Abstract
Detection of minor genetically distinct subpopulations within tumors is a key challenge in cancer genomics. Here we report STAR-FISH (Specific-To-Allele PCR – FISH), a novel method for the combined detection of single nucleotide and copy number alterations in single cells in intact archived tissues. Using this method, we assessed the clinical impact of changes in the frequency and topology of PIK3CA mutation and HER2/ERBB2 amplification within HER2+ breast cancer during neoadjuvant therapy. We found that the two genetic events are not always present within the same cell. Chemotherapy selects for PIK3CA mutant cells, a minor subpopulation in nearly all treatment-naïve samples, and modulates genetic diversity within tumors. Treatment-associated changes in spatial distribution of cellular genetic diversity correlated with poor long-term outcome following adjuvant trastuzumab therapy. Our findings support the use of in situ single-cell based methods in cancer genomics and imply that chemotherapy before HER2-targeted therapy may promote treatment resistance.
Cancer is a genetic disease, and thus the identification of somatic genetic alterations within tumors has been the focus of clinical oncology. Cancer genome sequencing studies have traditionally been performed on bulk tumors limiting their ability to detect minor subclones, which commonly drive therapy resistance1,2. Sequencing of bulk tumors also cannot accurately predict which mutations are present in the same versus in different cells. Sequencing of single cancer cells overcomes these limitations3,4, but currently this is still laborious, expensive and error-prone due the inefficiencies of whole genome amplification and thus, not yet suitable for the analysis of large patient cohorts. We developed a novel methodology termed STAR-FISH based on the combination of in situ PCR5-7 and fluorescence in situ hybridization (FISH)8-10 to enable the simultaneous detection of point mutations and copy number variation at the single cell level in intact formalin-fixed paraffin-embedded (FFPE) tissue samples.
We designed STAR-FISH for several commonly mutated genes in breast cancer focusing on clinically relevant mutational hotspots. PIK3CA is one of the most commonly mutated genes in breast cancer11. Mutations in PIK3CA12, predominantly occurring at two hotspots (p.Glu542Lys, and p.His1047Arg (c.3140A>G)), are present in about 20% of all breast tumors and in up to 40% of HER2+ subtype cancers. The PI3K/AKT pathway has been identified as a major determinant of resistance to trastuzumab, a HER2 (also known as ERBB2) targeting antibody13-19, implying that PIK3CA mutation may be used as a predictor of resistance. However, the significant heterogeneity for PIK3CA mutation both within different regions of the same tumor and also between different lesions in the same patient20,21 make its accurate detection challenging. We applied STAR-FISH to assess changes in intratumor cellular heterogeneity for HER2 amplification and PIK3CA His1047Arg mutation in a cohort of HER2+ breast cancer patients subjected to neoadjuvant chemotherapy followed by adjuvant trastuzumab, and correlated these changes with long-term clinical outcome.
RESULTS
STAR-FISH development and validation
The first step of STAR-FISH is an in situ PCR using mismatched primers designed to specifically amplify mutant and wild type alleles (Fig. 1a, Supplementary Figure 1a, Supplementary Table 1, Supplementary Note). The primers contain a 5’ overhang, a unique sequence not found in the human genome, which serves as a priming site in the second round of PCR. The use of a few amplification cycles in the first round and 30 cycles in the second round of PCR ensures proper amplification of the product with high specificity. PCR products are visualized by hybridization of fluorescently labeled probes complimentary to the 5’ overhang (Fig. 1a). The specificity of the primers for the PIK3CA His1047Arg mutation was first evaluated by PCR using genomic DNA isolated from human breast cancer cell lines with known PIK3CA mutation status (Fig. 1b). The sensitivity of the assay was tested by performing PCR on defined mixtures of DNA from MDA-MB-231 (PIK3CA wild type) and SUM-185PE cells (homozygous for PIK3CA His1047Arg mutation; Supplementary Figure 1b). Primers for the second round of PCR were tested in the same manner (data not shown). We also developed PCR assays for two other commonly occurring mutations in breast cancer, PIK3CA E542K and TP53 R175H mutations (Supplementary Figure 1c,d).
Figure 1.
