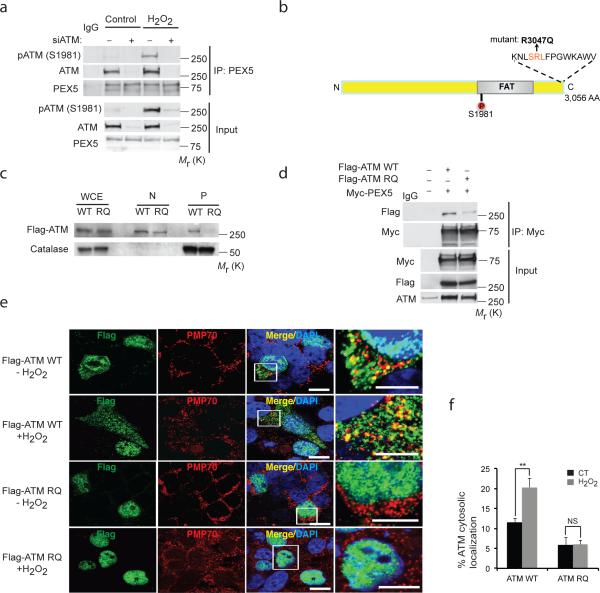
Peroxisomal proteins are targeted to this organelle by peroxisome import receptors, such as PEX528. ATM was co-immunoprecipitated with PEX5, and activated ATM (pATM) binding to PEX5 was increased by H2O2 (Fig. 2a). ATM has been reported to contain a putative PEX5 binding sequence (SRL) at its C-terminus23 (Fig. 2b). We introduced an arginine (R) to glutamine (Q) mutation into wild-type (WT) ATM at a.a. 3047 (R3047Q) (RQ-ATM) of the SRL (Fig. 2b). RQ-ATM localization to the peroxisome fraction of cells was greatly decreased (Fig. 2c), as was binding to PEX5 (Fig. 2d). Furthermore, while WT-ATM in the cytoplasm and punctate co-localization with the peroxisome membrane protein PMP70 increased in H2O2 treated cells, RQ-ATM remained primarily nuclear, and exhibited little co-localization with PMP70 (Fig. 2e,f).
Figure 2. PEX5 localizes ATM to the peroxisome.
(a) Immunoprecipitation with anti-PEX5 antibody in HepG2 cells transfected with control or ATM siRNA and treated with H2O2 (0.4 mM) for 3 h followed by imunoblotting for endogenous ATM. Inputs were immunoblotted using the indicated antibodies. (b) Schematic indicating the putative PEX5 binding region - SRL sequence (amino acid 3046, 3047 and 3048) at the C terminus of ATM. (c) Subcellular fractionation of HEK293 cells overexpressing Flag-ATM WT or Flag-ATM RQ (mutant). Immunoblotting was performed with indicated anti-Flag and catalase antibodies. WCE, whole cell extract; N, Nuclear; P, peroxisome fraction. (d) Immunoprecipitation was performed with anti-Myc antibody in HEK293 cells overexpressing Myc-PEX5 with Flag-ATM WT or Flag-ATM RQ and immunoblotted with Flag. Inputs were immunoblotted using the indicated antibodies. (e) Representative image of HEK293 cells overexpressing either Flag-ATM WT or Flag-ATM RQ and treated with or without H2O2 (0.4 mM) for 1 h and immunostained for Flag (ATM-green) and PMP70 (Peroxisome-red). Scale bar, 10 μm. High-magnification images of boxed areas are shown to the right (Scale bar, 5 μm). (f) Quantification of the percentage of ATM localized to the cytosol from Fig. 2e was performed on n=3 independent experiment, 100 (ATM transfected) cells were analyzed in each experiment. All error bars represent s.d., ** P < 0.01 (student's t test). Uncropped images of western blots are shown in Supplementary Fig. S9, Statistic source data for Fig. 2f can be found in Supplementary Table 1.
However, the intrinsic ability of this ATM mutant to be activated by ROS, and recognize DNA damage was not compromised. ATM is oxidized into an active dimer in response to H2O221. Both WT-ATM and the peroxisome localization-deficient RQ-ATM could be activated by H2O2 (Supplementary Fig. S1b) and in vitro kinase assays demonstrated that both WT-ATM and RQ-ATM were activated in the presence of the MRN complex and DNA (Supplementary Fig. S1c) and H2O2 (Supplementary Fig. S1d). To determine if peroxisome-localization deficient ATM could still respond to DNA damage, ATM-deficient cells (AT5, GM05849 and HepG2+ATMsiRNA) were reconstituted with either WT-ATM or RQ-ATM. ATM deficient cells (AT5, GM05849) formed a reduced number of γ-H2AX foci immediately after ionizing irradiation, and contained a higher number of residual foci post irradiation. This phenotype was rescued by RQ-ATM, which exhibited a γ-H2AX foci response equivalent to WT-ATM (Supplementary Fig. S1e, f); similar results were obtained using residual chromosome damage as an endpoint (Supplementary Fig. S1g, h). Therefore loss of peroxisomal localization did not affect the intrinsic kinase activity or ability of ATM to respond to DNA damage.
Peroxisomal ROS activates ATM to repress mTORC1 and induce autophagy
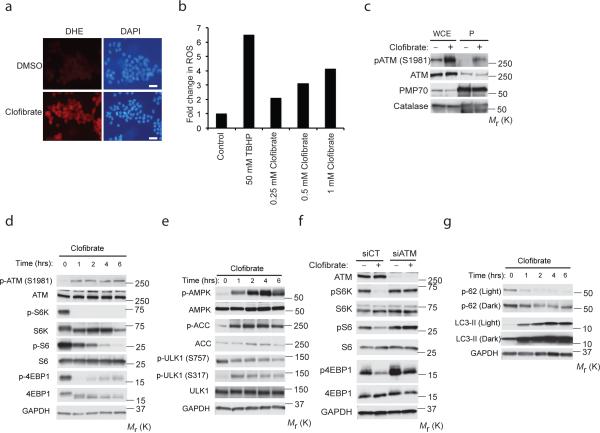
A role for ATM in peroxisome homeostasis would predict that this kinase would be activated by peroxisomal, as well as exogenous ROS. To test this prediction, we next asked if endogenous ROS produced at the peroxisome could activate ATM signaling. Rat liver FAO cells were treated with clofibrate, which activates PPAR (Supplementary Fig. S2a-c) and induces peroxisomal ROS, as shown by dihydroethidium (DHE) staining (Fig. 3a), and quantitation with DCFDA (Fig. 3b). Clofibrate also activated ATM at the peroxisome (Fig. 3c) and repressed mTORC1 signaling (Fig. 3d,e). Clofibrate also caused mTORC1 repression and induction of autophagy in GFP-LC3 MCF7 (Supplementary Fig. S2d, e) and HepG2 (Supplementary Fig. S2f) cells. Importantly, this response was abrogated in the absence of ATM (Fig. 3f) or TSC2 (TSC2−/− MEFs) (Supplementary Fig. S2g), demonstrating a requirement for both ATM and the TSC signaling node for mTORC1 repression by ROS, as previously shown25.
Figure 3. Peroxisomal ROS activates ATM to repress mTORC1 and induce autophagy.
