Abstract
Oscillatory gene expression is fundamental to mammalian development, but technologies to monitor expression oscillations are limited. We have developed a statistical approach called Oscope to identify and characterize the transcriptional dynamics of oscillating genes in single-cell RNA-seq data from an unsynchronized cell population. Applications to a number of data sets demonstrate the utility of the approach and also identify a potential artifact in the Fluidigm C1 platform.
Oscillatory gene expression is fundamental to mammalian development, homeostasis, and function1, yet technologies to monitor expression oscillations are limited. Recent advances in live-cell imaging have improved the sensitivity and specificity with which continuous measurements can be made within a single cell2, but due to limitations associated with reporters and detection channels, relatively few genes can be monitored in any given experiment. To study transcriptional oscillations on a genome-wide scale, mRNA microarray or RNA-seq time series experiments are often conducted3. Despite the benefits, heterogeneity in gene-specific frequency and phase make it difficult to identify an optimal sampling rate; and these methods require large quantities of synchronized starting material and consequently are limited to measurements of expression averaged over thousands of cells. Averaging over cells may miss or even misrepresent4 oscillations. Cell synchronization prior to profiling attenuates a number of these problems to enable study of a known oscillatory system (typically the cell cycle), but can dramatically alter the transcriptional dynamics of others, and does not facilitate de novo discovery.
Single cell RNA-seq (scRNA-seq) is a promising technology that allows for genome-wide expression profiling within a single cell, and thereby has the potential to capture a more precise representation of oscillation dynamics as well as unmask oscillations that are missed in bulk expression experiments. However, continuous monitoring within a cell is not possible, and high-resolution scRNA-seq time series experiments in distinct cells are prohibitive given the time required for sample preparation and sequencing. Even when scRNA-seq time series experiments become feasible, challenges associated with rate heterogeneity, sampling, and synchronization will remain.
Computational algorithms have been developed to address some of these challenges in both microarray5,7 and scRNA-seq studies4, but none are focused on identifying oscillating genes. Most are based on the recognition that different samples represent distinct states in a system, such as time points along a continuum or progression toward an endpoint. By obtaining multiple samples at a single5,7 or a few4 time points, and computationally reconstructing an appropriate order, temporal or other meaningful dynamics can be resolved. A key assumption that enables ordering is that genes do not change direction very often and thus samples with similar transcriptional profiles should be close in order.
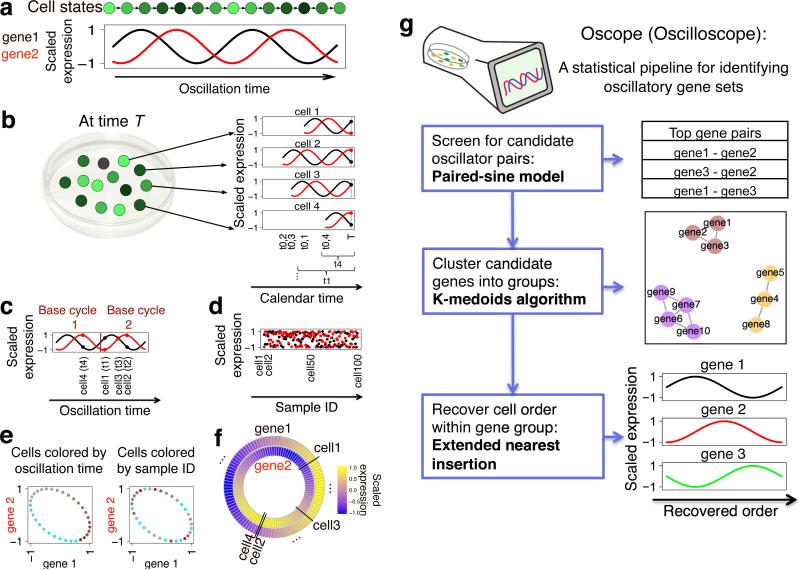
Oscillating genes pose challenges for these types of approaches since genes following the same oscillatory process need not have similar transcriptional profiles. Two genes with an identical frequency that are phase shifted, for example, will have little similarity (Fig. 1a). We have developed an approach called Oscope to identify oscillating genes in static, unsynchronized, scRNA-seq experiments. Like previous algorithms, Oscope capitalizes on the fact that cells from an unsynchronized population represent distinct states in a system. However, unlike previous approaches, we do not attempt to construct a linear order based on minimizing change among adjacent samples. Rather, Oscope utilizes co-regulation information among oscillators to identify groups of putative oscillating genes, and then reconstructs the cyclic order of samples for each group, defined as the order that specifies each sample's position within one cycle of the oscillation (referred to as a base cycle). As detailed below and in Online Methods, the reconstructed order aims to recover gene-specific cyclic profiles defined by the group's base cycle allowing for phase shifts between different genes. Importantly, for different groups of genes following independent oscillatory processes and/or having distinct frequencies, the cyclic orders of cells need not be the same (see Supplementary Fig. 1).
