Abstract
Translation in eukaryotes is surveilled to detect toxins and virulence factors and coupled to the induction of defense pathways. C. elegans germline-specific mutations in translation components are detected by this system to induce detoxification and immune responses in distinct somatic cells. An RNAi screen revealed gene inactivations that act at multiple steps in lipid biosynthetic and kinase pathways that act upstream of MAP kinase to mediate the systemic communication of translation-defects to induce detoxification genes. Mammalian bile acids can rescue the defect in detoxification gene induction caused by C. elegans lipid biosynthetic gene inactivations. Extracts prepared from C. elegans with translation deficits but not from wild type can also rescue detoxification gene induction in lipid biosynthetic defective strains. These eukaryotic antibacterial countermeasures are not ignored by bacteria: particular bacterial species suppress normal C. elegans detoxification responses to mutations in translation factors.
Introduction
Exposure of eukaryotes to chemical toxins induces the expression of detoxification enzymes and transporters that modify and excrete these xenobiotics1. Because inactivation by RNAi of genes that encode targets of natural toxins also induces detoxification responses, surveillance of the core cellular processes such as translation, electron transport, etc., rather than detection of toxins via their molecular signatures, may detect toxic and pathogen attacks and couple to the induction of defense responses2. Sentinel cells that detect xenobiotics could induce a protective systemic response. A prediction of this cellular surveillance model is that disruption of such core processes even by a host mutation in such components should be interpreted by this system as a toxic attack and cause induction of detoxification and immunity genes. Here, we report that a variety of mutation-induced defects that disrupt translation only in the C. elegans germline trigger the induction of detoxification and innate immune gene expression in the intestine, the organ most likely to encounter bacterial pathogens. Laser ablation of germline stem cells abrogates this xenobiotic response to germline translation-defective mutations, showing that germ cells are the signaling center. An RNAi screen for genes that are required for the induction of xenobiotic response genes after exposure to drugs that inhibit translation or in response to mutations that disable germline translation revealed a kinase cascade and a lipid biosynthetic pathway that generates systemic signals of impaired translation. Purified mammalian bile acids can rescue the signaling defects in the lipid biosynthetic gene-inactivated animals, suggesting that the signals of translational malaise are bile acid derivatives. Particular bacterial species from a panel that we tested can suppress these host surveillance and detoxification pathways, showing that these pathways are targets of bacterial modulation.
Results
Inactivation of C. elegans translation components by feeding the animals E. coli expressing specific dsRNAs targeting translation factor mRNAs induces the expression of xenobiotic detoxification genes, bacterial pathogen response genes, and aversion behavior2–4 (Fig. 1a–c; Supplementary Tables 1–3). Toxins such as the eukaryotic translation inhibitors G418, produced by the bacteria Micromonospora rhodorangea5, or hygromycin, produced by the soil bacteria Streptomyces hygroscopicus6, also induce these responses. Detoxification responses in animals include cytochrome P450’s (CYPs), UDP-glucuronosyltransferases (UGTs), glutathione S-transferases (GSTs), and p-glycoprotein transporters (PGPs) (Fig. 1a–e; Supplementary Tables 1–3). We chose a pgp-5::gfp fusion gene for assays of xenobiotic detoxification induction in response to G418, hygromycin, or ribosomal assaults via RNAi (Fig. 1a–e; Supplementary Tables 1–3) because of the robust response of this reporter gene and validation of this gene induction from microarray gene expression analysis in response to translational inhibition by toxins or RNAi4.
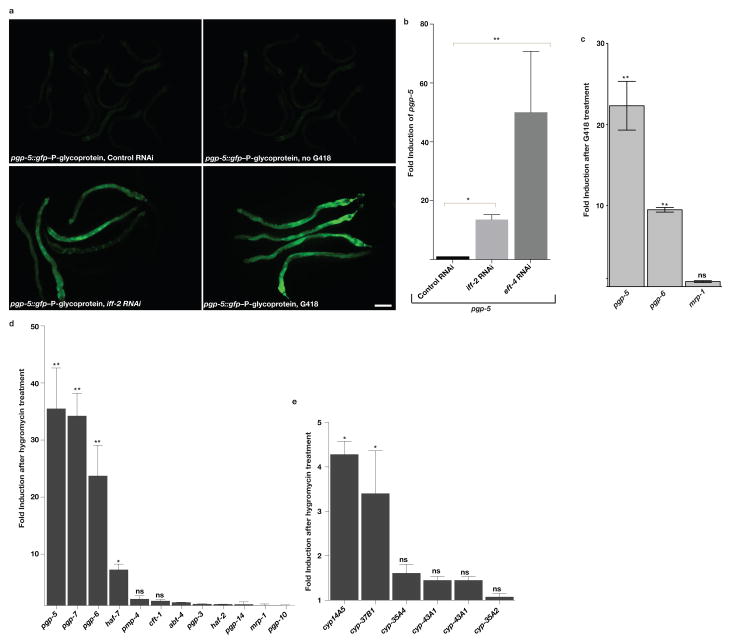
Figure 1. Translation inhibition using toxin or RNAi induces xenobiotic detoxification.
A) The toxin G418 or inhibition of translation by iff-2(translation initiation factor) RNAi induces pgp-5::gfp expression in the intestine as assessed using a transcriptional promoter fusion. Scale bar, 50μm.
B) RNAi of translation initiation factor (iff-2) or elongation factor (eft-4) induces pgp-5 mRNA as assessed by qRT-PCR. Fold change compared to control RNAi treated wildtype animals. **P<0.01.
C) G418 induces pgp-5 and pgp-6 mRNA from the chromosomal locus but not mrp-1–multidrug resistance protein homolog mRNA as assessed by qRT-PCR. Fold change compared to non-toxin-treated wildtype animals.
D) Hygromycin induces expression of particular xenobiotic efflux pump genes as assessed by qRT-PCR. Fold change compared to non-hygromycin-treated wildtype animals.
E) Hygromycin induces expression of particular xenobiotic detoxification genes as assessed by qRT-PCR. Fold change compared to no hygromycin-treated wildtype animals. B, C, D and E: Unpaired t test; **P<0.01, *P<0.05. ns; no significant difference. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d from n= 3 independent experiments is shown.
Systemic surveillance of translational inhibition
To test whether a mutational defect in translation in a single tissue is interpreted similarly to induce a systemic xenobiotic and innate immune response, we crossed the pgp-5::gfp fusion gene into mutants that are defective for translation only in the germline7. Some of the genes that encode protein translation components are duplicated in C. elegans, with one gene dedicated to translation in the germline and the other gene to somatic cell translation7. For example, C. elegans bears two translation initiation factor eIF-5A orthologues, iff-1 and iff-2, one specific to the germline and the other specific to somatic cells8. iff-1 is expressed only in the germline, and is required for its growth and proliferation; an animal homozygous for an iff-1 null allele is sterile due to a defect in germline translation, but has normal somatic iff-2 function and grows to adulthood at a normal rate8. In contrast, an iff-2 loss of function mutation is larval lethal8. Similarly, eft-3 and rpl-11.1 are required for translation in the germline while their duplicate genes eft-4 and rpl-11.2 mediate somatic cell translation7. Using GFP fusions to particular cytochrome p450 and ABC transporter detoxification genes, we observed high expression in the intestine in iff-1(tm483) homozygotes but almost no expression in iff-1(tm483)/+ heterozygous animals or wild type (Supplementary Fig. 1a). A GFP fusion to the innate immune response gene, irg-1, identified based on its strong response to the pathogen Pseudomonas aeroginosa PA143, was also activated in the iff-1(tm483) homozygous germline translation-defective mutant (Fig. 2a). Homozygous eft-3(q145) and rpl-11.1(ar228) sterile mutants also induced particular suites of detoxification genes, as assayed by RT-qPCR analysis or GFP fusion genes (Fig. 2a–d, and Supplementary Fig. 1b–c). Inactivation of the somatic homologues of these translation components also induced detoxification and innate immunity genes (Fig. 1a,b and Supplementary Fig. 1d). The germline translation mutation-induced xenobiotic defense response is not a general stress response because the mitochondrial stress response reporter gene hsp-60::gfp was not induced in the homozygous eft-3(q145) mutant (Supplementary Fig. 1e).