(a) Representative images of FAO cells treated with vehicle (DMSO) or 0.25 mM clofibrate for 1 h, superoxide production was detected using dihydroethidium (DHE) staining. Scale bar, 30 μm. (b) Representative data of DCFDA assay from two independent experiments depicting the levels of ROS in FAO cells treated with clofibrate at the indicated doses for 1 h. Tert-butyl hydroperoxide (TBHP) 50 μM was used as a positive control for the assay. (c) ATM activation was monitored at the peroxisome in response to clofibrate at 3 h (1 mM) as indicated by western blot analysis of whole cell extract (WCE) and peroxisomal fractions (P) obtained from FAO cells. (d and e) FAO cells treated with 1 mM clofibrate for the indicated time points, activation of ATM-AMPK-TSC2 signaling and suppression of mTORC1 was monitored by western analysis for p-ATM (S1981), ATM, pS6K (T389), S6K, pS6 (S235/236), S6, p4E-BP1 (T37/46), 4E-BP1, pAMPK (T172), AMPK, pACC (S79), ACC, pULK1 (S757, mTORC1 site), pULK1 (S317, AMPK site), and ULK1. (f) Western analysis of HEK293 cells transfected with control or ATM siRNA and treated with 0.5 mM clofibrate for 6 h using anti-pS6K (T389), S6K, pS6 (S235/236), S6, p4E-BP1 (T37/46) and 4E-BP1 antibodies. (g) FAO cells treated with clofibrate (1mM) for indicated times. Induction of autophagy was monitored by western analysis of p62 and LC3-II. Uncropped images of western blots are shown in Supplementary Fig. S9, Source data for Fig. 3b can be found in Supplementary Table 1.
ULK1, a kinase essential for autophagy in mammalian cells, is activated by AMPK (phosphorylation at Ser317) and inactivated by mTORC1 (Ser757) phosphorylation29. Peroxisomal ROS increased S317 and decreased S757 phosphorylation (Fig. 3e), consistent with activation of AMPK and suppression of mTORC1, respectively, and induction of autophagy by peroxisomal ROS. Concomitantly, p62 decreased and LC3II increased (Fig. 3g), which was blocked by bafilomycin A1 (Supplementary Fig. S2h). Clofibrate also increased formation of autophagosomes visualized as GFP-LC3 puncta (Supplementary Fig. S2e) and by electron microscopy (Supplementary Fig. S3a-f). Together, these data demonstrate that peroxisomal ROS activates ATM signaling to repress mTORC1 and induce autophagy.
ATM phosphorylates PEX5 at Ser141 in response to ROS
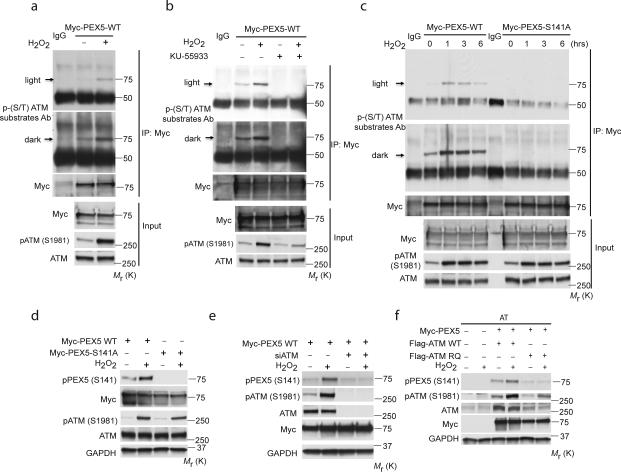
The minimal essential requirement for the substrates of ATM kinase is S*/T*Q, and hydrophobic amino acids at N-1 and N-3; negatively charged amino acids at N+1 are positive determinants for substrates phosphorylated by ATM30. Kinase prediction (scansite.mit.edu) identified a potential ATM S*/T*Q phosphorylation motif in PEX5 at Ser141 (Supplementary Fig. S4a,b). This highly conserved sequence in PEX5, similar to that of other ATM substrates, such as p53, matches this consensus ATM phosphorylation motif (Supplementary Fig. S4b). PEX5 was recognized by a pan-phospho-(S/T) ATM substrate antibody: immunoreactivity significantly increased with H2O2 (Fig. 4a), was blocked by the ATM inhibitor KU55933 (Fig. 4b), and lost in AT fibroblasts lacking ATM (Supplementary Fig. S4c), confirming the ATM-dependence of increased PEX5 immunoreactivity to this antibody in response to ROS.
Figure 4. ATM phosphorylates PEX5 at S141 in response to ROS.
(a) Immunoprecipitation of lysates from HEK293 cells overexpressing Myc-PEX5 and treated with H2O2 (0.4 mM) for 1 h, using an anti-Myc antibody followed by immunoblotting with a phospho-(S/T) ATM substrate antibody. Inputs were immunoblotted using the indicated antibodies. (b) Immunoprecipitation performed with anti-Myc antibody of lysates from HEK293 cells overexpressing Myc-PEX5, treated with H2O2 (0.4 mM) for 1 h in the presence/absence of an ATM inhibitor (KU-55933, 2 h pretreatment) followed by immunoblotting with phospho-(S/T) ATM substrate antibody. Inputs were immunoblotted using the indicated antibodies. (c) HEK293 cells transfected with either a Myc-PEX5-WT or Myc-PEX5-S141A mutant construct treated with H2O2 (0.4 mM) for the indicated times. Immunoprecipitation was performed with an anti-Myc antibody followed by immunoblotting with phospho-(S/T) ATM substrate antibody. Inputs were immunoblotted using the indicated antibodies. (d) HEK293 cells transfected with either a Myc-PEX5-WT or Myc-PEX5-S141A mutant construct treated with H2O2 (0.4 mM) for 1h. Western analysis was performed with anti- p-PEX5 (S141), Myc, p-ATM (S1981), ATM and GAPDH antibodies. (e) HEK293 cells transfected with Myc-PEX5-WT for 24 h following a siRNA knockdown of ATM for 48 h treated with H2O2 (0.4 mM) for 1h. Western analysis was performed with anti- p-PEX5 (S141) Myc, p-ATM (S1981), ATM and GAPDH antibodies. (f) AT (GM05849) fibroblast cells were transfected with Myc-Pex5 and Flag-ATM WT or Flag-ATM RQ mutant for 48 h and treated with H2O2 (0.4 mM) for 1h. Western analysis was performed with anti- p-PEX5 (S141) Myc, p-ATM (S1981), ATM and GAPDH antibodies. Uncropped images of western blots are shown in Supplementary Fig. S9.
Site-directed mutagenesis of serine 141 of PEX5 to alanine (S141A) revealed that while immunoreactivity with the pan-phospho-(S/T) ATM substrate antibody increased with H2O2 treatment in WT-PEX5, the S141A PEX5 mutant was not recognized by this antibody (Fig. 4c). A polyclonal antibody generated to the phospho-S141 PEX5 peptide confirmed that phosphorylation of PEX5 at S141 increased in response to ROS (Fig. 4d) and was dependent on ATM (Fig. 4e). While both WT-ATM and the RQ-ATM mutant retained kinase activity (see above), the RQ-ATM mutant failed to phosphorylate PEX5 at S141 (Fig. 4f). Together, these data demonstrate that PEX5 is phosphorylated on S141 in response to ROS, and this phosphorylation is ATM-dependent.
Ser141 regulates PEX5 ubiquitination at K209 in response to ROS
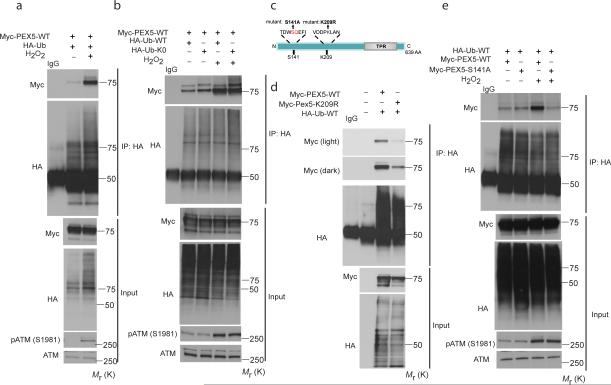
Ubiquitin on the peroxisome membrane surface is thought to target peroxisomes for degradation20, and membrane-bound PEX5 can be ubiquitinated31-35, although the regulation and specific site(s) of ubiquitination in peroxisomes targeted for pexophagy have not been elucidated. We observed dramatically increased PEX5 ubiquitination when ATM was activated by ROS (Fig. 5a). Because PEX5 is known to be both mono-and poly-ubiquitinated31-35, we examined PEX5 ubiquitination in response to ROS using HA-Ub-WT and HA-Ub-K0 (ubiquitin with all lysine residues mutated to arginine to prevent chain formation and poly-ubiquitination) co-expressed with Myc-PEX5. No decrease in the apparent molecular weight, nor amount of PEX5 ubiquitination in response to ROS using the HA-Ub-K0 construct (that can only be monoubiquitinated), compared to the HA-Ub-WT construct (Fig. 5b) was observed, indicating PEX5 was monoubiquitinated in response to ROS.