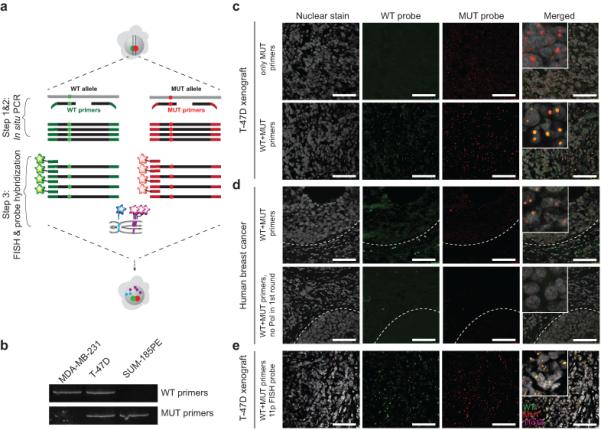
Outline of the STAR-FISH method and its validation. Scale bars represent 75 μm. (a) Schematic of the STAR-FISH protocol on a cell with heterozygous mutation. In step 1 & 2 in situ PCR with a mixture of wild-type (green) and mutant (red) primers is performed. Red and green dots represent the mutation site. In step 3, hybridization of fluorescent probe specific for WT and MUT PCR product is combined with hybridization of BAC (magenta) and CEP (blue) probes for genomic copy number variation detection. (b) PCR to test the specificity of PIK3CA H1047 primers using genomic DNA from breast cancer cell lines with known mutation status. MDA-MB-231 – PIK3CA WT, T-47D – heterozygous PIK3CA His1047Arg, and SUM185PE – homozygous PIK3CA His1047Arg mutation. (c) In situ PCR testing the specificity of primers for WT and His1047Arg MUT PIK3CA on T-47D breast cancer cell line xenografts. Upper panel – only mutant (MUT) primers were used in the first round of PCR and both primers were used in second round of PCR. Lower panel – both WT and MUT primers were used in both round of PCR. (d) In situ PCR for WT and His1047Arg MUT PIK3CA on a human primary breast tumor sample with known PIK3CA His1047Arg mutation. Upper panel – complete in situ PCR reaction. Dashed line – tumor-stroma border. Lower panel – in situ PCR without the polymerase in the first round of PCR. (e) STAR-FISH for WT (green) and His1047Arg MUT (red) PIK3CA in combination with FISH for 11q13x BAC probe (magenta) on T-47D xenografts.
Next, we performed in situ PCR on FFPE tissue slides of xenografts derived from MDA-MB-231 and T-47D breast cancer cell lines (Fig. 1c and Supplementary Figure 1e) followed by testing of primary human breast tumors with known PIK3CA His1047Arg mutation status (Fig. 1d). Signal for wild type and mutant PIK3CA was robustly detected within cancer cells whereas only wild type signal was visible in surrounding stromal cells (Fig. 1d). The false discovery rate (FDR) for PIK3CA mutation detection was equal 1 in 976 cells (FDR = 0.001) based on the analysis of MDA-MB-231 (PIK3CA wild type) cell line-derived xenografts. No signal was detected when the polymerase was omitted in the first round of PCR, confirming the specificity of the in situ PCR step (Fig. 1d). Similarly, in situ PCR was performed for PIK3CA E542K and TP53 R175H on histogel of BT-483 cells and xenografts derived from AU565 cells, respectively, that are known to contain these mutations (Supplementary Figure 1f,g).
To validate the sensitivity and specificity of the PIK3CA in situ PCR, we compared it to three independent methods: fluorescence-activated cell sorting (FACS), immunofluorescence (IF), and mass spectrometry22. For comparison with FACS and IF, we used xenografts derived from defined mixtures of wild-type and mutant cell lines expressing fluorescent tags. Mutant cell fraction determined by STAR-FISH was in accordance with the FACS and IF results and the variance between replicates was not significantly different between the various methods (Supplementary Figure 2a-d). For comparison with mass spectrometry, we performed PIK3CA His1047Arg in situ PCR on a cohort of breast tumors previously genotyped for this mutation by Sequenom MassARRAY23,24. We observed a strong correlation (linear regression y = 1.013x p < 0.0001, R2 = 0.9037 p < 0.0001) between the two methods (Supplementary Figure 3a-c). Interestingly, even in samples where MassARRAY did not detect any PIK3CA mutant alleles with STAR-FISH, we identified rare mutant cells below 8% frequency.
Lastly, we combined in situ PCR for the PIK3CA His1047Arg mutation with FISH for a chromosomal region commonly gained in breast cancer, and validated this method on a xenograft derived from the T-47D cell line that is heterozygous for PIK3CA His1047Arg mutation and has an 11q13 amplification (Fig. 1e). We were able to detect clear signals for 11q13 amplification and both wild type and mutant PIK3CA in individual cancer cells. These results confirmed the high specificity and sensitivity of the STAR-FISH assay for in situ detection of single nucleotide mutations in combination with genomic copy number variation.