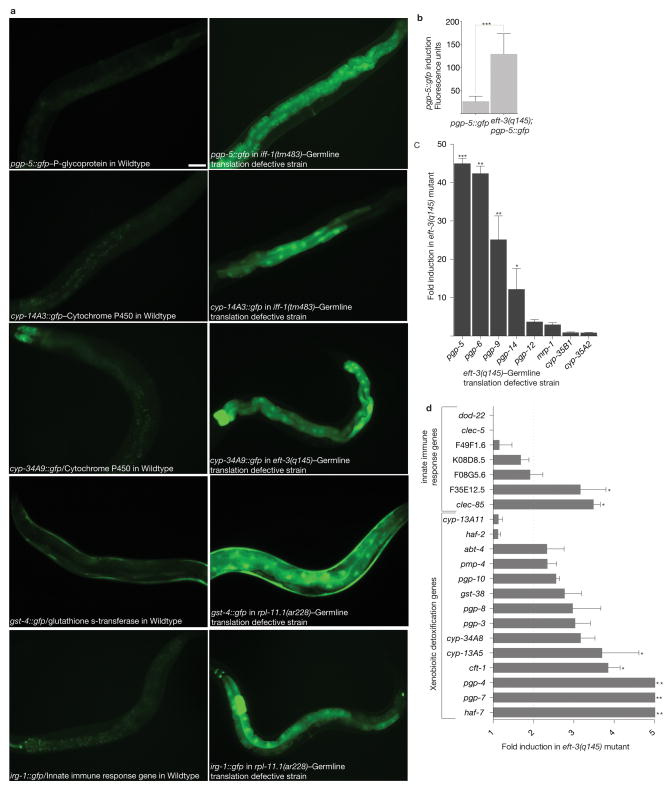
Figure 2. Translation defects in the germline induce systemic xenobiotic detoxification response.
A) Genetic defects in germline translation induce xenobiotic and innate immune response GFP fusion genes. iff-1, rpl-11.1(ribosomal protein L11) and eft-3 (an elongation factor 1-α ortholog) are expressed only in the germline and are required for translation in the germline only; they are not expressed in somatic cells or required for somatic translation7. Scale bar, 50 μm.
B) Quantification of pgp-5::gfp activation by germline translation defects. Fluorescence was measured using a COPAS Biosort. Unpaired t test; ***P<0.001. Mean ± s.d of n=413 and 1680 worms for pgp-5::gfp, and eft-3 (q145); pgp-5::gfp respectively. Data was consolidated from two independent experiments.
C) Genetic defects in germline translation induce xenobiotic efflux pump expression, as measured using PCR-based quantitation of mRNA levels of endogenous genes. Fold change compared to wildtype animals. Unpaired t test; ***P<0.001. **P<0.01. *P<0.05. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d from n=3 independent experiments is shown.
D) Genetic defects in germline translation induce xenobiotic and innate immune response genes, as measured using PCR-based quantitation of mRNA levels of endogenous genes. Fold change compared to wildtype animals. Statistical significance was determined using unpaired t test. **P<0.01. *P<0.05. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d from n=3 independent experiments is shown.
The translation-defective germline actively signals to the intestine to induce detoxification response genes; ablation of germ stem cells in eft-3(q145) pgp-5::gfp abrogated pgp-5::gfp induction in the intestine (Supplementary Fig. 1f–h). The systemic induction of detoxification by tissue-specific translation defects was not limited to the germline: tissue-specific translation deficits induced by RNAi in only muscle, neurons, hypodermis, using tissue-specific rescue of an rde-1 RNAi defective mutant2, can also induce a systemic detoxification in the intestine, showing that many cell types may be monitored by this system (Supplementary Fig. 1i,j).
To identify the signaling components that are required for induction of pgp-5::gfp or gst-4::gfp in the intestine in response to the iff-1(tm483) or rpl-11.1(ar228) germline-translation-defective mutations, we screened a 450-gene kinase and 600-gene transcription factor RNAi library (See Methods; Supplementary Tables 4,5). Positive hits from the screen were rescreened on the eft-3(q145);pgp-5::gfp germline translation-defective strain and most were required for pgp-5::gfp induction in eft-3(q145) (Fig. 3a,b). These gene inactivations also disrupted pgp-5::gfp or chromosomal pgp-5 induction by the translation-inhibitory drugs hygromycin or G418 (Fig. 3c–e). Several kinases including pmk-1–p38 MAPK, and its known upstream kinases, as well as the zip-2–bZIP, and skn-1–Nrf transcription factors were required for the intestinal induction of detoxification genes in the germline translation mutants (Supplementary Tables 4 and 5; Fig. 3a–e).
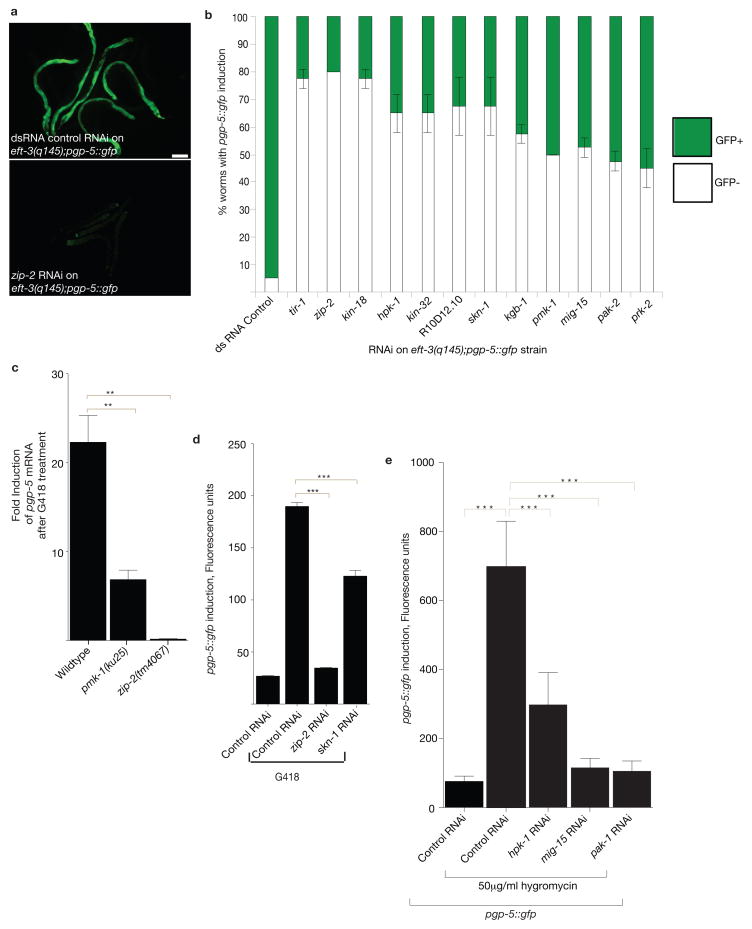
Figure 3. p38 MAPK signaling and zip-2–bZIP transcription factor are required for translation-inhibition-induced xenobiotic defense response.
A) RNAi of the zip-2–bZIP transcription factor gene disrupts the induction of pgp-5::gfp in response to germline translation defects in eft-3(q145);pgp-5::gfp. Scale bar, 50μm.
B) Kinase and transcription factor gene inactivations that disrupt pgp-5::gfp induction in response to germline translation defects in the eft-3(q145);pgp-5::gfp strain. n=60 worms. Mean ± s.d. n is consolidated from three independent experiments.
C) p38–pmk-1 and zip-2 are required for G418 induction of pgp-5 mRNA as assessed by qRT-PCR. Fold change compared to no drug wildtype animals. One-way ANOVA; **P<0.01. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d from n=3 independent experiments is shown.
D) zip-2 and skn-1 transcription factors are required for G418-induced pgp-5::gfp expression. Fluorescence was measured using a COPAS Biosort. Statistical significance was determined using one-way ANOVA. ***P<0.001. Mean ± s.e.m of n=400 worms for control RNAi and n=399, 563, and 99 worms, respectively for G418 in combination with dsRNA control, zip-2 RNAi and skn-1 RNAi treatments. Data was consolidated from two independent experiments.
E) hpk-1,mig-15,and pak-1 are required for hygromycin-induced induction of pgp-5::gfp. One-way ANOVA; ***P<0.001. Mean ± s.d. of n=136 worms for control RNAi and n=397, 268,123, 143 worms respectively for hygromycin treatment in combination with control RNAi, hpk-1 RNAi, mig-15 RNAi and pak-1 RNAi. Data was consolidated from two independent experiments.
Using an antibody to PMK-1–p38 MAPK protein to analyze which tissues activate PMK-1, as determined by its relocalization to the nucleus9, we observed that the PMK-1 kinase is activated in the intestine of germline translation-defective mutants (Fig. 4a,b). This immunostaining was absent in pmk-1(km25) null mutant animals demonstrating the specificity of the antibody (Supplementary Fig. 2a–c). Transgenic animals that express mCherry-labeled PMK-1 protein in the intestine under the vha-6 promoter confirmed the intestinal activation of PMK-1 observed with the antibody: mCherry-labeled PMK-1 protein was nuclearly-localized in eft-3(q145);pmk-1(km25) compared to pmk-1(km25) with similar levels of mCherry::PMK-1 expression (Supplementary Fig. 2d). Nuclear mCherry::PMK-1 in the intestine of eft-3(q145);pmk-1(km25) colocalized with the phospho-p38 antibody staining, suggesting that the nuclear PMK-1 corresponds to activated PMK-1–p38 MAPK (Supplementary Fig. 2e). Because expression of p38 MAPK only in the intestine of the eft-3(q145);pmk-1(km25) mutant, missing PMK-1 in all other cells types, is sufficient to induce p38 MAPK activation and subsequent nuclear localization in the intestine, the PMK-1–p38 MAPK acts in the intestine to transduce the signal of germline translation defects to somatic xenobiotic response.