Figure 5. S141 regulates PEX5 ubiquitination at K209 in response to ROS.
(a) HEK293 cells expressing Myc-PEX5-WT and HA-Ub treated with H2O2 (0.4 mM) for 6 h, were co-immunoprecipitated using anti-HA and blotted using anti-Myc antibody. The inputs were immunoblotted using the indicated antibodies. (b) HEK293 cells expressing Myc-PEX5-WT and HAUb-WT or HA-Ub-K0 (ubiquitin with all lysine residues mutated to arginine) constructs were treated with H2O2 (0.4 mM) for 6 h, and Myc-PEX5 co-immunoprecipitated using anti-HA and blotted using anti-Myc antibody. Inputs were immunoblotted using the indicated antibodies. (c) Schematic representation of the N-terminus of PEX5 showing the putative ATM phosphorylation site (S141) and ubiquitination site (K209). (d) HEK293 cells expressing HA-Ub-WT and Myc-PEX5-WT or Myc-PEX5-K209R plasmids were treated with H2O2 (0.4 mM) for 6 h, and Myc-PEX5 coimmunoprecipitated using anti-HA antibody, and blotted using anti-Myc antibody. Input lysates were immunoblotted using the indicated antibodies. (e) HEK293 cells expressing HA-Ub-WT and Myc-PEX5-WT or Myc-PEX5-S141A constructs were treated with H2O2 (0.4 mM) for 6 h, and Myc-PEX5 co-immunoprecipitated using anti-HA antibody and blotted using anti-Myc antibody. Input lysates were immunoblotted using the indicated antibodies. Uncropped images of western blots are shown in Supplementary Fig. S9.
Prediction of ubiquitination sites on PEX5 using PhosphoSite (www.phosphosite.org) and a series of constructs generated by site-directed mutagenesis were used to identify lysine 209 as a major site for PEX5 ubiquitination (Fig. 5c, 5d and Supplementary Fig. S5a). A search of mass spectrometry databases (www.phosphosite.org) revealed that K209 of PEX5 had been identified by mass spectrometry as being ubiquitinated36, confirming K209 as a bona fide site for PEX5 ubiquitination.
Phosphorylation can modulate protein ubiquitination by regulating E3 ligase recognition or subcellular localization of substrates37, leading us to ask whether phosphorylation of PEX5 by ATM regulated PEX5 ubiquitination in response to ROS. Mutation of the S141 ATM phosphorylation site in PEX5 abrogated increased ubiquitination of PEX5 in response to ROS (Fig. 5e), indicating that phosphorylation at this site regulated ROS-induced PEX5 ubiquitination. In agreement with these data, ROS-induced ubiquitination of endogenous PEX5 was deficient in ATM-null cells (Supplementary Fig. S5b) and following ATM knock down with siRNA (Supplementary Fig. S5c). Low background PEX5 ubiquitination was seen with both WT and S141A PEX5 (Fig. 5e), suggesting there may be other sites of ubiquitination that are not dependent on ATM phosphorylation at S141, nor ROS-responsive.
The RING peroxins PEX2, PEX10 and PEX12 are part of a peroxisome-localized E3 ligase responsible for polyubiquitination of PEX534, and as expected, siRNA knockdown of these peroxins reduced polyubiquitination of PEX5 (Supplementary Fig. S5d). Knockdown of these E3 ligases also reduced monoubiquitination of PEX5, suggesting this peroxisomal E3 ligase also participates in PEX5 monoubiquitination (Supplementary Fig. S5e). Collectively, these data indicate that PEX5 is ubiquitinated in response to ROS, and that this ubiquitination is regulated by ATM phosphorylation.
Ubiquitinated PEX5 binds to p62 to target peroxisomes for pexophagy
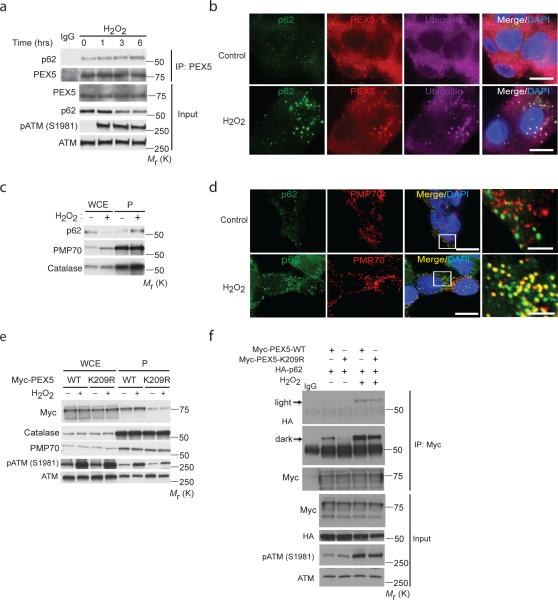
We next asked whether the autophagy adaptor p62, which contains both LIR (autophagosome) and UBA (ubiquitin) binding domains, recognizes ubiquitinated PEX5 to mediate pexophagy. The binding of endogenous PEX5 to p62 was enhanced by ROS (Fig. 6a), and although the majority of the PEX5 import receptor is cytosolic38, co-localization of PEX5 in peroxisomes with p62 and ubiquitin was increased by H2O2 (Fig. 6b). Whereas total p62 decreased in response to H2O2, p62 association with peroxisomes increased dramatically in response to ROS (Fig. 6c). p62 binding to peroxisomes was confirmed by co-localization of p62 with the peroxisome marker PMP70 in liver cells (Fig. 6d). Moreover, co-localization of PMP70 with FIP200 (ULK1 complex component) or ATG16L1 (ATG5 complex component), both essential for autophagosome formation39, 40, significantly increased with H2O2 (Supplementary Fig. S6a).
Figure 6. Ubiquitinated PEX5 binds with autophagy adaptor protein p62 in response to ROS.
(a) HepG2 cells treated with H2O2 (0.4 mM) for the indicated time points were immunoprecipitated with anti-PEX5 and immunoblotted with anti-p62 antibodies. Inputs were immunoblotted using the indicated antibodies. (b) Representative image of HepG2 cells treated with H2O2 (0.4 mM) for 3h and immunostained with p62 (green) , PEX5 (red) and ubiquitin (purple). Scale bar, 10 μm. (c) Western analysis of whole cell extracts (WCE) and peroxisome fractions (P) of HepG2 cells treated with 0.4 mM of H2O2 for 1 h immunoblotted using anti-p62, anti-PMP70 and catalase antibodies. (d) Representative images of FAO cells treated with 0.4 mM of H2O2 for 1 h and immunostained with p62 (green) and PMP70 (peroxisomes-red). Scale bar, 10 μm. High-magnification images of boxed areas are shown to the right (Scale bar, 2.5 μm). (e) Subcellular fractionation of HEK293 cells overexpressing Myc-PEX5 WT or Myc-PEX5 K209R (mutant) treated with 0.4 mM of H2O2 for 3 h. Immunoblotting was performed with indicated anti-Myc, catalase, PMP70, phospho-ATM and ATM antibodies. WCE, whole cell extract; P, peroxisome fraction. (f) HEK293 cells transfected with HA-p62 and Myc-PEX5-WT or Myc-PEX5-K209R, and treated with 0.4 mM of H2O2 for 6h. Lysates were immunoprecipitated with anti-Myc and immunoblotted with anti-HA antibodies. Inputs were immunoblotted using the indicated antibodies. Uncropped images of western blots are shown in Supplementary Fig. S9.