STAR-FISH analysis of HER2+ breast tumor samples
We applied STAR-FISH for the analysis of PIK3CA His1047Arg mutation and HER2 amplification status in a cohort of 22 HER2+ breast cancer patients undergoing neoadjuvant chemotherapy followed by adjuvant trastuzumab treatment (Supplementary Figure 4, Supplementary Table 2). For each patient we analyzed a treatment-naïve biopsy sample and surgical excision of the residual tumor after neoadjuvant treatment. In each tissue sample, we evaluated 3-5 distinct areas of the tumor to account for regional heterogeneity. Using STAR-FISH, we quantified copy numbers for HER2 and chromosome 17 centromeric (CEP17) probes, and PIK3CA wild type and mutant signal in individual nuclei (on average 289 nuclei per area; Fig. 2a,b). Cells with nuclei < 200 pixels were excluded as they likely correspond to infiltrating leukocytes. HER2 amplification was defined using the HER2/CEP17 ratio ≥ 2.0 as cutoff25. To facilitate further statistical and intratumor diversity analyses based on these single cell measurements, each cell with evaluable signal was assigned to one of the following categories: WT – a cell with wild type PIK3CA signal and no HER2 amplification, WT+AMP – a cell with wild type PIK3CA signal and HER2 amplification, MUT – a cell with mutant PIK3CA signal and no HER2 amplification, MUT+AMP – a cell with mutant PIK3CA signal and HER2 amplification, and AMP – a cell with no PIK3CA signal and HER2 amplification (Fig. 2c and Supplementary Table 3). A subset of cells with no detectable signal for any of the four probes analyzed (“NA” cells) and cells with no PIK3CA signal and no HER2 amplification (“HER2noAmp” cells) were excluded from further analyses, as these cells likely represent non-specific technical outliers (see Supplementary Note and Supplementary Figure 5).
Figure 2.
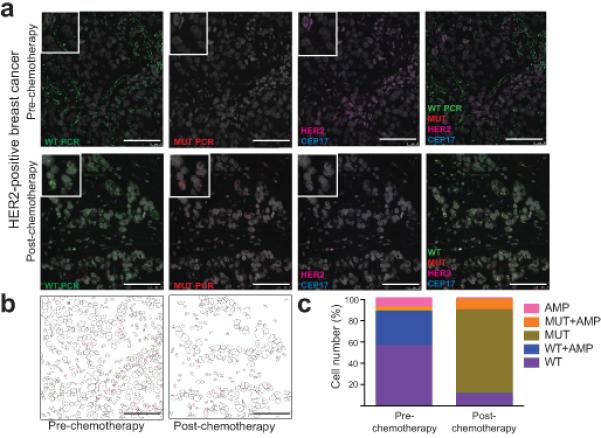
STAR-FISH analysis of breast cancer. Scale bars represent 75 μm. (a) STAR-FISH performed on a HER2+ tumor before and after neoadjuvant chemotherapy. WT PIK3CA - green, MUT PIK3CA – red, CEP17 – blue, HER2 – magenta, nuclear stain (To-Pro-3) – gray. (b) Automated quantification of STAR-FISH signal in single cells. Images represent tumor topology before and after chemotherapy based on computational segmentation of confocal images of the nuclei. Each nuclear outline present in the picture was used as a region of interest (single nucleus) in which STAR-FISH signals were quantified. (c) Graphs depict the frequencies of cells within each of the five defined cell categories (“species”) before and after treatment.