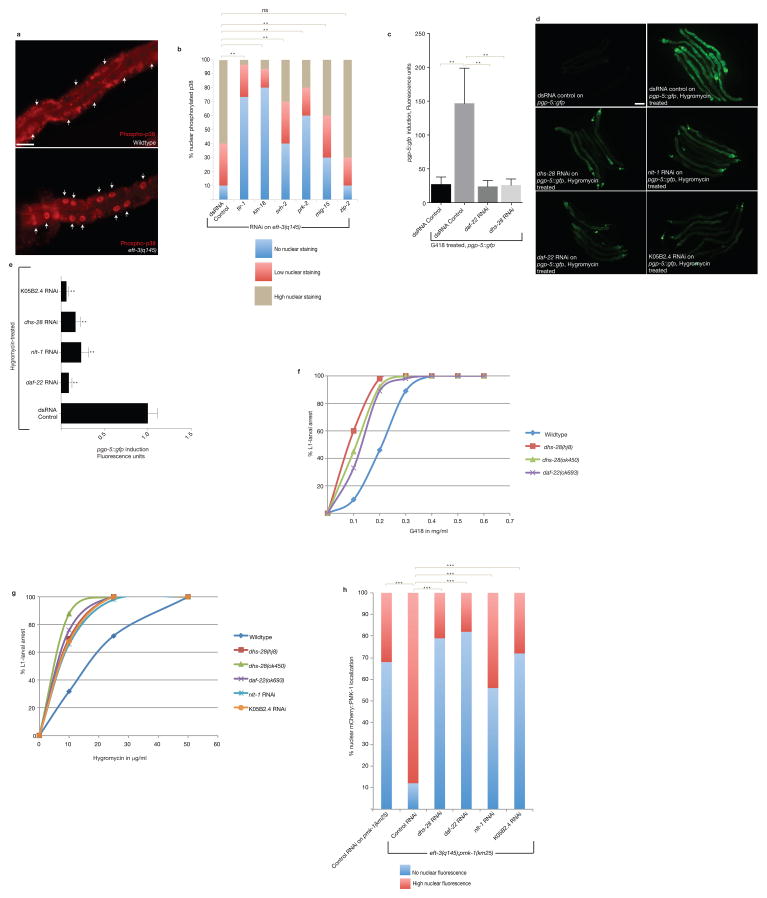
Figure 4. Bile acid-like biosynthetic signaling is required for translation-defective-induced xenobiotic defense response.
A) Germline translation defects in the eft-3(q145);pgp-5::gfp strain causes p38 MAPK phosphorylation and nuclear translocation of active p38 PMK-1 in the intestine. Arrows indicate the nucleus. Scale bar, 20μm.
B) tir-1, kin-18, svh-2, mig-15,and prk-2 are required for germline translation-defect-induced activation of p38 in the intestine. One-way ANOVA; **P<0.01. ns, P>0.05. n=50 (dsRNA control),45 (tir-1 RNAi), 50 (kin-18 RNAi),28 (svh-2 RNAi), 40 (prk-2 RNAi), 50 (mig-15 RNAi) and 32 (zip-2 RNAi) nuclei, respectively. Data represent one out of two independent experiments.
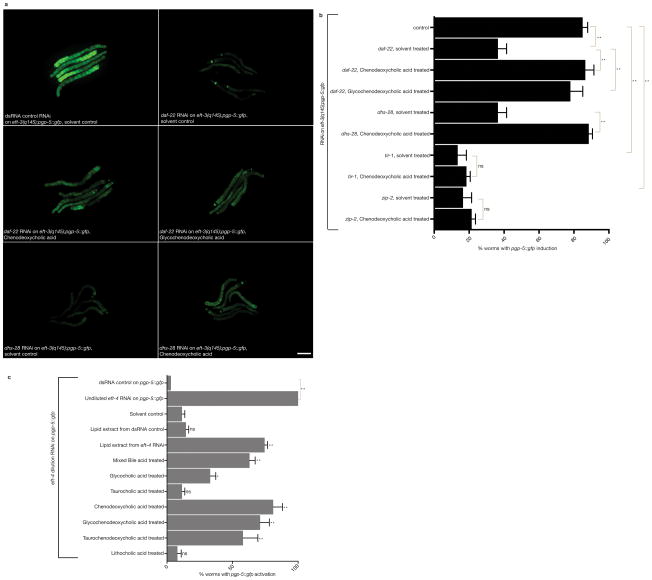
C) Inactivation of lipid and bile acid biosynthetic genes disrupts the induction of pgp-5::gfp in response to G418. Error bars represent SD. One-way ANOVA; **P<0.01. n=413 (dsRNA control non-G418 treated), n=2271(dsRNA control G418 treated), n=239 (daf-22 RNAi),n=514 (dhs-28 RNAi) worms. Data was consolidated from two independent experiments.
D) Lipid and bile acid biosynthetic genes are required for hygromycin-induced pgp-5::gfp. Scale bar, 50μm.
E) Inactivation of genes required for lipid–bile acid biosynthesis disrupts the induction of pgp-5::gfp in response to hygromycin. Error bars represent SD. One-way ANOVA; **P<0.01. n=10 worms for each condition. Data represent one out of two independent experiments.
F) While about 40% of control RNAi treated wildtype animals treated with 0.2 mg/ml G418 grew to adulthood, RNAi of daf-22 and dhs-28 cause >70% of animals treated with 0.2 mg/ml G418 to arrest at L1-larval stage. n=20 worms for each condition. Data represent one out of two independent experiments.
G) Mutations in genes required for lipid–bile acid biosynthesis cause hypersensitivity to G418. n=20 worms for each condition. Data represent one out of two independent experiments.
H) Lipid and bile acid biosynthetic genes are required for germline translation-defective-induced p38 nuclear localization. One-way ANOVA; ***P<0.001. n=50 nuclei (control RNAi on pmk-1 (km25)) and n=42 (control RNAi),32 (dhs-28 RNAi), 18 (daf-22 RNAi), 35 (nlt-1RNAi), and 50 (K05B2.4 RNAi) intestinal nuclei, respectively for eft-3 (q145); pmk-1 (km25). Data represent one out of two independent experiments.
We monitored PMK-1 nuclear localization after gene inactivation of the other hits in the screen, to order them upstream or downstream of PMK-1 activation and nuclear localization. Inactivation of prk-2 (Pim oncogene related kinase), kin-18 (TAO1 kinase), svh-2 (hepatocyte growth factor receptor related) and mig-15 (Nck-interacting kinase) as well as the known upstream MAPK component tir-1 caused decreased nuclear PMK-1 in the intestine of eft-3(q145) homozygotes (Fig. 4b), mapping these kinase activities upstream of PMK-1 nuclear relocalization. By contrast, RNAi of zip-2 had no effect on PMK-1 nuclear relocalization in eft-3(q145) homozygotes (Fig. 4b), showing that the ZIP-2 transcription factor acts downstream or in parallel to this kinase cascade.
Genome-wide RNAi screen for translational surveillance regulators
A genome-wide RNAi screen identified 170 gene inactivations that disrupt pgp-5::gfp response to G418 (Supplementary table 8). We tested the 170 primary screen candidate gene inactivations in duplicate for disruption of pgp-5::gfp induction in eft-3(q145) homozgyous animals (Supplementary table 9); 71 gene inactivations also disrupted normal pgp-5::gfp induction in response to a germline translation defect (Supplementary table 9; Figure 4c–e). Inactivation of many of the genes from the RNAi screen caused hypersensitivity to G418 or hygromycin (Supplementary Fig. 2g, Fig. 4f,g), suggesting that when detoxification response is decoupled from the detection of translation defects, these drugs show increased potency. DAVID analysis revealed a strong enrichment of kinase signaling in this genome-wide screen, including the MAP kinase pathway implicated in innate immune responses2,4 (Supplementary table 9) and several of the genes that identified in the kinase and transcription factor RNAi library screens (Supplementary Fig. 2f). In addition, multiple steps in a dual peroxisomal fatty acid or bile acid-like biosynthetic pathway disrupted pgp-5::gfp induction in response to eft-3(q145) germline translational defects, G418 or hygromycin (Supplementary table 9; Fig. 4d,e). For example, daf-22 and nlt-1 encode conserved protein domains contained in mammalian SCPx, which mediates bile acid and branched chain fatty acid metabolism10,11 which are bisected into two C. elegans genes. Similarly, dhs-28 encodes a protein that is orthologous to next upstream enzyme in the mammalian bile acid and branched chain fatty acid biosynthetic pathway, peroxisomal multifunctional protein 210,11.