Ubiquitination of PEX5 to tether p62 to the peroxisome was further supported by decreased K209R PEX5 in the peroxisome fraction (Fig. 6e) and decreased p62 binding to K209R versus WT-PEX5 (Fig. 6f). Similarly, WT-PEX5 binding to p62 was increased by ROS (Supplementary Fig. S6b) and decreased with the K209R PEX5 mutant in HEK293 cells (Supplementary Fig. S6c). However, while the K209R mutation decreased basal PEX5 binding to p62, and attenuated the increase in p62 binding to PEX5 observed in response to ROS (Fig. 6f), some p62 binding to K209R-PEX5 could be detected, albeit at much lower levels, suggesting that other sites of ubiquitination on PEX5 may also be recognized by this autophagy adaptor, perhaps in response to other stimuli. Together, these data show that the autophagy adaptor protein p62 binds to PEX5, and that ubiquitination at K209 by ROS increases this interaction.
Induction of pexophagy by ROS
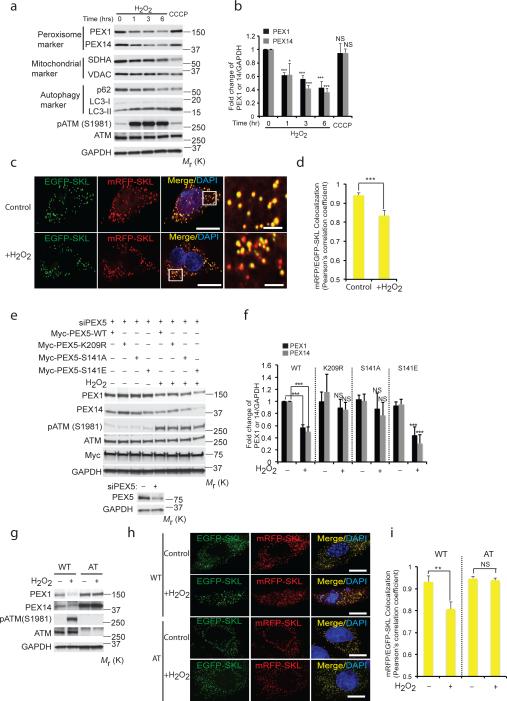
Pexophagy is a naturally occurring process for regulation of peroxisome number, which can also lead to selective destruction of peroxisomes in response to metabolic signals15, 18. In cells stably co-expressing GFP-LC3 (an autophagosome marker) and DsRed-SKL (a peroxisome marker), autophagosomes were visualized to sequentially tether and engulf peroxisomes in response to ROS (Supplementary Fig. S7a and Supplementary Video 1), with a concomitant decrease in peroxisomal proteins PEX1 and PEX14 (Fig. 7a,b). Bafilomycin A1 blocked this decrease, and dramatically increased p-PEX5 (S141) in the peroxisome fraction, confirming ROS had increased autophagic flux/pexophagy (Supplementary Fig. S7 b-d). Interestingly and in contrast, little mitophagy was seen in response to ROS (Fig. 7a,b). ROS-induced pexophagy was also demonstrated by using peroxisome-localized EGFP-SKL and mRFPSKL41. In response to ROS, co-localization of the relatively stable mRFP signal with the more labile EGFP signal decreased as peroxisomes engulfed by autophagosomes fused with lysosomes in cells undergoing pexophagy (Fig. 7c,d and Supplementary Video 2). Increased pexophagy in response to ROS was further confirmed by electron microscopy (Supplementary Fig. S3a-f and S8a-d, respectively); no decrease of PEX1 and PEX14 protein levels was observed in response to ROS in p62 knockdown cells (Supplementary Fig. S7e,f).
Figure 7. Induction of pexophagy by ROS.
(a) HepG2 cells treated with either H2O2 (0.4 mM) for indicated times or CCCP (50 μM, 6 h). Western analysis of peroxisome proteins PEX1 and PEX14 (SDHA and VDAC as mitochondrial markers, p62 and LC3-II as autophagy markers). (b) Quantification of PEX1 and PEX14 intensity normalized to GAPDH from Fig. 7a. (mean ± s.d., n = 3 independent experiments, student's t test). * P < 0.05 and *** P < 0.001, NS= not significant. (c) Representative images using HepG2 cells transfected with an mRFP-EGFP-SKL construct and treated with H2O2 for 6h. Scale bar, 10 μm. (d) Pearson's correlation coefficient for colocalization between mRFP-SKL and EGFP-SKL was calculated from Fig. 7c. n=4 independent experiment, 10 cells were analyzed in each experiments. All error bars represent s.d., (student's t test) *** P < 0.001. (e) HEK293 cells transfected with Myc-PEX5-WT or Myc-PEX5-K209R or Myc-PEX5-S141 for 24 h following prior siRNA knockdown of PEX5 for 48 h, treated with H2O2 (0.4 mM) for 6 h. Western analysis was performed using anti- PEX1, PEX14, phosphor-ATM (S1981), ATM, MYC and GAPDH antibodies. Corresponding immunoblots for HEK293 cells transfected with control or siRNA PEX5 showing levels of PEX5. (f) Quantification of PEX1 and PEX14 intensity normalized to GAPDH from Fig. 7e. (mean ± s.d., n = 4 independent experiments, student's t test). *** P < 0.001, NS= not significant. (g) WT (GM08399) and AT (GM05849) fibroblasts were treated with H2O2 (0.4 mM) for 6 h and immunoblotted with PEX1, PEX14, phospho-ATM, ATM and GAPDH antibodies. (h) Representative images using WT (GM08399) and AT (GM05849) fibroblasts transfected with a mRFP-EGFP-SKL construct and treated with H2O2 for 6h. Scale bar, 15 μm. (i) Pearson's correlation coefficient for colocalization between mRFP-SKL and EGFP-SKL was calculated from Fig. 7h. Quantification was performed from Fig S6a on n=3 independent experiments, 10 cells were analyzed in each experiment. All error bars represent s.d., (student's t test) ** P < 0.01, NS= not significant. Uncropped images of western blots are shown in Supplementary Fig. S9, Statistic source data for Fig. 7b,d,f and i can be found in Supplementary Table 1.
A requirement for ATM signaling to induce pexophagy and maintain peroxisome homeostasis would predict that cells that were ATM- or TSC2-deficient (downstream effector for suppression of mTORC1) would have elevated levels of ROS due to an inability to maintain peroxisome homeostasis. We and others have found that both ATM-deficient human lymphoblasts from patients with A-T disease and TSC2-deficient MEFs have elevated levels of ROS relative to wild-type cells3. Following knockdown of PEX5, reconstitution of cells with either WT-PEX5, S141A (ATM-phosphorylation deficient), S141E (phosphomimetic) or K209R (ROS-induced ubiquitination-deficient) PEX5 mutants suggested pexophagy (decreased PEX1 and PEX14) was attenuated in cells expressing S141 or K209R PEX5 mutants, but not the S141E phosphomimetic PEX5 mutant (Fig. 7e,f). However, the S141E mutation did not induce pexophagy, indicating phosphorylation (and ubiquitination) was not sufficient to induce pexophagy (Fig. 7e,f). Pexophagy (decreased PEX14-positive puncta, PEX1 and PEX14 protein level, and co-localization of EGFP and mRFP) was observed in response to ROS in ATM-proficient cells, but not ATM-deficient AT cells (Fig. 7g, h, i and Supplementary Fig. S7g,h). Similarly, ROS-induced pexophagy was observed in ATM-deficient cells reconstituted with WT-ATM, but not kinase-dead ATM (Supplementary Fig. S7i) and was abrogated by knockdown of the PEX2/10/12 E-3 ligase (Supplementary Fig. S7j), confirming the key-role played by ATM in signaling pexophagy in response to ROS.