Figure 1.
Overview of Oscope. (a) Shown are an oscillating gene group with two genes and corresponding cell state. (b) In an unsynchronized scRNA-seq experiment, mRNA is collected at time T from cells in varying states. t0,i and ti show cell i's oscillation start time and oscillation time, respectively. (c) The same genes and cells as in b, where cells are ordered by the genes’ oscillation times. (d) Expression for 100 unsynchronized cells. (e) Scatter plots of gene 1 vs. gene 2, which are independent of order. Cells are colored from cyan to brown following the x-axes of c and d, respectively. (f) Results of base cycle reconstruction for the 100 cells shown in d. (g) Flowchart of the Oscope pipeline (see Online Methods).
Oscope is developed to identify oscillators in the scenario where a single cell's mRNA expression is oscillating through cell states. An overview is provided (Fig. 1a-g). Specifically, shown is a schematic of a single-cell oscillating through cell states (Fig. 1a). For simplicity, states are defined there by oscillations in just two genes. In a typical scRNA-seq experiment, cells are collected at the same calendar time T. However, without synchronizing, cells will be in different states at time T and consequently will have different gene expression values (Fig. 1b). If it were possible to sort cells by the oscillation times of genes, defined as the amount of calendar time the cell has been oscillating prior to collection time T, identifying oscillating genes and characterizing their dynamics would be straightforward (Fig. 1c). However, oscillation time is unobserved in an scRNA-seq experiment. With this type of snapshot data, gene expression of oscillating genes is indistinguishable from random noise (Fig. 1d) and therefore existing methods8,9 for identifying cyclic features are not applicable here. Recognizing that a scatter plot of expression values for genes oscillating with similar frequency will form an ellipse independent of order (Fig. 1e), Oscope fits a 2-dimensional sinusoidal function to all gene pairs and chooses those with high scores. Note the elliptical shape is preserved when the oscillation has varying speed and/or when partial synchronization happens (see Supplementary Fig. 2). Once candidate genes are identified, the K-medoids algorithm is applied to cluster genes into groups with similar frequencies, but possibly different phases. Then, for each group, Oscope recovers the cyclic order which places cells by their position within one cycle of the oscillatory process underlying the group. Given static data, it is not possible to reconstruct multiple cycles of an oscillatory process since the dynamics of late cycles are identical to those of earlier cycles, by definition. For example, the gene expression values in cells 2 and 4 are identical (Fig. 1b), even though cell 2 has passed through a full cycle but cell 4 has not. Here we define the base cycle as the minimal cycle that is repeated in an oscillatory process (an example is shown in Fig. 1c). Oscope uses an extended nearest insertion algorithm to order cells with respect to their position in a base cycle without specifying a direction of time (Fig. 1f).