Because the lipid–bile acid pathway could synthesize a small molecule endocrine signal of translational malaise that might be conserved between species, we studied this pathway in more detail. Using the nuclear localization of PMK-1 p38 MAPK as a molecular signature of its activation, we found that the lipid–bile acid signaling components are required for germline translation-defect activation of nuclear PMK-1–p38 MAPK in the intestine (Fig. 4h; Supplementary Fig. 2h). Thus, lipid signaling functions upstream of intestinal p38 MAPK signaling.
To explore the types of lipids that the dhs-28, daf-22 pathway generates to regulate the induction of pgp-5::gfp expression in translation-defective animals, we tested whether defects in dhs-28 or daf-22-mediated lipid–bile acid biosynthesis could be rescued for pgp-5::gfp induction by addition of purified mammalian bile acids (Supplementary Fig. 2i). Addition of bovine chenodeoxycholic acid or glycochenodeoxycholic acid partially rescued the defect in induction of pgp-5::gfp caused by the C. elegans daf-22 or dhs-28 lipid biosynthesis gene inactivations (Fig. 5a–b) but did not rescue the defect in induction of pgp-5::gfp caused by tir-1 or zip-2 (Fig. 5b). Deoxycholic acid weakly rescued but lithocholic acid did not rescue. Thus a bile acid product of the daf-22 and dhs-28 pathway is required for signaling translational malaise rather than other lipids. Treatment of wild type animals carrying pgp-5::gfp with these purified mammalian bile acids did not induce pgp-5::gfp, suggesting that the bile signal is not sufficient to signal ribosomal deficiency (Supplementary Fig. 3a,b). However addition of mammalian bile acids enhanced the induction of pgp-5::gfp in worms subjected to mild inhibition of translation via dilution of rpl-7 RNAi (Supplementary Fig. 3c). Bile acid addition was sufficient to induce xenobiotic detoxification if the MAP kinase signaling was enhanced by a mutation in vhp-1, which encodes a phosphatase that negatively regulates the JNK and p38 MAPK12,13 (Supplementary Fig. 3d). Thus both activation of PMK-1 MAPK signaling and production of the bile acids may be required for induction of detoxification response pathways. Bile acid treatment does not induce hsp-4::gfp (a ER stress response chaperone gene) or gpdh-1::gfp (osmotic stress response gene) (Supplementary Fig. 3e,f), showing that it is not a general stress inducer.
Figure 5. Bile acid signaling couples translation defects to the induction of xenobiotic defense genes.
A) Mammalian bile acids rescue the defect in pgp-5::gfp induction caused by daf-22 or dhs-28 bile biosynthetic gene inactivations in the eft-3(q145);pgp-5::gfp strain with a germline translation elongation factor mutation. Scale bar, 50μm.
B) Quantitation of the rescue by mammalian bile acids of the defect in pgp-5::gfp induction caused by RNAi inactivation of C. elegans lipid–bile acid biosynthetic genes. Error bars represent SD. One-way ANOVA; *P<0.01. ns, P>0.05. n=100 worms. Data represent one out of three independent experiments.
C) Exogenous bile acids or lipid extracts from translationally-challenged C. elegans enhance the induction of pgp-5::gfp expression in response to mild translational inhibition by diluted eft-4 RNA. Data are from two independent experimental replicates. N=100 worms. Error bars represent SD. Statistical significance was calculated by comparing to solvent control treated animals on eft-4 dilution RNAi using one-way ANOVA. **P<0.01. ns, P>0.05. Data represent one out of three independent experiments.
If bile acid-like signals are produced by C. elegans in response to ribosomal deficiency, we expected that a crude lipid extract from C. elegans with translation defects would induce or enhance induction of pgp-5::gfp in another animal. A lipid extract from translationally-disrupted eft-4 RNAi treated-animals did not induce pgp-5::gfp in wild type animals but lipid extracts from these animals enhanced the response to mild inhibition of translation (Fig. 5C; Supplementary Fig. 3g,h). Lipid extracts from wild type animals did not show this activity (Fig. 5C). Lipid extracts from animals undergoing translational inhibition either by iff-2 RNAi or rpl-7 RNAi showed the same activity as eft-4 RNAi extracts.
Lipid signalling mediates translational surveillance
C. elegans produces bile acid-like steroids 10,11 from nutritionally derived cholesterol. In addition to their role in bile acid biosynthesis, daf-22 and dhs-28 encode peroxisomal proteins that also mediate lipid modifications to a secreted dauer arrest pheromone14; however we found no evidence of a coupling of translation-defects to dauer pheromone production (Supplementary Fig. 3i). C. elegans bile-like dafachronic acids also act at the most downstream outputs of this endocrine system to regulate dauer arrest via the DAF-12 nuclear hormone receptor15, but this pathway is distinct from the daf-22, ntl-1, and dhs-28 pathway because inactivation of the daf-12 nuclear hormone receptor gene or addition of purified dafachronic acids does not affect pgp-5::GFP induction in response to ribosomal defects (Supplementary Fig. 3j).
Modulation of translational surveillance by bacterial species
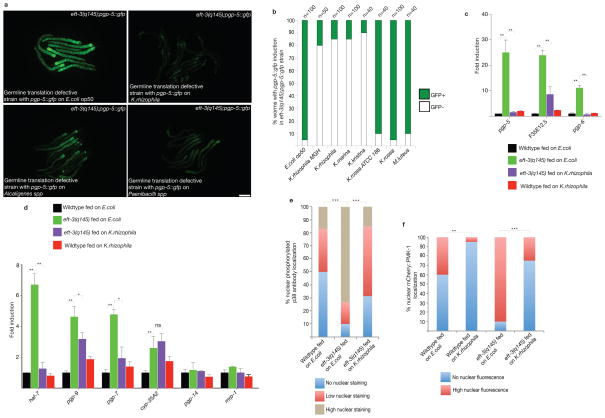
If a microbial secondary metabolite or virulence factor targets the same surveillance and xenobiotic response gene pathways, host surveillance and response to translation deficits would be compromised, rendering other toxins or virulence factors more effective. We screened a diverse collection of bacterial species (Supplementary Table 6) for disruption of intestinal pgp-5::gfp induction by a germline translation mutation when these bacterial species were fed to the eft-3(q145); pgp-5::gfp strain. Three bacterial species out of 40 tested strongly suppressed the induction of pgp-5::gfp in the homozygous eft-3(q145); pgp-5::gfp strain: Kocuria rhizophila, Alcaligenes spp., and Paenibacillus spp. (Fig. 6a,b; Supplementary Fig. 3k). Kocuria spp are human skin microflora16–18. Kocuria rhizophila19,20 and Alcaligenes spp21–23 are human opportunistic pathogens while Paenibacillus spp. is a soil bacteria24–26 associated with root knot nematodes and suppresses their virulence on plants27. Kocuria rhizophila also suppresses detoxification responses as measured by qRT-PCR to monitor endogenous genes (Fig. 6c,d). Other K. rhizophila isolates or the closely related K. kristinae and K. marina also abrogated induction of pgp-5::gfp in the germline translation mutant animals but feeding on other close taxons, K. rosea or Micrococcus luteus did not disrupt pgp-5 induction (Fig. 6b). The unfolded protein response was normal in upr-1(zc6);hsp-4::gfp animals grown on Alcaligenes spp., Paenibacillus spp. or Kocuria rhizophila, as it is on E. coli (Supplementary Fig. 3l), suggesting that the suppression of the activation of pgp-5::gfp by these bacterial strains is a specific regulation of detoxification and immunity.
Figure 6. Bacterial suppression of the induction of xenobiotic responses by a germline-translation mutation.
A) C. elegans grown on lawns of Paenibacilli, Kocuria or Alcaligenes bacteria fail to induce pgp-5::gfp in response to the germline-translation-defects in eft-3(q145);pgp-5::gfp. Scale bar, 50 μm.
B) C. elegans grown on various Kocuria species fail to induce pgp-5::gfp in response to the germline translation defects in eft-3(q145);pgp-5::gfp. Data represent one of two independent experiments. n denotes number of worms.
C) C. elegans grown on K.rhizophila fail to induce pgp-5 and pgp-6 in response to germline translation defects in eft-3(q145). Error bars represent SD. Unpaired t test; **P<0.01. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d is from n=3 independent experiments.
D) C. elegans grown on K.rhizophila fail to induce various chromosomal xenobiotic and innate immune response genes in response to germline translation defects in eft-3(q145). Error bars represent SD. Unpaired t test; **P<0.01. *P<0.05. ns, not significant. ~300 worms per condition were washed off 1 plate for each experiment. Mean ± s.d, n=3 independent experiments.
E) Feeding on K.rhizophila disrupts both basal and constitutive p38 MAPK phosphorylation as assessed using an antibody that recognizes active phosphorylated PMK-1 protein. One-way ANOVA. ***P<0.001. For wildtype on E.coli, n=20, for eft-3(q145) n=32 (E.coli) and 40 (K. rhizophila) nuclei, respectively. Data represent one out of two independent experiments.