DISCUSSION
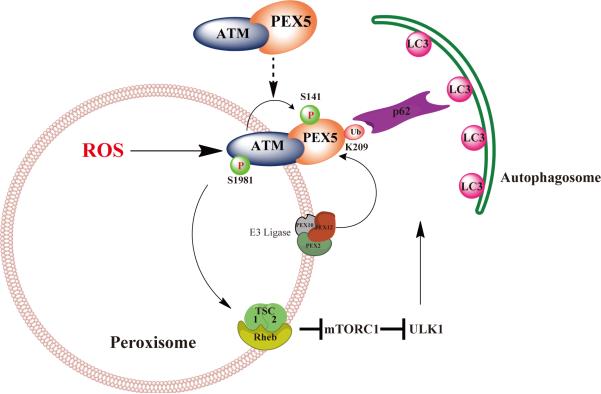
ATM-mediated suppression of mTORC1 and phosphorylation of PEX5 by ROS provides an attractive mechanism where by ROS acts as a rheostat to induce pexophagy to prevent oxidative damage to the cell from excessive or defective peroxisomes. As shown in the model in Figure 8, we demonstrated that in addition to suppressing the autophagy inhibitor mTORC13, the ATM kinase triggers pexophagy by inducing PEX5 phosphorylation and subsequent ubiquitination, resulting in p62 binding and targeting of peroxisomes for degradation.
Figure 8.
Working model for peroxisomal ATM signaling to the TSC signaling node to repress mTORC1, phosphorylation of PEX5 to induce ubiquitination by PEX2/10/12 E3 ligase, and recognition of Ub-PEX5 by p62 to induce pexophagy in response to ROS.
Selective autophagy has been observed for many cellular organelles. During selective autophagy, autophagy receptors play a key role in targeting of the autophagosome to organelles. In mammals, the most prevalent autophagy-targeting signal is the modification of cargo by ubiquitination42. For example, selective autophagy of damaged mitochondria (mitophagy) occurs after the mitochondrial kinase PINK1 phosphorylates the mitochondrial outer membrane protein mitofusin 2, inducing its ubiquitination by recruiting the E3 ligase parkin43, 44, or directly phosphorylating the E3 ligase parkin to trigger mitophagy45. However, for other organelles, including the peroxisome, the molecular mechanism(s) underlying cargo recognition and trafficking during selective autophagy has been lacking42, 46.
We show that one mechanism for inducing selective autophagy of peroxisomes is activation of ATM, phosphorylation and ubiquitination of PEX5, and binding of the autophagy adapter p62. Interestingly, in P. pastoris, PEX5 has been shown to be a redox regulated protein, where H2O2 decreases import of PTS1 proteins into peroxisomes47, 48. PEX14 has also been reported to bind LC3-II under conditions of amino acid starvation49 and overexpression of peroxisomal membrane protein PMP34, fused with an ubiquitin on the cytosolic face of peroxisomes, is sufficient to trigger turnover of peroxisomes20. Whether these peroxisomal proteins are also targets of the ATM kinase, or regulated by other, yet to be identified mechanisms, is unknown. There is also evidence that in addition to p62, the autophagy adapter NBR1 can also participate in mammalian pexophagy20, 50, suggesting other pathways in addition to p62 binding to PEX5 for selectively targeting peroxisomes for autophagy may also exist.
PEX5 as a target for the ATM kinase is particularly attractive. PEX5 is known to be ubiquitinated after docking at the peroxisome membrane31-33, 35, becoming either polyubiquitinated and targeted for proteosome-mediated degradation, or monoubiquitinated for recycling back to the cytosol31, 34, 35. Our data reveal novel role for PEX5 as a target of the ATM kinase, which when phosphorylated at S141, becomes ubiquitinated at K209 and serves as a target for the autophagy adaptor p62, providing yet another role (pexophagy) for ubiquitination of PEX5 at the peroxisome.
Our data show that ATM signaling at the peroxisome participates in pexophagy via two pathways. The first is activation of AMPK and TSC2, leading to repression of mTORC1. mTORC1 is a well-known inhibitor of authophagy, and relief of this repression via AMPK activation and phosphorylation of ULK1 at S317 would increase autophagic flux. The second is phosphorylation of PEX5, triggering ubiquitination of this peroxisomal protein, and binding of the autophagy adapter protein, p62, targeting peroxisomes for pexophagy. Data that the phosphomimetic S141E PEX5 mutation alone was unable to induce pexophagy in the absence of ATM activation by ROS suggests both mTORC1 repression and PEX5 phosphorylation are important, and phosphorylation (and ubiquitination) of PEX5 may be necessary, but not sufficient, to induce pexophagy.
To date, studies on the role of cell signaling in peroxisome homeostasis have primarily focused on the role of cell signaling pathways in peroxisome biogenesis via regulation of transcription of genes required for peroxisome biogenesis18. For example, drugs such as hypolipidemic fibrates that act as PPARα ligands, transcriptionally up-regulate genes that promote peroxisome biogenesis. Importantly, in response to PPARα activation, genes for peroxisome-localized metabolic processes that generate ROS are disproportionately up-regulated relative to those for ROS scavengers, resulting in increased ROS generation. The resultant oxidative stress is thought to contribute to the hepatocarcinogenecity of PPARα ligands in rodents51. Reactive intermediates generated at the peroxisome include free radicals such as superoxide and H2O2, and reactive nitrogen species (RNS). Many free radical scavengers, including catalase and superoxide dismutase (SOD), are specifically targeted to the peroxisome to protect the cell from peroxisomal ROS and RNS. Interestingly, we have recently shown that ATM can also be activated by RNS to repress mTORC1 to induce autophagy52. This suggests the interesting possibility that ATM may also be activated by RNS produced by peroxisomes, and that both ROS and RNS could act as rheostats for cellular sensing of excessive or aberrantly functioning peroxisomes and induction of pexophagy to maintain homeostasis.
Because ROS can be produced by other organelles, it will be interesting to determine if ROS produced at other sites activates ATM and induces pexophagy, or if mechanisms exist to prevent peroxisomes from being targeted for pexophagy in response to ROS produced elsewhere in the cell. There are several possible mechanisms by which cells could regulate pexophagy in response to ROS to provide organelle-specificity. For example, when oxidized by ROS, ATM forms an active dimer21. We do not know at this time if PEX5 recognizes and binds ATM as a monomer or a dimer. Whether ROS produced by other organelles can lead to pexophagy and/or ATM-mediated phosphorylation/ubiquitination of PEX5 at the sites we identified (S141 and K209) is also not known. While S141 appears to be necessary for ROS-induced ubiquitination, we do not know if there are other sites/modifications that occur on PEX5 (or other peroxisomal proteins) that contribute to specificity, modifications that could occur only at peroxisomes, or specifically in response to peroxisomal ROS/RNS. These interesting hypotheses, now await further testing.
While ATM's role in the DNA damage response in the nucleus is well known, cytoplasmic functions for ATM are now emerging. Interestingly, an ATM R3047X mutation with a truncated C-terminus lacking the last 10 amino acids including SRL sequence has been identified in several A-T patients21, 53, and in contrast to the RQ-ATM, this mutant cannot be activated by ROS21. Together with the data presented here, a picture emerges of ROS activation and localization to peroxisomes, as essential characteristics of ATM. ATM plays a role in preventing lysosome accumulation, and ATM−/− mice exhibit an increase in lysosome numbers54. The ATM kinase also been reported to localize to mitochondria55. At the mitochondria, ATM is activated by mitochondrial dysfunction, with loss of ATM resulting in increased mitochondrial content, ROS and oxygen consumption, suggesting that the ATM kinase plays an important role in maintaining mitochondrial homeostasis55 as well as peroxisome homeostasis. As a result, a picture is rapidly emerging of ATM as an important component of the oxidative stress response in the cytoplasm3, 25 as well as DNA repair in the nucleus56, with important roles protecting the cell from both DNA and oxidative damage.