The nearest insertion algorithm10 was developed to address the traveling salesman problem. Given pairwise distances among cities, the nearest insertion algorithm provides a computationally efficient way to order cities so that overall distance travelled is minimized. Here, we extend the nearest insertion algorithm to order cells within an oscillatory gene group so that distance between each gene's expression and its gene-specific base cycle profile is minimized on average over all genes in the group. Once the recovered order for each group is obtained, if it is of interest to estimate phase or further characterize oscillations, subsequent algorithms developed for time course analysis may be applied (e.g. Fourier transformation, spline fitting, etc.).
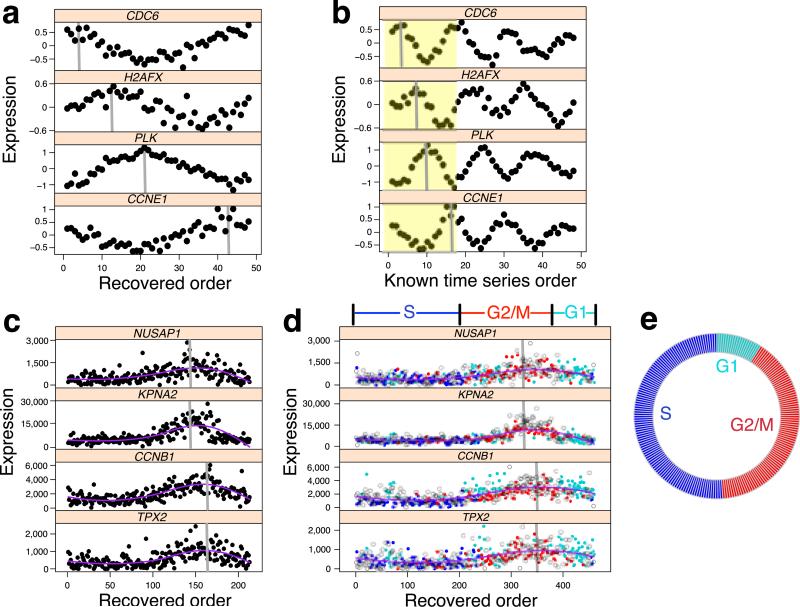
To evaluate the ability of Oscope to identify oscillating groups of genes and reconstruct the cyclic order underlying their base cycles, we consider a time series experiment where oscillating genes were of interest8 and applied Oscope after permuting the sample order. The top gene group identified by Oscope had 151 genes, 116 of which were validated as oscillating in earlier work8. Shown are the reconstructed base cycles for four of the genes identified by Oscope (Fig. 2a) compared with the known time series order (Fig. 2b). The results demonstrate that Oscope successfully recovered the base cycle profile of each gene; phase shifts are also correctly inferred (Supplementary Fig. 3 shows all 151 genes). The results of simulation studies and additional case studies provide further insights into the operating characteristics of the approach (Supplementary Note, Supplementary Fig. 4-10 and Supplementary Table 1).
Figure 2.
Oscope uncovers oscillatory signals in case study datasets. (a) Four genes in the time series data from Whitfield et al. 20028 with profiles ordered by Oscope; the peak of the base cycle is marked in gray. (b) The same four genes following the known order over time with the peak of the first base cycle (shown in yellow) marked in gray. (c) Four genes from a 29 gene group identified by Oscope using scRNA-seq data from 213 unlabeled hESCs. Shown are Oscope recovered profiles. (d) The same four genes ordered using 460 cells (213 unlabeled and 247 H1-Fucci cells are shown as open circles and dots, respectively). The Fucci labels (ignored prior to applying Oscope) are shown in different colors for the 247 cells. Phase boundaries defined by the reconstructed order are shown above the plots. (e) The proportion of unlabeled cells that fall into each phase defined by the boundaries in d.