Analyzing the frequency of cell types in all cases demonstrated that the majority of the untreated tumors were composed of cells with HER2 amplification and wild type PIK3CA, with very small fractions of cells with mutant PIK3CA (Fig. 3a and Supplementary Figure 6, and 7a). In contrast, residual tumors after neoadjuvant therapy showed a dramatic increase in the relative frequency of PIK3CA mutant cells and a less pronounced decrease in the fraction of cells with HER2 amplification (Fig. 3a and Supplementary Figure 6 and 7a,b). These results demonstrate that cells with PIK3CA mutation pre-exist in HER2+ tumors but represent a minor population prior to therapy. The high sensitivity of STAR-FISH is reflected in our ability to detect these cells in almost all (20/22) untreated biopsy samples. Based on the TCGA breast cancer study12, PIK3CA His1047Arg mutation is only detected in 17% of HER2+ tumors. This seeming discrepancy can be explained by the fact that in 17 out of 22 (77.3%) cases, the cells with PIK3CA His1047Arg mutation constitute less than 5% of all cells within a tumor, and therefore would not be detected by bulk tumor sequencing at the depths of coverage attained in the TCGA breast cancer study. To validate our STAR-FISH results by an independent, highly sensitive mutation-detection method, we performed droplet digital PCR26,27 using DNA extracted from FFPE slides adjacent to the ones used for STAR-FISH (see Supplementary Note and Supplementary Figure 8a-d). PIK3CA mutant cell detection with the two methods was highly concordant for samples with more than 20% tumor cell content (linear regression y = 0.6835x, p < 0.0001, correlation R2 = 0.901, p = 0.0003). These results confirm that STAR-FISH allows for the detection of rare mutant cells with high specificity (FDR =0.001), which is critical for the identification of probable resistance mechanisms.
Figure 3.
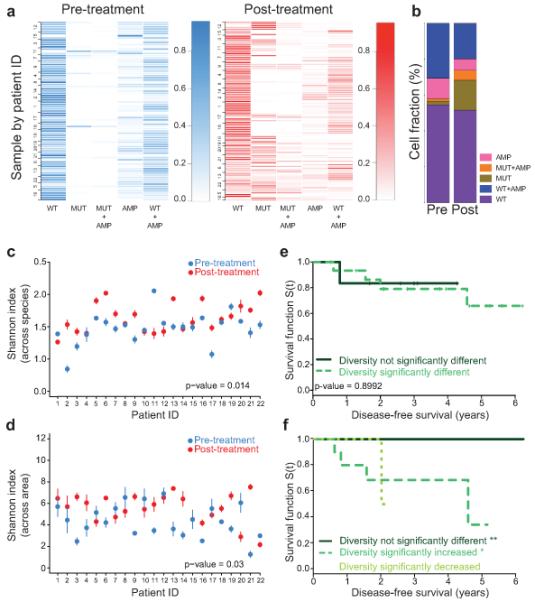
Changes in intratumor heterogeneity and patient outcomes. (a) Frequency of each cell type before and after treatment. Rows - single areas of a tumor (numbers are patient IDs). Columns - cell types (Supplementary Table 3). Samples clustered based on the frequency of cell types. Scale bar represents color intensity gradient indicating frequency. (b) Summary of cell type frequencies in all patients combined before (Pre) and after (Post) therapy. Graph depicts the mean percentage of each cell type in all areas and samples combined. (c, d) Shannon index of diversity based on counts of five cell types combining all areas in each sample - across species comparison (c) and across different areas of the same tumor - across area comparison (d). Confidence intervals defined by nonparametric bootstrap resampling. Global difference in diversity before and after treatment for all patients was calculated using the paired Wilcox signed-rank sum test. Error bars represent two times the standard error obtained from 1,000 bootstrap samples. (e, f) Changes in diversity and clinical outcomes. Associations between changes in cellular diversity after chemotherapy across species (e) and across area (f). Patients grouped based on changes in diversity after treatment: (e) solid line – no significant change (n = 6), dashed line – significant change (n = 16); log-rank test p-value 0.8992 and (f) solid line – no significant change (n = 9), dashed line – significant increase (n = 11); dotted line - significant decrease (n = 2); log-rank test p-values: 0.0178 (**), 0.0596 (*), and 0.359 (ns), respectively.
Our single cell analysis of the frequency of cells with HER2 amplification by STAR-FISH revealed substantial heterogeneity and changes due to treatment in line with results of prior studies28-39. In pre-treatment samples only 32.5%±18% of all cells contained HER2 amplification, which decreased to an average of 23.5%±23.4% cells after neoadjuvant therapy with significant variation among patients. Assuming that the heterogeneity of the primary tumor is reflective of that of disseminated cancer cells targeted by adjuvant therapy, the decrease in the frequency of HER2 amplified cells after neoadjuvant therapy may contribute to the suboptimal efficacy of HER2-targeted therapies in some patients. Our results also imply that it would be beneficial to re-evaluate tumors following recurrence and design subsequent therapies based on this knowledge.