Methods
Antibodies
Antibodies against phospho-S6 (S235/236; #2211, 1:4,000 western blotting (WB)), S6 (#2217, 1:4,000 WB), phospho-4E-BP1 (T37/46; #2855, 1:2,000 WB), 4E-BP1 (#9644, 1:2,000 WB), phospho-p70-S6K (T389; #9205, 1:500 WB), p70-S6K (#9202, 1:500 WB), phospho-AMPK (T172; #2531, 1:500 WB), AMPK (#2532, 1:500 WB), phospho-ULK1 (S757; #6888 1:1000 WB), phospho-ULK1 (S317; #6887 1:1000 WB), ULK1 (#8054 1:1000 WB), lamin A/C (#2032, 1:1,000 WB), VDAC (#4866, 1:1,000 WB), LC3B (#2775, 1:1,000 WB), HA (#3724, 1:2000 WB); TSC2 (#4308, 1:2000 WB); phospho-(S/T) ATM/ATR substrates (#2851,1:1000 WB); phospho-ACC (#3661L, 1:1000 WB); ACC (#3662S, 1:1000 WB); were purchased from Cell Signaling Technology. phospho-ATM S1981(ab81292, 1:1,000 WB, 1:200 Immunofluorescence (IF)); Catalase (ab1877, 1:5,000 WB, 1:500 IF); ACAA1 (ab84635, 1:2,000 WB), HA (ab9110, 0.5μg per 100μg of total protein for immunoprecipitation (IP)), Catalase (ab1877, 1:500 IF) and PEX14 (ab109999, 1:1,000 WB) were purchased from Abcam. c-Myc (9E10) (sc-40, 1:2,000 WB, 2μg per 100μg of total protein for IP); EHHADH (sc-99388, 1:2,000 WB); p62 (sc-28359, 1:1,000 WB); PEX1 (SC-21957, 1:500 WB); GAPDH (SC-25778, 1:5000 WB); ATM (SC-23921 1:1000 WB) and SDHA (SC-27992, 1:500 WB) were purchased from Santa Cruz Biotechnology. Other antibodies used in this study are as follows: phospho-PEX5 (S141) was generated by Covance; ATG16L (AP1817b, 1:50 for IF) was purchased from ABGENE; FIP200 (10043-2-AP) was purchased from Proteintech; Flag (Sigma F3165, 1:4,000 WB, 1:500 IF, 1.25 μg per 100 μg of total protein for IP); PMP70 (Sigma SAB4200181, 1:5,000 WB, 1:500 IF); ATM (GeneTex, GTX70103, 1:1,000); LDH (Chemicon AB1222, 1:2,000 WB); PEX5 (ProteinTech #12545-1-AP, 1:1,000 WB, 1:200 IF); β-integrin (BD Transduction Laboratories, #610468, 1:2,000 WB), Ubiquitin (Sigma, U5379, 1:100 IF); PEX5 (Santa Cruz, SC-23188 1:300 IF); p62 (ARP 03-GP62-C, 1:100). Secondary antibodies (1:2,000) conjugated to horseradish peroxidase were purchased from Santa Cruz Biotechnology. Secondary antibodies for immunofluorescence staining, anti-mouse, -goat and -rabbit Alexa fluor 488, 546 and 633 were obtained from molecular probes (1:1,000). Reagents: Hydrogen peroxide solution (323381-25ML); Clofibrate (C6643-1G), Bafilomycin A1 were purchased from Sigma and Ku55955 was purchased form Tocris Bioscience.
Cell culture and Transfection
Control (GM15871) and Zellweger (GM13267) cells, wild type (WT) human fibroblast (GM08399) and ataxia telangiectasia (AT) patients fibroblast (GM05849) cells were obtained from Coriell Cell Repositories and grown in MEM supplemented with 15% fetal bovine serum (FBS). TSC2+/+ and TSC2−/− MEFs (kind gifts from Dr. Kwiatkowski (Harvard Medical School, USA)), and HEK 293 (ATCC) cells were cultured in DMEM supplemented with 10% FBS. HepG2 (ATCC) cells were cultured in MEM supplemented with 10% FBS. FAO (Sigma) cells were cultured in F12 media supplemented with 10% FBS and 2mM L-glutamine (Life technologies). MCF7 cells were grown in modified IMEM media supplemented with 10% FBS. MCF7-GFP-LC3 were provided by Dr. Gordon Mills (U.T.M.D. Anderson Cancer Center, USA) and grown in RPMI 1640 media supplemented with 10% FBS. RPE cells stably expressing GFP-LC3 and RFP-SKL were grown in DMEM media supplemented with 10% FBS. All media and FBS were obtained from Life Technologies and Sigma respectively. Transient transfections were performed using the Lipofectamine 2000 reagent (Invitrogen) according to manufacturer's instructions. All the cell lines were authenticated by STR profiling using the Charaterized Cell Line Core facility at the U.T. M.D. Anderson Cancer Center. All the cell lines were also tested and confirmed negative for mycoplasma.
Plasmids and mutagenesis
hPEX5 is carried out by forward primer (5’-CGGTCGACCATGGCAATGCGGGAG-3’) and reverse primer (5’-GTCGACTCACTGGGGCAGGCCAAAC-3’) which include SalI restriction sites, to get the full-length hPex5 and ESTs clone as template (Expressed-sequence tags, GE Healthcare). hPEX5 PCR product cloned into pJET1.2/blunt cloning vector to amplify full-length hPex5. hPex5 is cut out by SalI from pJET1.2/blunt cloning vector insert it into pCMV-Myc expression vector. A series of mutant constructs of PEX5 were generated by QuikChange Lightning Multi Site-Directed Mutagenesis Kit. The primers for different sites were designed as follow.
| K28R | (Fw 5’-CCCAGGACAGGGCCCTTCG-3’; |
| Rv 5’-CGAAGGGCCCTGTCCTGGG-3’), | |
| K52R | (Fw 5’-GGCAGCCTCCAGGCCTTTGGGAG-3’; |
| Rv5’-CTCCCAAAGGCCTGGAGGCTGCC-3’), | |
| K170R | (Fw5’-CAATCAGAGGAGAGGCTGTGGCTGGGAG-3’; |
| Rv5’-CTCCCAGCCACAGCCTCTCCTCTGATTG-3’), | |
| K204R | (Fw 5’-CTTTGTGGCCAGAGTGGATGAC-3’; |
| Rv5’-GTCATCCACTCTGGCCACAAAG-3’), | |
| K209R | (Fw5’-GATGACCCCAGATTGGCTAATTC-3’; |
| Rv 5’-GAATTAGCCAATCTGGGGTCATC-3’), | |
| K292R | (Fw5’-GAGGAGATGGCAAGACGGGATGCTGAG-3’; |
| Rv5’-CTCAGCATCCCGTCTTGCCATCTCCTC-3’), | |
| S141A | (Fw5’-GAGACTGACTGGGCCCAAGAATTCATC-3’; |
| Rv5’-GATGAATTCTTGGGCCCAGTCAGTCTC-3’). | |
| S141E | (Fw5’-GAGACTGACTGGGAGCAAGAATTCATC-3’; |
| Rv5’-GATGAATTCTTGCTCCCAGTCAGTCTC-3’). |
Flag-ATM-WT (#31985) and HA-p62 (#28027) plasmids were purchased from Addgene. Flag-ATM R3047Q mutant constructs was generated by QuikChange Lightning Multi Site-Directed mutagenesis Kit (Agilent Technologies).
| R3047Q: FW 5’-GACCCCAAAAATCTCAGCCAACTTTTCCCAGGATGGAAAGC-3’; |
| RV 5’-GCTTTCCATCCTGGGAAAAGTTGGCTGAGATTTTTGGGGTC-3’). |
HA-Ub (#18712) and HA-Ub-K0 (#17603) plasmids were purchased from Addgene. DsRed-PTS1 was provided by Dr. Michael Mancini (Baylor College of Medicine, USA). pAT-003 mRFP-EGFP-SKL was provided by Dr. Suresh Subramani (University of California, San Diego, USA). Specificity of mutagenesis was confirmed by direct sequencing using the Sequencing and Microarray Core at U.T. M.D. Anderson Cancer Center, Houston, Texas.