To further evaluate Oscope on scRNA-seq data, we profiled single undifferentiated human embryonic stem cells (hESCs)11. We applied Oscope to three replicate scRNA-seq experiments on H1 hESCs (n=213). One of the top groups identified by the K-medoids algorithm in Oscope contained 29 genes (Supplementary Table 2), 21 of which are annotated as belonging to the Gene Ontology Cell Cycle biological process (GO:0007049). The reconstructed base cycle is characterized by peaked expression of genes known to be involved in G2 phase progression (e.g. NUSAP1 and KPNA2) and M phase progression (e.g. CCNB1 and TPX2)12 (Fig. 2c and Supplementary Fig. 11). To confirm whether the recovered profiles were associated with cell cycle phasing, we performed additional scRNA-seq experiments (n=247) on H1 hESCs harboring the fluorescent ubiquitination-based cell cycle indicators13 (H1-Fucci, see Online Methods) in which cells were identified as being in G1, S, or G2/M phase. We combined the H1 and H1-Fucci data sets and applied Oscope. The reconstructed order using the same 29 genes largely recapitulates the three phases of the cell cycle (Fig. 2d and Supplementary Fig. 12). The phase boundaries defined by the reconstructed order classified 72% of H1-Fucci hESCs into the correct phase. Since the H1-Fucci data set does not provide an unbiased estimate of the number of cells in each phase, we classified the unlabeled H1 hESCs by the phase boundaries and estimated the proportion in each phase. The proportion of unlabeled H1 cells in each phase is consistent with the notion of a shortened G1 phase in undifferentiated hESCs14(Fig. 2e). Out of the eight genes that were not annotated as belonging to the cell cycle pathway, six of them have been shown to be associated with cell cycle in a previous publication12. All eight genes, including the two less well-characterized oscillatory genes CALM2 and ZNF165, show cell cycle related base cycle profiles (Supplementary Fig. 13).
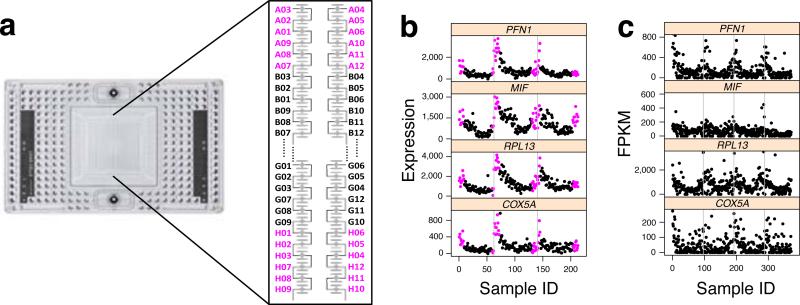
A second top group of genes identified by Oscope showed an oscillatory pattern related to the capture site and output well positions on the Fluidigm C1 chip (Supplementary Table 3). In particular, these genes all have increased expression in cells captured in sites with small or large plate output IDs, across all three replicate hESC scRNA-seq experiments. The capture sites involving increased gene expression are physically located close to each other on the chip (Fig. 3a, the capture sites’ corresponding plate output IDs are labeled following the recommendation by the manufacture user guide). To examine this potential artifact, we developed an ANOVA-based artificial trend detection algorithm (Online Methods), and applied the algorithm on the combined data from the three H1 experiments. We found that 403 genes show strong artificial trends (Supplementary Fig. 14 and Supplementary Table 4), consistent in each experiment (Fig. 3b). To further investigate the artifact and to rule out biases that may be due to sequencing, we estimated expression via qPCR on select genes (Supplementary Fig. 15) and found the trend already present in the full-length single-cell cDNA libraries. We also see this trend in publicly available datasets from other labs using various cell types (Fig. 3c and Supplementary Fig. 16).
Figure 3.
Oscope uncovers dynamic signals of technical origin in scRNA-seq datasets. (a) Default plate output ID layouts of the capture sites on the C1 chip. (b) Expression of four genes with potential ordering effects. Cells are ordered by the C1 plate output ID (A01-A12, B01-B12, ..., H01-H12). Cells from the colored capture sites in a are also shown in magenta. Three replicate hESC experiments are separated by gray lines. (c) The same four genes for a data set obtained from Trapnell et al., 2014, ordered following the cell order listed in their supplementary data4. The four experiments are separated by gray lines. The y-axes are limited to 98th quantile of gene-specific FPKMs for better visualization.