F) Feeding on K.rhizophila disrupts the nuclear translocation of active phosphorylated p38 MAPK. Unpaired t test.. ***P<0.001. **P<0.01. For wildtype, n=50 (E.coli) 40 (K.rhizophila) nuclei, and for eft-3 (q145) n=55 (E.coli) and 48 (K. rhizophila) nuclei, respectively. Data represent one of two independent experiments.
Heat-killed Kocuria spp. did not disrupt pgp-5::gfp induction, showing that it is not a nutritional insult to C. elegans (Supplementary Fig. 3m). Supernatants from stationary cultures of Kocuria spp. did not suppress pgp-5::gfp induction in the germline- translation-defective C. elegans mutant (Supplementary Fig. 3m), suggesting that no stable toxin or virulence factor is secreted into the growth media. While PMK-1 is nuclearly localized in intestinal nuclei of eft-3(q145) homozygous animals fed on E.coli, weak or no nuclear staining and a reduction in intestinal nuclear mCherry::PMK-1 was seen in eft-3(q145) homozygous animals fed on K. rhizophila (Fig. 6e,f; Supplementary Fig. 2d). These data suggest that Kocuria produces, for example, a virulence factor that is transferred to the C. elegans intestine to silence the response to germline translation defects upstream of the PMK-1 MAP kinase cascade.
Discussion
We have shown that mutations that disrupt translation in particular tissues are misapprehended as a bacterial attack by an innate immunity and detoxification system that responds with gene expression countermeasures to microbial toxins. The induction of this pathway by germline mutations in translation factor genes is a strong support for the hypothesis2 that toxins and virulence factors are detected by their inhibition of core cellular machinery rather than by their chemical detection in the mixture of much more abundant cellular biochemicals. This system of surveillance has some similarities to effector triggered immunity system of plant pathogens in that for both systems virulence factors are detected and an immune response ensues28. But effector triggered immunity posits a dedicated host protein receptor, such as the nucleotide-binding–leucine-rich-repeat proteins, for each virulence factor, whereas the cellular surveillance system monitors host cellular pathways in parallel for any inhibition by toxin or virulence factor2. This surveillance model has the advantage of using the toxicity to host pathways of a toxin, which may interact with high affinity with its conserved cellular target, as the sensitive detector and trigger for induction of detoxification and immunity.
The disruption of core cellular processes in specific tissues such as the germline induce xenobiotic defense response in distant unaffected tissues. This finding although analogous to the mitochondrial dysfunction-induced systemic stress response29,30 or the systemic heat shock response31 involves distinct set of xenobiotic detoxification genes and signaling pathways, suggesting that distinct cellular insults trigger distinct response pathways. While the intestine and skin are the most likely tissues to first encounter bacterial toxins and virulence factors, the germline is also subject to bacterial infection: Wolbachia infection of the germline is endemic across arthropods32 as well as the nematode pathogen Brugia malayi33.
While the MAP kinase pathway has a well-established role in pathogen defense, our screen shows that this pathway also surveils for small chemical toxins. In addition, we could order the action of newly identified genes, such as the bile acid signaling pathway genes and less studied kinases, such as the kin-18 RIO1 kinase, upstream of the MAP kinase pathway genes. In mammals, RIO1 kinase binds to ribosomes where it mediates ribosomal assembly34; kin-18–RIO1 activity in ribosome assembly or on the mature ribosome where it remains bound may also play a role in translational surveillance signaling to the MAPK pathway.
Our screen for gene inactivations that disrupt this surveillance pathway revealed many hits in a C. elegans lipid biosynthetic pathway, suggesting that particular lipids constitute a major axis of toxin and bacterial pathogen signaling. The partial rescue of the signaling defects of gene inactivations such as daf-22 and dhs-28 with 95% pure mammalian chenodeoxycholic acid or glycochenodeoxycholic acid, supports the model that the C. elegans lipid signal is a bile acid. However, it is possible that other lipids in those 95% pure mammalian bile salts actually mediate the rescue. Mammalian bile acids are conventionally thought to aid digestion as emulsifiers of fat, but a role in various systemic endocrine hormone-like functions has emerged35. Bile acids in mammals are synthesized from cholesterol in the liver as primary bile acids and, significantly for our proposal that they constitute signals of microbial attack, are metabolized by particular mammalian gut microbes to secondary bile acids36. Bile acids in mammals regulate metabolic pathways by activation of Farnesoid X receptor as well as the G-protein-coupled receptor (GPCRs) such as TGR536,37. The defect in xenobiotic responses that we observe after inactivation of the C. elegans nhr-267 nuclear hormone receptor gene may be a homologous response to mammalian bile salt FXR responses, although nhr-267 is not an FXR orthologue (Supplementary table 8&2). Conversely, mammalian bile acids detected by FXR and LXR may be internal signals of bacterial attack that couple via these nuclear hormone receptors to systemic detoxification. In this sense, these nuclear hormone receptors do not surveil for the infinity of possible chemical and protein toxins with their ligand binding domains, only for internally generated bile acid or other signals of distress produced by the system of surveillance of core cellular machinery2.
Our screen for bacterial activities that suppress xenobiotic surveillance found that 10% of the disparate bacterial strains tested suppressed xenobiotic detoxification responses to a C. elegans germline transation mutation. While this hit rate seems remarkably high, bacterial strains can each produce dozens of virulence factors and toxins, many of which have evolved to disrupt eukaryotic biology. So this screen may have tested hundreds of toxins and virulence factors, many under selection in bacteria for their anti-eukaryotic activities, for suppression of the C. elegans xenobiotic surveillance and detoxification system. The Kocuria antisurveillance activity was not found in taxonomically related bacteria and depends on continued contact with live bacteria, suggesting that a transferred protein virulence factor(s) rather than a stable secreted small molecule mediates the activity. The microbial modulation of animal surveillance of core cellular components has important implications for the behavior of pathogenic and commensal bacteria. It suggests that bacteria may produce drugs and transferred proteins that silence eukaryotic surveillance systems to in turn suppress eukaryotic countermeasures to toxins and virulence factors. The removal of such innate immunity silencing activities by disruptions of the microbiome with antibiotics for example could trigger stronger immune reactions including autoimmunity and inappropriate immune reaction to normally benign microbes or chemicals.
Materials and methods
N2 Bristol was the wildtype strain used.
The strains and mutant alleles used are in Supplementary Table 7.
RT-qPCR experiments
RNA was isolated using TRI Reagent (Sigma), followed by chloroform extraction and isopropanol precipitation. Briefly ~ 300 day 1 adults treated with or without drugs were collected. RNA was DNAase treated using the TURBO DNA-free kit (Applied Biosystems). cDNA was prepared using the First strand cDNA synthesis kit from Invitrogen. qRT-PCR was performed with an iCycler machine (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). All reactions were done in triplicate and on at least 2–4 biological replicates. All the values are normalized to ama-1 as internal control as well as to the transcript levels in untreated wildtype.