RNAi knockdown
On-TARGETplus ATM SiRNA (L-003201-00-0005); Si GENOME human PEX2, PEX10 and PEX12 SiRNA (M-006548-02, M-006545-00, and M-019337-02); si SMARTpool human PEX5 (M-015788-00); si SMARTpool human SQSTM1 (M-010230-00); were purchased from Dharmacon. siRNAs were resuspended in 1X siRNA buffer (GE Dharmacon) to obtain a 20 μM stock, aliquots were then frozen to prevent repeated freeze thawing. HEK 293 cells were transfected with indicated Si RNA at 10 nM final concentrations with DharmaFECT 1 or DharmaFECT 4 (GE Dharmacon) according to manufacturer's instruction, for 72 h before collection of lysates.
Cellular fractionation
Cytosolic, nuclear and membrane fractions were obtained as describe previously (ref 57). Peroxisomes were isolated using Peroxisome Isolation Kit (Sigma) as per manufacturer's instructions. Briefly, cells (~2×108) were homogenized in 1X peroxisome extraction buffer, and centrifuged at 1,000g for 10 minutes. The supernatant was centrifuged at 2,000g for 10 minutes, and collected and furthered centrifuged the sample at 25,000g for 20 minutes. The pellet re-suspended in 400μl 1X peroxisome extraction buffer, and subjected to density gradient centrifugation at 100,000 for 90 minutes. The peroxisome-enriched fraction was collected from the bottom layer of the density gradient.
Immunoprecipitation and western blotting assay
The cells were washed in PBS 3 times and lysed directly using CST lysis buffer (20mM Tris-HCl (pH 7.5), 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100, 2.5mM Na pyrophosphate, 1mM β-glyceropphosphate for 30 minutes at 4°C. The lysis buffer contained 1X protease inhibitor cocktail and phosphatase inhibitor cocktail 2 and 3 (Sigma). Lysates were microcentrifuged at 4°C at max speed (13.2rpm) for 10 minutes. The supernatant was subjected to BCA Protein Assay (Thermo Scientific) to quantify protein levels. For immunoprecipitation, the cell lysates were incubated with the indicated antibodies and Magnetic A/G beads (Thermo Scientific) overnight at 4 °C. The beads were pelleted and washed with lysis buffer 5 times and were heated in 1X denaturing loading buffer for 10 minutes at 95 °C before resolving by SDS-PAGE. The cell lysates were separated on a 4-15% gel (Bio-Rad), transferred to PVDF membranes and probed with antibodies. Densitometric analysis for quantification of expression levels was performed using ImageQuantTL software and data were normalized with GAPDH expression. Student's t-test (two tailed) was performed on at least three biological repeats using GraphPad Prism software. Error bars represent standard deviations of normalized fold changes.
Protease protection assay
The crude peroxisomal fraction was isolated using Peroxisome Isolation Kit (Sigma). The fractionation sample was separated into two groups: Group 1, Proteinase K (Roche) 0.1μg/ml; Group 2, Proteinase K 0.1μg/ml and 1% Triton X-100. Both Groups were incubated on ice for 5, 15 and 30 minutes respectively. PMSF was used to stop the reaction and samples processed by western blot assay.
γ-H2AX foci analysis
Cells were cultured in chamber slides followed by fixation, and immunostaining for detection of phosphorylated H2AX as previously described (refs 58-60). Fluorescent images of foci from 100 cells for each experiment were captured as described previously (ref 59-60). Nuclear sections were captured, and images obtained by projection of the individual sections as described previously (ref 58).
Chromosome aberrations analysis
Ionizing radiation (IR)-induced chromosomal aberrations were analyzed at metaphase. Cells in exponential phase were irradiated with 3 Gy, incubated for 9 h post irradiation, treated with colcemid for 3 hr then fixed stained with Giemsa. Metaphase chromosomes were analyzed as described previously (refs 58, 61). Categories of asymmetric chromosome and chromatid aberrations scored included dicentrics, centric rings, interstitial deletions-acentric rings, terminal deletions, breaks, gaps and exchanges. For each experiment fifty metaphases were analyzed and each experiment was repeated three times.
ATM kinase assay
Kinase assays were performed in kinase buffer (50 mM HEPES, pH 7.5, 50 mM potassium chloride, 5 mM magnesium chloride, 10% glycerol, 1 mM ATP, and 1 mM DTT) for 90 min at 30 °C in a volume of 40 μl. Kinase assays with oxidation were performed in the absence of DTT with 817 μM H2O2, 0.4 nM or 0.8 nM ATM, 100 nM GST-p53 substrate.
Electron microscopy
The cells were plated on chamber slides for Electron Microscopy, and treated with vehicle or Clofibrate or H2O2. The samples were fixed using 2% glutaraldehyde in 100 mM sodium cacodylate with 2mM CaCl2 at room temperature. Images were obtained at the High Resolution Electron Microscopy Facility at U.T. M.D. Anderson Cancer Center and Baylor College of Medicine.
Immunofluorescence microscopy
Cells were plated on coverslips and maintained at 37 °C and 5% CO2 for 24 hours before staining. Cells were washed with 1×phosphate-buffered saline (PBS three times and fixed in 4% paraformaldehyde for 15 minutes, permeabilized in 0.5% Triton X-100 for 10 minutes, blocked with 3.75% BSA in PBS for 1 h at room temperature, and incubated with primary antibody overnight at 4 °C. Secondary antibodies were applied for 1 h at 37 °C, stained with DAPI for 2 minutes and mounted using SlowFade® Gold Antifade reagent (Life Technologies). Images were captured using either a Deltavision Deconvolution Microscope (DeltaVision Elite,GE) or a Nikon confocal system. Live cell imaging was performed using Deltavision Deconvolution Microscope-equipped with sCMOS camera, and a temperature controlled CO2 incubation chamber. Images were acquired with a 60X/1.42 oil objective (Olympus). SoftWoRx software was used for acquisition of image stacks, time-lapse and deconvolution. For time-lapse, the cells were plated on glass bottom microwell dishes (MatTek Corporation) for 24 hours ahead of time and then immediately treated with H2O2 before image acquisition on the stage. . The images were acquired every 3 minutes with Z-stacks at 37 °C and 5% CO2. The video of stacked images was acquired every 3 minutes. Images were quantified using ImageJ software. For co-localization analysis, Pearson's Correlation Coefficient was calculated using Imaris software V.7.6.1 (Bitplane AG). The numbers of PEX14-positive vesicles were calculated using ImageJ. At least 100 cells per condition in 4 independent experiments were used for quantification.
ROS Measurement by DCFDA assay and Dihydroethidium (DHE) staining
FAO cells were plated in 96 well plates (black bottom) for 24h and maintained at 37 °C and 5% CO2. Cells were treated with (0.25 mM, 0.5 mM and 1mM) Clofibrate (Sigma) or DMSO (vehicle control) for 1h. Tert-butyl hydroperoxide (TBHP) served as a positive control in this experiment. Cells were stained with DCFDA for 30 minutes and followed by measurement of the absorbance using a fluorescent plate reader (Synergy H1 Hybrid, BioTek) with excitation wavelength at 485 nm and emission wavelength at 535 nm. For DHE staining, FAO cells were plated on chamber slides for 24 h and maintained at 37 °C and 5% CO2. Cells were treated with 0.25 mM Clofibrate (Sigma) for 1 h or DMSO (vehicle control). The cells were incubated with 5 μM DHE (in PBS) for 30 minutes at 37 °C in a dark chamber, fixed for 10 minutes in 4% paraformaldehyde and images promptly captured using an Olympus BX40 fluorescence microscope.