The scRNA-seq technology offers an unprecedented ability to snapshot genome-wide transcription in single cells, but is not amenable to longitudinal studies that monitor changes in individual cells in situ. Oscope allows investigators to identify and characterize oscillating gene groups. Applications in a number of settings should improve our understanding of known oscillators, as well as facilitate the discovery of novel ones. Furthermore, adjusting for oscillators using the characterization provided by Oscope should increase the power to investigate other signals associated with differentiation and/or subpopulations15.
ONLINE METHODS
Oscope: paired-sine model
An oscillatory gene group is a group of genes having the same frequency with phase shifts that may vary among pairs but are preserved across cells. For example, if ψgi,gj,s denotes the phase shift between gi and gj in cell s, then ψgi,gj,s needs not equal ψgj,gk,s, but ψgi,gj,1 = ψgi,gj,2 = ··· = ψgi,gj,S. Oscillation time is the difference between cell collection time T and the start of oscillation.
For a pair of genes g1 and g2, denote the matched gene expression (rescaled to [−1, 1]) in S cells as (Xg1,1, Xg2,1), (Xg1,2, Xg2,2), ... , (Xg1,S, Xg2,S). If the two genes follow a sinusoidal process with a phase shift, then the following equations hold for each cell s in 1,2, ··· , S: Xg1,s = sin(ts = φg1) and Xg2,s = sin(ts + φg1 + ψg1,g2), where ts indicates oscillation time of cell s; φg1 indicates the starting phase of gene 1; and ψg1,g2 indicates the phase shift between the two genes where the subscript s is dropped since ψg1,g2 is assumed common to all cells.
By trigonometric identities,
Given this, the following equation holds for any cell:
and there exists an optimal ψg1,g2 for which the error term is zero, where
To search for gene pairs with associated dynamic changes, Oscope linearly rescales gene-specific gene expression measurements to range between −1 and 1, and estimates the optimal ψgi,gj for all gene pairs (gene i, gene j) defined as that which minimizes . With this metric, gene pairs are rank ordered by ; and candidate oscillatory genes are those genes in the top gene pairs (Oscope's default is the top 5%; this threshold may be changed by users based on the empirical distribution of the 's).
Oscope: K-medoids clustering
To cluster the candidate oscillatory genes detected from the paired-sine model into distinct groups, we use the K-medoids algorithm with as the dissimilarity metric. With this metric, gene pairs with small 's are more likely to be clustered together. The optimal K is picked by maximizing the Silhouette distance. To avoid detecting gene groups with a purely linear relationship, only groups having within-group phase differences are further considered in order recovery. Specifically, for any pair of genes gi,gj within a group, we define the phase-shift residual as υgi,gj = min((π – ηgi,gj), ηgi,gj), in which ηgi,gj = (ψgi,gj mod π). Oscope's default takes groups whose 90th quantile of υgi,gj's is greater than π/4 for further order recovery.
Oscope: Extended Nearest Insertion
We developed an extended nearest insertion (ENI) algorithm to recover the cyclic order for each oscillatory group defined in the K-medoids clustering step. Cells are ordered cyclically according to their position within one cycle of the oscillation, referred to as a base cycle. The ENI starts with three randomly selected cells and forms a loop (undirected graph). A 4th cell is chosen at random and inserted into the three cell-cell gaps on the loop. This forms three candidate orders. We evaluate each order using the aggregated mean squared error (MSE) of a sliding polynomial regression (SPR). For a given order, SPR is fitted to the expression of each gene. To capture cyclic features of the data, SPR fits m polynomial regression models starting with m evenly distributed points on the loop. The largest MSE among the m models is defined as the MSE of the SPR for this gene. For each order, the aggregated MSE of an oscillatory gene group is defined as the summation of the MSE's among all genes. The optimal order of the first 4 cells is then selected as the one that minimizes the aggregated MSE. This process is repeated to insert the fifth cell and so on, until all cells are in the loop. A 2-opt algorithm is then applied to avoid finding local maxima.