Primer sequence include
pgp-5F: TGTTCGAGCACTTAACATGGA
pgp-5R: CCCTGAATTACAGCTCGTTTG
pgp-6F: CGTTGGAGACAACACGACAT
pgp-6R: CAAATGTCCAGTCGACACGC
cyp-34A9F:ATTCATGCCTGTTTGGGTGCTC
cyp-34A9R:GCATCCACAAAGTCTTCACCCT
cyp-35B1F:TGCTTCTGACCACATTTCTTGCTG
cyp-35B1R: CAGGAGGGAGAAAGCGATCTTTG
ama-1F: CGGTCAGAAAGGCTATCGAG
ama-1R: CCAACCTCCTGACGATTGAT
cyp-14A3F: CTGTTCTTGATTTCTGGCTTGCTGG
cyp-14A3R: GACAGCTTGTGTGTACGGCA
pgp-12F: TCGACTGAGGATGTCGGTTCCATTG
pgp-12R: CAATGATTCCTAAACCCGGCTGCG
mrp-1F: GCAACAATTCCAACATTCCGCAAG
mrp-1R: CTGCATATTGCACGAGGTCACCA
pgp-9F: CCGGTACACAAGCACAGTAGATC
pgp-9R: CTGCCGCCGGAATCTATTTGACA
pgp-14F: CGAGCGACCTACCCGCAAATTAC
pgp-14R: GCACAAATGGTTCCAATGAACAGG
cyp-35A2F: AAGTATCAAGACTACCACCAGGTCC
cyp-35A2R: GACATCTCTCATAACCGGCGCATG
cft-1F: AATTCTGGAAGTCTTTCGCTGGAG
cft-1R: CCATTTCCACGCTGTCTTCTCCAAC
pgp-4F: GCGATTGATCACCGACTCGC
pgp-4R: CAATTGGTGCCATACTCCAGC
pgp-3F: GTTCACTTATCCAACTCGCCCAG
pgp-3R: GCTTCCTCGATGTCTTGATCCGTG
pgp-7 F: GATTACACGACACCACTCATAGCTC
pgp-7 R: CATTCCGCGTTCATACTGCTCATA
cyp25A1F: GGTTTAACGAGCTTCACGAGCAG
cyp25A1R: CAGATCGTGTATGCTTCCAACCA
cyp13A11F: TTATTCTCAGCTCAAGGCCATCG
cyp13A11R: CCATTGCTATGCGTCCAATCACGT
cyp44A1F: ACATTCTCCGAAATTCCTGGTCC
cyp44A1R: GCAGACGATACCATTCAGGACCATT
cyp13A7F: AGTCAAAGGGCCTAGAGGTCTAC
cyp13A7R: CTTCCATCTATGGCCTTGAGCAG
cyp14A1F: AGATGAAGCAAACGGGAAATCCTGG
cyp14A1R: GACATTGATGGCAAACGTGCTG
cyp34A8F: CGGGTCAGGAGACCACATCTAC
cyp34A8R: CTGGTACGGGCTGACTATCAATCGT
cyp13A5F: TTGGAAGCGGTTGAGAACTCTG
cyp13A5R: CGAAGGCTGGTTGTATAGATGGGAA
pgp-8F: GATTTCGAGAGGCGTTCAACG
pgp-8R: GAAGCAAGTGTCCAGCTTGT
pgp-10F: AGCTGATGGCGATGGGTCTG
pgp-10R: CATCGCAATCGACAGTCCCAA
K08D8.5F: ATGCTACCGAATGCTTTTCC
K08D8.5R: TCCTTGGGTGTAGTTTCCAA
F35E12.5F: ACACAATCATTTGCGATGGA
F35E12.5R: GGTAGTCATTGGAGCCGAAA
gst-38F: TCCAATGCTCGAGGTAGATGGCAA
gst-38R: ACGAGCCTCCGCGTAATAGTCTTT
clec-85 F: GTCTTTGGTCAATGCACACG
clec-85 R: CGACTGAGCATCCGTAAACT
clec-5 F: GCTGCTGAGAGCAAATGTAC
clec-5 R: CCATAGGAGGGAAACTGAACTG
F08G5.6F: TGGACAACCCAGATATGCAA
F08G5.6R: GTATGCGATGGAAATGGACA
pmp-4F: TTTGTGGCTCAACTTGAAGG
pmp-4R: GCTAATCCCAGGTAAGACTCA
haf-2F: ACAGTGAGGGATACGGAAAC
haf-2R: CCACAGTACGAGAGAAGACG
Haf-7F: AAGGAGCTGAATACGAGAGG
Haf-7R: GCACTGAGCACAATTTGACA
Abt-4F: CTGTCGCGGCATACATAAAG
Abt-4R: GAGTCGGATTGTGTAGGTGA
F49F1.6F: CTTGTGGAATATGCCATCAG
F49F1.6R: GGGCATTGTATCTTAACAGC
T28D8.5F: GTCGACTCAAGACCATCATGC
T28D8.5R: GAGTATCGGTAACGCAGACACC
Generation of pvha-6::mCherry::pmk-1 transgenic worms
pvha-6::mcherry:pmk-1::tbb-2 3′UTR was constructed in the pCFJ151 (MosSCI ttTi5605) vector backbone using the Gibson Isothermal Assembly method using the following primers:
ZED1: GTGCGCGCGATGCATTCGAAGATCTGCCCAGATATTGCCAGCATGCTCAACGTTG
ZED2: TATATGATCCATTGTTGTCTGTGGAAACATCTTATACAATTCATCCATGCCACCTGTC
ZED3: CGACAGGTGGCATGGATGAATTGTATAAGATGTTTCCACAGACAACAATGGATC
ZED4: GAAGGGAATGCTTGAAAGGATCTTGCATCTACGATTCCATTTTCTCCTCATCTTCC
ZED5: GAAGATGAGGAGAAAATGGAATCGTAGATGCAAGATCCTTTCAAGCATTCCC
ZED6: ATCTTACTTGCACTTATAATACGACTCATGATCCACGATCTGGAAGATTTCCAAC
The resultant result plasmid were injected into the wildtype worms at 5 ng/ml concentration along with myo-2::NLSgfp co-injection marker to generate transgenic extrachromosomal array worms.
Drug treatment
G418 (GoldBio) or hygromycin (GoldBio) was diluted in M9 to the desired concentration and added onto preseeded OP50 E.coli bacteria containing NGM plates. The plates were dried uncovered in the hood for at least 1 hour before seeding the worms. Synchronized L1-stage animals were dropped onto the drug containing plates and scored later. For drug resistance assays synchronized L1 larval stage animals of the appropriate genotype were fed with either control RNAi or with RNAi clones targeting bile acid biosynthetic pathways until they reach day two of adulthood. Subsequently, the RNAi-treated animals were egg-prepped to obtain synchronized L1 larvae. ~100 L1 larvae were added to the plates containing G418. 100nM of Δ4-Dafachronic Acid or (25S)-Δ7-Dafachronic Acid from Cayman Chemical were used in the daf-12 experiments. In the purified bile experiments, bile from bovine and ovine (Mixed bile acids), chenodeoxycholic acids, glycochenodeoxycholic acid, glycocholic acid, taurocholic acid, lithocholic acid, and taurochenodeoxycholic acid from Sigma were diluted in M9 to the desired concentration (100μM) and added onto preseeded OP50 E.coli bacteria containing NGM plates. The plates were dried uncovered in the hood for at least 1 hour before seeding the worms.
Screen methods used for identifying genetic pathways that mediate translation mutant-induced xenobiotic defense response
For the kinase and transcription factor cherry picked library primary screen, iff-1(tm483)/+ ; pgp-5::gfp heterozygous adults were fed on RNAi-media containing plates seeded with E. coli expressing dsRNA corresponding to a C. elegans kinase or transcription factor gene and screened for whether the ¼ of the progeny of the genotype iff-1(tm483)/iff-1(tm483); pgp-5::gfp show decreased pgp-5::gfp expression compared to no gene inactivation control animals. Scoring for GFP induction was done only on iff-1(tm483)/ iff-1(tm483) homozygotes, which could be scored using absence of fluorescence from an RFP fusion gene integrated on the balancer chromosome. The screen was conducted at 22°C. Secondary screens on kinase or transcription factor hits from the iff-1(tm483)/iff-1(tm483); pgp-5::gfp initial screen were similarly done on the eft-3(q145);pgp-5::gfp strain. The cherry-picked libraries included mostly transcription factors and kinases libraries but they also included some genes annotated to act in the kinase pathways that are not actually kinases (such as tir-1). The screen for identifying genes required for rpl-11.1(ar228) germline translation-inhibition induced gst-4::gfp activation was conducted by feeding rpl-11.1(ar228)/+ ; gst-4::gfp heterozygous adults on RNAi-media containing plates seeded with E. coli expressing dsRNA corresponding to a C. elegans kinase or transcription factor gene and screened for whether the ¼ of the progeny of the genotype rpl-11.1(ar228)/ rpl-11.1(ar228); gst-4::gfp show decreased gfp expression visually compared to no gene inactivation control animals. Scoring for GFP induction was done only on rpl-11.1(ar228)/ rpl-11.1(ar228) homozygotes, which could be scored using another dominant Unc marker from the balancer chromosome.
Staining of dissected intestine and gonad
Germline and intestine were dissected in M9 using 22-gauge hypodermic needles by cutting either near the pharynx or the tail region. The germline and intestine are released from the worm because of pressure and were separated from the worm carcass. The dissected germline and intestine were fixed with 1% formaldehyde in M9 for 10 min and subsequently fixed in ice-cold methanol for 5 min at room temperature. The dissected germline and intestine were washed three times by centrifugation at 2000 RPM for 1 minute with 1X PBS containing 0.1% Tween 20 (PBST). Following the fixation, the dissected germline and intestine were blocked with PBST containing 1mg/ml BSA for one hour at room temperature. After blocking, the dissected germline and intestine were stained with Anti-ACTIVE® p38 pAb, Rabbit, (pTGpY) from Promega (Catalog no: V1211) diluted 1:1000 in PBST containing 1mg/ml BSA for 2 hour at room temperature. The dissected germline and intestine were washed three times in PBST and then stained with Goat Anti-Rabbit IgG (Catalog no:111-165-144) secondary antibody from Jackson Research diluted in 1:1000 in PBST containing 1mg/ml BSA for 2 hour at room temperature. The dissected germline and intestine were washed three times in PBST and then mounted on 2% agarose pad on glass slide with Vectashield® containing DAPI (Catalog no: H-1200) from Vector Labs for imaging. For staining the pmk-1::mcherry transgenic worms, the worms were dissected, fixed and blocked as above. The dissected intestine was incubated with Living Colors® mCherry Monoclonal Antibody from Clontech (Catalog no:v 632543), and Anti-ACTIVE® p38 pAb, Rabbit, (pTGpY) from Promega (Catalog no: V1211) diluted 1:2000 and 1:1000 respectively in PBST containing 1mg/ml BSA for 2 hours. Following the incubation, the dissected intestine was washed three times in PBST and stained with Anti-mouse IgG (Catalog no: 515165-003) and Anti-Rabbit IgG (Catalog no: 111-095-144) secondary antibodies from Jackson Research diluted to 1:1000 in PBST containing 1mg/ml BSA for 2 hour at room temperature. The dissected intestine was washed three times in PBST and then mounted on 2% agarose pad on glass slide with Vectashield containing DAPI from Vector Labs for imaging.