RNA extraction and quantitative RT-PCR
RNA was extracted using RiboPure Kit (Life technologies). Briefly, the procedure is as follow: Cells were plated in 6 wells plates and cultured at 37 °C and 5% CO2 for 24 hours. The cells were washed with PBS three times before scrapping in 1 ml TRI Reagent solution (Ambion). 1 ml of the homogenate was transferred to 1.5 ml centrifuge tube. 200 μl of chloroform was added and vortexed at maximum speed. Following a 5 minute incubation at room temperature, the samples centrifuged at maximum speed for 10 minutes. 400 μl of the aqueous phase was transferred to a new tube followed by addition of 200 μl 100% ethanol. The sample was transferred to Filter Cartridge-Collection and centrifuged 1 minute at maximum speed. The column was washed with 70% ethanol and a 100 μl of elution buffer added to elute RNA. We used Taq Man Fast Universal Kit for quantitative RT-PCR assay on an ABI Viia7. The primers were purchased from Life Technologies and human GAPDH was used as normal control.
Statistical analysis
The data are shown as mean ±s.d. Student's t-test (two-tailed) was performed for the comparisons between two groups and P < 0.05 considered statistically significant. All experiments were repeated at least three times and the representative data shown as indicated.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Suresh Subramani (University of California, San Diego, CA) for critical advice and providing the mRFP-EGFP-SKL plasmids, Dr. Michael Mancini (Baylor College of Medicine, Houston, TX) for providing DsRed-SKL plasmid. We are also grateful for the assistance of Kenneth Dunner (U.T.M.D. Cancer Center, Houston, TX) and Debra Townley (Baylor College of Medicine, Houston, TX) in electron microscopy image acquisition. This work was supported by National Institutes of Health (NIH) Grant R01 CA143811 to C.L.W., a Robert A. Welch Endowed Chair in Chemistry (BE-0023) to C.L.W., and NIH grants CA129537 and CA154320 to T.K.P. J.J. was a recipient of China Scholarship Council (CSC).
Footnotes
AUTHOR CONTRIBUTIONS
J.Z., D.N.T., J.J. and C.L.W. designed research; J.Z., D.N.T., J.J., J.K., A.A., R.T.P., R.D., J.T-M., J-H.L., R.K.P. and V.K.C. performed research; J.Z., D.N.T., J.J., R.D., T.T.P., T.P., M.B.K., and C.L.W. analyzed data; J.Z., D.N.T. and C.L.W wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annual review of nutrition. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 2.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Alexander A, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010;107:4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 5.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nature reviews. Molecular cell biology. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 6.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 8.Santos MJ, et al. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. The Journal of biological chemistry. 2005;280:41057–41068. doi: 10.1074/jbc.M505160200. [DOI] [PubMed] [Google Scholar]
- 9.Singh I, Singh AK, Contreras MA. Peroxisomal dysfunction in inflammatory childhood white matter disorders: an unexpected contributor to neuropathology. Journal of child neurology. 2009;24:1147–1157. doi: 10.1177/0883073809338327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JJ, Aitchison JD. Peroxisomes take shape. Nature reviews. Molecular cell biology. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weller S, Gould SJ, Valle D. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet. 2003;4:165–211. doi: 10.1146/annurev.genom.4.070802.110424. [DOI] [PubMed] [Google Scholar]
- 12.Reuber BE, et al. Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nature genetics. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 13.Fransen M, Nordgren M, Wang B, Apanasets O. Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim Biophys Acta. 2012;1822:1363–1373. doi: 10.1016/j.bbadis.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Dunn WA, Jr., et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 15.Farre JC, Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Iwata J, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. The Journal of biological chemistry. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. International journal of cell biology. 2012;2012:512721. doi: 10.1155/2012/512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci U S A. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 22.Lim DS, et al. ATM binds to beta-adaptin in cytoplasmic vesicles. Proc Natl Acad Sci U S A. 1998;95:10146–10151. doi: 10.1073/pnas.95.17.10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watters D, et al. Localization of a portion of extranuclear ATM to peroxisomes. The Journal of biological chemistry. 1999;274:34277–34282. doi: 10.1074/jbc.274.48.34277. [DOI] [PubMed] [Google Scholar]
- 24.Benjamin D, Hall MN. TSC on the peroxisome controls mTORC1. Nat Cell Biol. 2013;15:1135–1136. doi: 10.1038/ncb2849. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 27.Kastan MB, Lim DS. The many substrates and functions of ATM. Nature reviews. Molecular cell biology. 2000;1:179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 28.Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. The Journal of biological chemistry. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 31.Brown AI, Kim PK, Rutenberg AD. PEX5 and ubiquitin dynamics on mammalian peroxisome membranes. PLoS computational biology. 2014;10:e1003426. doi: 10.1371/journal.pcbi.1003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho AF, et al. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. The Journal of biological chemistry. 2007;282:31267–31272. doi: 10.1074/jbc.M706325200. [DOI] [PubMed] [Google Scholar]
- 33.Grou CP, et al. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. The Journal of biological chemistry. 2008;283:14190–14197. doi: 10.1074/jbc.M800402200. [DOI] [PubMed] [Google Scholar]
- 34.Platta HW, et al. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Molecular and cellular biology. 2009;29:5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thoms S, Erdmann R. Peroxisomal matrix protein receptor ubiquitination and recycling. Biochim Biophys Acta. 2006;1763:1620–1628. doi: 10.1016/j.bbamcr.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 36.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Molecular & cellular proteomics : MCP. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Molecular cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Miyata N, Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Molecular and cellular biology. 2005;25:10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nature structural & molecular biology. 2013;20:144–149. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nazarko TY, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014;204:541–557. doi: 10.1083/jcb.201307050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 45.Shiba-Fukushima K, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Scientific reports. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green DR, Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apanasets O, et al. PEX5, the shuttling import receptor for peroxisomal matrix proteins, is a redox-sensitive protein. Traffic. 2014;15:94–103. doi: 10.1111/tra.12129. [DOI] [PubMed] [Google Scholar]
- 48.Ma C, Hagstrom D, Polley SG, Subramani S. Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. The Journal of biological chemistry. 2013;288:27220–27231. doi: 10.1074/jbc.M113.492694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Experimental cell research. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Deosaran E, et al. NBR1 acts as an autophagy receptor for peroxisomes. Journal of cell science. 2013;126:939–952. doi: 10.1242/jcs.114819. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez FJ, Shah YM. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 52.Tripathi DN, et al. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci U S A. 2013;110:E2950–2957. doi: 10.1073/pnas.1307736110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ditch S, Paull TT. The ATM protein kinase and cellular redox signaling: beyond the DNA damage response. Trends in biochemical sciences. 2012;37:15–22. doi: 10.1016/j.tibs.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlow C, et al. ATM is a cytoplasmic protein in mouse brain required to prevent lysosomal accumulation. Proc Natl Acad Sci U S A. 2000;97:871–876. doi: 10.1073/pnas.97.2.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentin-Vega YA, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nature reviews. Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 57.Cai SL, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandita RK, et al. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol. 2006;26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hunt CR, et al. Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status. Cancer Res. 2007;67:3010–3017. doi: 10.1158/0008-5472.CAN-06-4328. [DOI] [PubMed] [Google Scholar]
- 60.Gupta A, et al. MOF Phosphorylation by ATM Regulates 53BP1-Mediated Double-Strand Break Repair Pathway Choice. Cell reports. 2014 doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhar S, Squire JA, Hande MP, Wellinger RJ, Pandita TK. Inactivation of 14-3-3sigma influences telomere behavior and ionizing radiation-induced chromosomal instability. Mol Cell Biol. 2000;20:7764–7772. doi: 10.1128/mcb.20.20.7764-7772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.