Whitfield data and statistical analysis
Microarray gene expression data were downloaded from http://genome-www.stanford.edu/Human-CellCycle/HeLa/. In total, five experiments were available at this site from Whitfield et al., 20028; experiment 3 was used here as it has the largest sample size. For this experiment, double thymidine block was used to synchronize HeLa cells and expression was profiled for 9,559 genes at 48 time points following synchronization. To minimize the effect of outliers, gene-specific values > 95th (< 5th) quantile of expression were imputed to the 95th (5th) quantile. Oscope was applied on the data with permuted sample order (Supplementary Table 5). After applying the paired-sine model to all genes, the top 5% were used as input for the K-medoids algorithm. Using the 151 genes in the top cluster (Supplementary Table 6), the ENI algorithm was applied with m = 4 and the degree of freedom of SPR was set to 3. To obtain the optimal order, the 2-opt algorithm was applied with 20,000 iterations (Supplementary Table 7). 874 genes were defined as periodic by the auto-regression model in Whitfield et al., 20028. We used these 874 genes as a validation set in our evaluation.
H1 hESC cell culture
Undifferentiated H1 human embryonic stem cells (hESCs) were cultured in E8 medium16 on Matrigel-coated tissue culture plates with daily media feeding at 37 °C with 5% (vol/vol) CO2. Cells were split every 3-4 days with 0.5 mM EDTA in 1 × PBS for standard maintenance. Immediately before preparing single cell suspensions for each experiment, hESCs were individualized by Accutase (Life Technologies), washed once with E8 medium, and resuspended at densities of 5.0-8.0 × 105 cells/mL in E8 medium for cell capture. The H1 hESCs is registered in the NIH Human Embryonic Stem Cell Registry with the Approval Number: NIHhESC-10-0043. Details of the H1 cells can be found online (http://grants.nih.gov/stem_cells/registry/current.htm?id=29). All the cell culture performed in our laboratory have been routinely tested negative for mycoplasma contamination and authenticated by cytogenetic tests.
H1 hESC single cell capture and single-cell cDNA library preparation
Single-cell loading, capture, and library preparations were performed following the Fluidigm user manual “Using the C1 Single-Cell Auto Prep System to Generate mRNA from Single Cells and Libraries for Sequencing.” Briefly, 5,000-8,000 cells were loaded onto a medium size (10-17 μm) C1 Single-Cell Auto Prep IFC (Fluidigm), and cell-loading script was performed according to the manufacturer's instructions. The capture efficiency was inspected using EVOS FL Auto Cell Imaging system (Life Technologies) to perform an automated area scanning of the 96 capture sites on the IFC. Empty capture sites or sites having more than one cell captured were first noted and those samples were later excluded from further library processing for RNA-seq. Immediately after capture and imaging, reverse transcription and cDNA amplification were performed in the C1 system using the SMARTer PCR cDNA Synthesis kit (Clontech) and the Advantage 2 PCR kit (Clontech) according to the instructions in the Fluidigm user manual. Full-length, single-cell cDNA libraries were harvested the next day from the C1 chip and diluted to a range of 0.1-0.3 ng/μL. Diluted single-cell cDNA libraries were fragmented and amplified using the Nextera XT DNA Sample Preparation Kit and the Nextera XT DNA Sample Preparation Index Kit (Illumina). Libraries were multiplexed at 24 libraries per lane, and single-end reads of 67-bp were sequenced on an Illumina HiSeq 2500 system.
H1 hESC: read mapping and quality control
Reads were mapped against the Hg19 Refseq reference via Bowtie 0.12.817 allowing up to two mismatches and up to 20 multiple hits. The expected counts and TPM's were estimated via RSEM 1.2.318. Cells having less than 5,000 genes with TPM > 1 were removed in quality control. 62, 78 and 73 cells passed the quality control in three replicate hESC experiments for a total of 213 H1 hESCs.