Microscopy
Nematodes were mounted onto agarose pads and images were taken using a Zeiss AXIO Imager Z1 microscope fitted with a Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software. Fluorescent images were converted to 16-bit images, thresholded and quantified using ImageJ. Student’s t test and ANOVA was used determine statistical significance.
Genome-wide RNAi screen
RNAi bacteria expressing the dsRNA corresponding to each worm gene were grown in LB media with 25μg/ml carbenicillin overnight and seeded onto RNAi agar plates containing 1mM IPTG. The following RNAi libraries were used for screening: Ahringer RNAi library, Vidal supplemental RNAi clone library and the new supplemental clones RNAi Library from Source Bioscience. The plates were allowed to dry in a laminar flow hood and incubated at room temperature overnight to induce dsRNA expression. 20 synchronized L1 larvae pgp-5::gfp expressing animals were placed onto RNAi-containing agar plates, allowed to develop at 15°C for 3 days to the L4-stage when 0.3 mg/ml of G418 in M9 was added to each RNAi well. The plates were allowed to dry in a laminar flow hood and incubated at 20°C. In the primary screen, the plates were screened visually for changes in GFP fluorescence and worm developmental defects. Any RNAi clones that caused lethality or decreased size or dramatically slowed development were excluded from the analysis. All the positive clones were retested at least twice and the clones were verified by DNA sequencing. The 170 hits from the G418 screen were also tested on the iff-1(tm483)/iff-1(tm483); pgp-5::gfp and the eft-3(q145);pgp-5::gfp strains; 71 gene inactivations also disrupted activation of pgp-5 by these germline translation factor gene mutations. DAVID analysis was done on the 170 hits from the primary screen as well as on the 71 hits from the secondary screen with the germline translation defective mutation. From this analysis of the full genome screen, kinases were by far the most enriched, at a probability of 10−17. Wormbase version WS247 was used to assign gene names to the hits.
Lipid extract preparation
A lipid extraction protocol14 was used with the following modifications. Bleach-prepared embryos from wildtype were allowed to hatch without bacterial food and then fed from the L1-stage until adulthood in benign dsRNA control RNAi bacteria, eft-4 RNAi, iff-1 RNAi and rpl-7 RNAi bacteria on 60 large RNAi plates at 22°C. These worms were washed off the plates and resuspended in M9. The worms were washed at least three times to remove the bacteria. Subsequently, the worms were allowed to settle and the supernatant was removed. 5 ml of packed worms were lyophilized separately and kept at −80°C until use. The worm pellets were powdered in 0.1 M NaCl and liquid nitrogen using a mortar. The powdered pellets were extracted with 95% ethanol at 22°C for 24 h. The extracts were filtered using Whatman GF/A glass filter and were evaporated to dryness and resuspended in methanol.
Coculturing pgp-5::gfp animals with eft-3(q145) worms to test for animal to animal signaling of ribosomal stress
~2000 L1-larval stage eft-3(q145) homozygotes were sorted onto a 10cm NGM plate preseeded with E.coli OP50 using COPAS Biosorter®. ~100 L1-larval stage pgp-5::gfp worms were subsequently dropped onto the same plate and incubated at 20°C for 60 hours. The pgp-5::gfp worms on the plate were differentiated from the eft-3(q145) homozygotes on the basis of fertility. The fertile pgp-5::gfp worms on the plate was scored visually for pgp-5::gfp induction. In another experimental trial, the worms were sorted onto 10-cm plates seeded with dead OP50 instead of live E.coli OP50. In a third experimental trial, the worms were sorted onto 3-cm plates seeded with dead OP50 instead of live E.coli OP50. In a fourth experimental trial, the different concentrations (10–100 μm) of mixed bile acids were added to the worms were sorted onto 3-cm plates seeded with dead OP50. In all the cases, the fertile pgp-5::gfp worms on the plate was scored visually for pgp-5::gfp induction. We did not find any evidence for GFP induction in the non-eft-3 mutant animals. The result of one such experiment is shown in Supplementary figure 3I.
Testing for non-specific effects of bile acids
Synchronized L1-larval stage hsp-4::gfp transgenic animals were grown to L4-larval stage at 20°C and treated with 1 μg/ml of DTT alone or with chenodeoxycholic acid 100μM for 24 hours. In experimental trials we also used DTT concentration renging from 1 to 7.5 μg/ml with and without different concentrations of mixed bile acids. In all cases, we did not observe induction of hsp-4::gfp or disruption of DTT induction of hsp-4::gfp by bile acids. One such experiment is shown in Supplementary figure 3e. In this experiment, synchronized L1-larval stage hsp-4::gfp transgenic worms were grown to L4-larval stage at 20°C and treated with 1 μg/ml of DTT alone or with chenodeoxycholic acid 100μM for 24 hours and imaged. 10 μg/ml of DTT induces hsp-4::gfp expression and it was used as positive control.
Growth and handling of microbes used
16S ribosomal sequence was amplified using 16S-specific primers and sequenced to identify the microbes. Brain Heart Infusion media as well as plates were used for culturing and testing the effect of Enterococcus faecalis and E. faecium on worms. For Sacchromyces boulardi, YPD media was used for growing the yeast and a concentrated culture was added to SK media. For Lactobacillus spp, Lactobacilli broth was used for growing and then a concentrated culture was added to SK media. SK media was used for all the other microbes.
RT-qPCR experiments with worms fed on Kocuria
For the RT-PCR experiments involving Kocuria, synchronized L1-larval stage wildtype or eft-3(q145) homozygotes were fed on E.coli OP50 until L3-larval stage. The worms were subsequently washed several times in M9 to remove the adhering E.coli and then the worms were dropped onto SK medium plates containing either E.coli OP50 or Kocuria. The plates were incubated at 20°C for 40 hours and the worms were washed off the plates using M9. Following several washes to remove the bacteria, the worms were resuspended in M9. ~ 300 worms were used to isolate RNA using TRI Reagent (Sigma), followed by chloroform extraction and isopropanol precipitation. RNA was DNAase treated using the TURBO DNA-free kit (Applied Biosystems). cDNA was prepared using the First strand cDNA synthesis kit from Invitrogen. qRT-PCR was performed with an iCycler machine (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). All reactions were done in triplicate and on at least 2–4 biological replicates. All the values are normalized to ama-1 as internal control as well as to the transcript levels in untreated wildtype. The Primer sequences used are given above.
Preparation of Kocuria extracts
K.rhizophila was grown in LB media for 5 days at 37°C and the culture were centrifuged at 4000 RPM for 20 min at 4°C. The supernatant was passed through a 0.45micron filter twice and the eluate was collected. 400μl of the eluate was added to 10-cm preseeded OP50 E.coli bacteria containing NGM plates. After the liquid has evaporated, ~100 L1-larval stage eft-3(q145);pgp-5::gfp–germline translation defective mutant worms were added to the plates and incubated at 20°C. When the worms reached adult stage (~60 hours after seeding onto plates), they were scored visually for the pgp-5::gfp induction. Supernatants prepared from OP50 grown in LB media using the same protocol mentioned above was used as control.
Preparation of heat-inactivated Kocuria
For preparing heat killed K.rhizophila, 5 ml overnight culture of K.rhizophila grown in LB media was centrifuged at 4000 RPM for 20 min at 4°C. The supernatant was removed and the pelleted bacteria were resuspended in 1 ml of M9. 500μl of the bacteria was heat inactivated at 70°C for 2 hours while the rest was used as control. 100 μl of the heat-inactivated culture was streaked onto LB media plates to ensure that bacteria are inviable. Following heat inactivation, 200 μl bacteria were seeded onto SK-media plates, dried and ~100 L1-larval stage eft-3(q145);pgp-5::gfp–germline translation defective mutant worms were added to the plates and incubated at 20°C. When the worms reached adult stage (~60 hours after seeding onto plates), they were scored visually for the pgp-5::gfp induction. 200 μl bacteria of the control non heat-inactivated bacteria prepared above were used as controls.