H1 hESC: statistical analysis
Expression within each cell was normalized following median normalization19 implemented in EBSeq 1.5.420. Gene means and variances were also estimated using EBSeq after adjusting for library sizes. High mean and high variance genes were selected prior to applying Oscope. Specifically, we took genes with mean expected count greater than 100 as genes with high mean. To define high variance genes, we fit a linear model on log(variance) ~ log(mean) + c. Genes with variance above the fitted line were defined as high variance genes. Genes with mappability scores18 less than 0.8 were further eliminated. Applying these steps to the 213 H1 hESCs gave 2,376 genes to which Oscope was applied (Supplementary Table 8). To minimize the effect of outliers, gene-specific values > 95th (< 5th) quantile of expression were imputed to the 95th (5th) quantile. After applying the paired-sine model, the top 5% of genes were used as input for the K-medoids algorithm. Using the 29 genes in the cell cycle cluster, the ENI module was applied with m = 4 and the degree of freedom of SPR was set to 3. To obtain the optimal order, the 2-opt algorithm was applied with 20,000 iterations (Supplementary Table 9).
H1-Fucci hESC cell line
Fluorescent ubiquitination-based cell cycle indicator (Fucci) H1 hESCs were generated by PiggyBAC insertion of a cassette encoding an EEF1A promoter-driven mCherryCDT1-IRES-EgfpGMNN double transgene (custom ordered from GenScript). Individual clones were isolated by sorting double-positive single cells by fluorescence activated cell sorting (FACS) and maintained as described above. The H1-Fucci cell line provides a two color fluorescence labeling system allowing single-cell suspensions from G1, S or G2/M cell-cycle phases to be isolated by FACS, followed by loading single-cell suspensions onto the Single-Cell Auto Prep IFC using a medium size (10-17 μm) chip. FACS was performed on the FACSAria IIIu instrument and using FACSDiva software version 6.1.3 (both from Becton Dickinson). Unlabeled H1 cells or cells stained with single fluorochromes served as controls for fluorescence gating. Libraries and sequencing reads were processed in the same manner as described above.
H1-Fucci: read mapping, quality control and statistical analysis
Reads were processed in the same way as in the H1 hESC data. A total of 91, 80 and 76 cells in G1, S and G2/M, respectively, passed our quality control criteria as defined in the H1 hESC read mapping and quality control section. Statistical analysis on H1 and H1-Fucci combined data was carried out as described in H1 hESC statistical analysis. Based on the recovered order (Supplementary Table 10), the phase boundaries (Fig. 2d) are defined as the boundaries that give the smallest misclassification rate between three cell cycle phases based on the reconstructed order.
Statistical model to identify genes with ordering effects
We used an ANOVA model to identify genes with potential ordering effects. Within each H1 hESC experiment, we grouped cells into eight groups defined by capture site. Recall that capture sites are labeled as A01, ... , A12, B01, ..., B12, ..., H01, ..., H12 to match their corresponding position in the output wells (Fig. 3a and Supplementary Table 11). We grouped cells from sites with the same starting letters. For each gene, we applied an ANOVA model on the combined data set from all three H1 hESC experiments. The model tests for differences in mean expression across the eight cell groups. A total of 403 genes were identified (p-value < 0.005) using this ANOVA approach.
Single-cell real-time quantitative PCR (qPCR)
Single-cell cDNA harvested from the Fluidigm C1 IFC was transferred to a 96-well plate and subsequently quantified and diluted according to the Fluidigm user manual. Two microliters of the diluted single-cell cDNA were subsequently used in replicated qPCR reactions with individual 1 × TaqMan Gene Expression assays and 1 × TaqMan Universial PCR Master Mix II (Life Technologies) in a total volume of 10.0 μL. qPCR was performed using ViiA™ 7 System; and data analysis was performed using ExpressionSuite™ (all from Life Technologies). TaqMan Gene Expression assays (Life Technologies) were used for two genes: PFN1 (Hs00748915_s1), MIF (Hs00236988_g1), with GAPDH (Hs02758991_g1) as an internal control. Although the TaqMan Gene Expression assay are compliant with the MIQE guidelines for publications, the actual sequences of the primers and probes are not released for each assay. The amplicon context sequence for each assay can be identified as following:
>PFN1: 223 bps (5’-to-3’)
ccaccttcggcgttcccagtactgacctcgtctgtcccttccccttcaccgctccccacagctttgcacccctttcctccccatacac acacaaaccattttattttttgggccattaccccataccccttattgctgccaaaaccacatgggctgggggccagggctggatgg acagacacctccccctacccatatccctcccgtgtgtggttggaaaact
>MIF: 83 bps (5’-to-3’)
ctgtgcggcctgctggccgagcgcctgcgcatcagcccggacagggtctacatcaactattacgacatgaacgcggccaatgt
>GAPDH: 110 bps (5’-to-3’)
ccctggccaaggtcatccatgacaactttggtatcgtggaaggactcatgaccacagtccatgccatcactgccacccagaaga ctgtggatggcccctccgggaaactg
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health GM102756, 4UH3TR000506-03, 5U01HL099773-06, the Charlotte Geyer Foundation, and the Morgridge Institute for Research. N.L. was supported by the Shapiro Fellowship. C.B. was supported by the Canadian Institutes of Health Research Banting Postdoctoral Fellowship. We thank M. Probasco and N. Propson for their assistance of sorting cells by FACS; J. Bolin, A. Elwell, and B.K. Nguyen for the preparation and sequencing of the RNA-seq samples. We thank A. Gitter, K. Korthauer and R. Bacher for comments that helped improve the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
N.L., L.C., R.M.S., J.A.T. and C.K. designed research, analyzed data and wrote the manuscript; C.B. generated the H1-Fucci cell line; Y.L., J.C. and X.L. contributed to the simulation studies; and P.J. performed RNA-seq read mapping, quantification and quality control.
COMPETING FINANCIAL INTERESTS
J.A.T. is a founder, stockowner, consultant, and board member of Cellular Dynamics International (CDI).
ACCESSION CODES
Gene expression omnibus: GSE64016
Code availability. The R package R/Oscope is available at https://www.biostat.wisc.edu/~kendzior/OSCOPE/
REFERENCE
- 1.Aulehla A, Pourquie O. Current opinion in cell biology. 2008;20:632–637. doi: 10.1016/j.ceb.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Shin I, et al. Nucleic acids research. 2014;42:e90. doi: 10.1093/nar/gku297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Joseph Z, Gitter A, Simon I. Nature Reviews Genetics. 2012;13:552–564. doi: 10.1038/nrg3244. [DOI] [PubMed] [Google Scholar]
- 4.Trapnell C, et al. Nature biotechnology. 2014 doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu P, Gentles AJ, Plevritis SK. PLoS computational biology. 2011;7:e1001123. doi: 10.1371/journal.pcbi.1001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, Bar-Joseph Z. Computational Biology and Bioinformatics, IEEE/ACM Transactions on. 2008;5:172–182. doi: 10.1109/TCBB.2007.70233. [DOI] [PubMed] [Google Scholar]
- 7.Magwene PM, Lizardi P, Kim J. Bioinformatics. 2003;19:842–850. doi: 10.1093/bioinformatics/btg081. [DOI] [PubMed] [Google Scholar]
- 8.Whitfield ML, et al. Molecular biology of the cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Lichtenberg U, et al. Bioinformatics. 2005;21:1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 10.Rosenkrantz DJ, Stearns RE, Lewis PM., II SIAM journal on computing. 1977;6:563–581. [Google Scholar]
- 11.Thomson JA, et al. science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 12.Santos A, Wernersson R, Jensen LJ. Nucleic acids research. 2014;43:D1140–D1144. doi: 10.1093/nar/gku1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaue-Sawano A, et al. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 14.Becker KA, et al. Journal of cellular physiology. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 15.Buettner F, et al. Nature biotechnology. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, et al. Nature methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biology. 2010;R25 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Dewey CN. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leng N, et al. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.