Phospho-PMK-1 activation staining assay with worms fed on Kocuria
Synchronized L1-larval stage wildtype or eft-3(q145) homozygotes were fed on E.coli OP50 until L3-larval stage. The worms were subsequently washed several times in M9 to remove the adhering E.coli and then the worms were dropped onto SK medium plates seeded with either E.coli OP50 or Kocuria. The plates were incubated at 20°C for 40 hours and ~100 worms were picked onto glass dish containing M9. The intestine of the worms was dissected in M9 using 22-gauge hypodermic needles by cutting either near the pharynx or tail region. The intestine is released from the worm because of pressure and is gently separated from the worm carcass. The dissected intestine was fixed with 1% paraformaldehyde in M9 for 10 min and subsequently fixed in ice-cold methanol for 5 min at room temperature. The dissected intestine was washed three times by centrifugation at 2000 RPM for 1 minute with 1X PBS containing 0.1% Tween 20 (PBST). Following the fixation, the dissected intestine was blocked with PBST containing 1mg/ml BSA for one hour at room temperature. After blocking, the intestine was stained with Anti-ACTIVE® p38 pAb, Rabbit, (pTGpY) from Promega (Catalog no: V1211) was diluted 1:1000 in PBST containing 1mg/ml BSA for 2 hour at room temperature. The dissected intestine was washed three times in PBST and then stained with Goat Anti-Rabbit IgG (Catalog no:111-165-144) secondary antibody from Jackson Research diluted in 1:1000 in PBST containing 1mg/ml BSA for 2 hour at room temperature. The intestine was washed three times in PBST and then mounted on 2% agarose pad on glass slide with Vectashield® containing DAPI from Vector Labs for imaging.
PMK-1 nuclear localization assay with worms fed on Kocuria
Synchronized L1-larval stage pmk-1::mCherry carrying transgenic worms or eft-3(q145); pmk-1::mCherry carrying transgenic worms and were fed on E.coli OP50 until L3-larval stage. The worms were subsequently washed several times in M9 to remove the adhering E.coli and then the worms were dropped onto SK medium plates seeded with either E.coli OP50 or Kocuria. The plates were incubated at 20°C for 40 hours. The worms were mounted onto 1% agarose pads and imaged using a Zeiss AXIO Imager Z1 microscope fitted with a Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software.
Kocuria feeding suppresses pgp-5::gfp activation in response to germline translation inhibition
Synchronized L1-larval stage eft-3(q145); pgp-5::gfp worms were fed on E.coli OP50 until L3-larval stage. The worms were subsequently washed several times in M9 to remove the adhering E.coli and then the worms were dropped onto SK medium plates seeded with either E.coli OP50 or Kocuria. The plates were incubated at 20°C for 40 hours. The worms were mounted onto 1% agarose pads and imaged using a Zeiss AXIO Imager Z1 microscope fitted with a Zeiss AxioCam HRm camera and Axiovision 4.6 (Zeiss) software. Similar experimental procedure was used to test other Kocuria species and Micrococcus luteus.
Supplementary Material
Acknowledgments
Strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and Shohei Mitani of the Japanese National Bioresources Project. This work was supported in part by the National Institutes of Health Grant AG043184-16A (to G.R.). Thanks to David Coil and Jonathan Eisen for providing Kocuria species.
Footnotes
Author contributions
J.A.G. and G.R conceived the project. E.J. and E.M. provided the microbes used in the experiments. E.J. performed the bacterial suppression experiments. J.L.F. helped with the data collection and analysis. X.Z. conducted the qRT-PCR experiments. P.B. constructed transgenic pmk-1::mcherry lines. J.A.G. and G.R wrote the paper.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Xu CC, Li CYTC, Kong ANTA. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. 2005;28:249–268. doi: 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- 2.Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunbar TLT, Yan ZZ, Balla KMK, Smelkinson MGM, Troemel EREC. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11:375–386. doi: 10.1016/j.chom.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwan DLD, Kirienko NVN, Ausubel FMF. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waitz JA, Sabatelli F, Menzel F, Moss EL. Biological activity of antibiotic G-418, a new micromonospora-produced aminoglycoside with activity against protozoa and helminths. Antimicrob Agents Chemother. 1974;6:579–581. doi: 10.1128/aac.6.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pittenger RC, Wolfe RN, Hoehn PN, Daily WA, McGuire JM. Hygromycin. I. Preliminary studies on the production and biologic activity of a new antibiotic. Antibiot Chemother (Northfield Ill) 1953;3:1268–1278. [PubMed] [Google Scholar]
- 7.Maciejowski JJ, et al. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanazawa M, et al. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech Dev. 2004;121:213–224. doi: 10.1016/j.mod.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Mertenskötter A, Keshet A, Gerke P, Paul RJ. The p38 MAPK PMK-1 shows heat-induced nuclear translocation, supports chaperone expression, and affects the heat tolerance of Caenorhabditis elegans. Cell Stress Chaperones. 2013;18:293–306. doi: 10.1007/s12192-012-0382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferdinandusse S, et al. Mutations in the gene encoding peroxisomal sterol carrier protein X (SCPx) cause leukencephalopathy with dystonia and motor neuropathy. Am J Hum Genet. 2006;78:1046–1052. doi: 10.1086/503921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Autio KJ, et al. Role of AMACR (α-methylacyl-CoA racemase) and MFE-1 (peroxisomal multifunctional enzyme-1) in bile acid synthesis in mice. Biochem J. 2014;461:125–135. doi: 10.1042/BJ20130915. [DOI] [PubMed] [Google Scholar]
- 12.Kim DH, et al. Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci U S A. 2004;101:10990–10994. doi: 10.1073/pnas.0403546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuno T, et al. The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 2004;23:2226–2234. doi: 10.1038/sj.emboj.7600226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher RA, et al. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci U S A. 2009;106:1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Motola DL, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 16.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillion M, et al. Comparative study of normal and sensitive skin aerobic bacterial populations. Microbiologyopen. 2013;2:953–961. doi: 10.1002/mbo3.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeeuwen PL, et al. Microbiome dynamics of human epidermis following skin barrier disruption. Genome Biol. 2012;13:R101. doi: 10.1186/gb-2012-13-11-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker K, et al. Kocuria rhizophila adds to the emerging spectrum of micrococcal species involved in human infections. J Clin Microbiol. 2008;46:3537–3539. doi: 10.1128/JCM.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moissenet D, et al. Persistent bloodstream infection with Kocuria rhizophila related to a damaged central catheter. J Clin Microbiol. 2012;50:1495–1498. doi: 10.1128/JCM.06038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azzopardi EAE, Azzopardi SMS, Boyce DED, Dickson WAW. Emerging gram-negative infections in burn wounds. J Burn Care Res. 2011;32:570–576. doi: 10.1097/BCR.0b013e31822ac7e6. [DOI] [PubMed] [Google Scholar]
- 22.Obata T, Goto Y, Kunisawa J, Sato S. Indigenous opportunistic bacteria inhabit mammalian gut-associated lymphoid tissues and share a mucosal antibody-mediated symbiosis. Proc Natl Acad Sci U S A. 2010;107:7419–7424. doi: 10.1073/pnas.1001061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnenberg GF, et al. Innate Lymphoid Cells Promote Anatomical Containment of Lymphoid-Resident Commensal Bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright MR, Griffin CT. Specificity of Association between Paenibacillus spp. and the Entomopathogenic Nematodes, Heterorhabditis spp. Microb Ecol. 2004;48:414–423. doi: 10.1007/s00248-003-0166-0. [DOI] [PubMed] [Google Scholar]
- 25.Montalvo-Katz S, Huang H, Appel MD. Association with Soil Bacteria Enhances p38-Dependent Infection Resistance in Caenorhabditis elegans. Infect Immun. 2013;81:514–520. doi: 10.1128/IAI.00653-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Hou A. Host-Microbe Interactions in Caenorhabditis elegans. ISRN microbiol. 2013;2013:356451. doi: 10.1155/2013/356451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son SH, Khan Z, Kim SG, Kim YH. Plant growth-promoting rhizobacteria, Paenibacillus polymyxa and Paenibacillus lentimorbus suppress disease complex caused by root-knot nematode and fusarium wilt fungus. J Appl Microbiol. 2009;107:524–532. doi: 10.1111/j.1365-2672.2009.04238.x. [DOI] [PubMed] [Google Scholar]
- 28.Stuart LM, Paquette N, Boyer L. Effector-triggered versus pattern-triggered immunity: how animals sense pathogens. Nat Rev Immunol. 2013;13:199–206. doi: 10.1038/nri3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saridaki A, Bourtzis K. Wolbachia: more than just a bug in insects genitals. Curr Opin Microbiol. 2010;13:67–72. doi: 10.1016/j.mib.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Fenn K, Blaxter M. Wolbachia genomes: revealing the biology of parasitism and mutualism. Trends Parasitol. 2006;22:60–65. doi: 10.1016/j.pt.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Widmann B, et al. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol Biol Cell. 2012;23:22–35. doi: 10.1091/mbc.E11-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 36.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.