Changes in Intratumor heterogeneity after chemotherapy
Neoadjuvant chemotherapy appears to enrich for PIK3CA mutant cells, potentially increasing the risk of resistance to subsequent HER2-targeted adjuvant therapies, both conferred by the PIK3CA mutation but also by the enrichment for cells that lack the HER2 amplicon. Analysis of each individual tumor area (Fig. 3b) revealed an overall increase in the diversity of cellular subpopulations, as the majority of treatment-naïve samples were composed of only three cell populations, whereas post-treatment tumors often contain all five cell types (Supplementary Figure 6 and 7a). Moreover, in many samples there was also a significant variation among different areas of the same tumor, and this variance increased in many cases after chemotherapy (Supplementary Figure 6). These results demonstrate that neoadjuvant chemotherapy changes the relative frequencies of genetically distinct cell populations in treatment-resistant residual tumors and suggest that some of these changes may have an unfavorable impact on the efficacy of subsequent adjuvant HER2-targeted therapies.
To further investigate global changes in intratumor cellular genetic heterogeneity for PIK3CA and HER2, we calculated Shannon and Simpson’s indices40 in each sample before and after neoadjuvant treatment. These diversity measures allow for the characterization of each sample by a single value, which reflects the relative abundances of cells in each of the five categories described above. The diversity index was significantly different between pre- and post-treatment samples (Fig. 3c and Supplementary Figure 7c,d). This significant shift in diversity was also present when we compared different areas of the same tumor (Fig. 3d). These results imply that neoadjuvant treatment impacts intratumor cellular genetic diversity not only due to changes in cell populations, but also by increasing regional heterogeneity within tumors.
Three of the patients received targeted therapy, trastuzumab, in addition to chemotherapy in the neoadjuvant setting (patients 20, 21, and 22). Interestingly, two of them had a decrease in diversity after neoadjuvant therapy, suggesting the presence of and selection for a preexisting subpopulation resistant to treatment (Supplementary Figure 9). However, a larger number of samples will be necessary to demonstrate the generality of this finding.
Because PIK3CA mutations are thought to be associated with treatment resistance in HER2+ tumors15-19 and the changes in intratumor diversity were in part due to variations in the fraction of PIK3CA mutant cells, we analyzed potential associations between changes in diversity and patient outcome. Interestingly, the difference in cellular diversity between pre- and post-neoadjuvant chemotherapy samples did not impact disease-free survival following adjuvant trastuzumab treatment (Fig. 3e). Similarly, changes in the fraction of PIK3CA mutant cells by themselves were not predictive of recurrence (data not shown). However, when the diversity index was calculated based on cell types across different areas of the same tumor, a significant change in diversity was associated with shorter disease-free survival (Fig. 3f). Because long-term survival is largely defined by progression to metastatic disease, our results imply a potential role for regional differences in the primary tumor microenvironment in selecting for treatment-resistant cancer cells capable of migration and metastatic dissemination. Surprisingly, both an increase and a decrease in the diversity of cell populations across different areas of the same tumor were associated with shorter disease-free survival (decrease in diversity did not reach significance likely due to low sample number, n = 2). Both patients showing a decrease in diversity across tumor areas after treatment received trastuzumab, a HER2-targeting therapy, in the neoadjuvant setting in addition to the adjuvant trastuzumab (Supplementary Figure 3). Cell type frequency plots in patients treated with neoadjuvant chemotherapy+trastuzumab (patients 20, 21 and 22) showed that the decrease in post-treatment diversity is due to the high similarity among the areas tested with all five cell types present in similar proportions. Thus, including trastuzumab in neoadjuvant treatment seems to promote spatial homogeneity, but it does not prevent the increase in post-treatment cellular heterogeneity in patients who did not achieve pathologic complete response. Taken together, these results demonstrate that heterogeneity within primary tumors is an important determinant of long-term clinical outcome and that therapeutic resistance may be associated with changes both in cellular genetic and regional heterogeneity. These results also highlight the need for analyzing tumors at the single cell level in situ and the high clinical value of assessing primary tumors before and after neoadjuvant therapy and designing follow up therapies based on this knowledge.
Co-ocurrence of PIK3CA mutation and HER2 amplification
STAR-FISH allows for the detection of genetic alterations in single cells, and thus can be used to investigate the co-occurrence of two or more genetic events. The frequency of cells with single or combined genetic alterations can be used to predict the relative order of these changes to decipher the probable course of tumor evolution41. Thus, we used our STAR-FISH data to explore the relative order of HER2 amplification and PIK3CA mutation. If the two genetic events arise independently, then the probability of having both events present in the same cell is equal to the product of probabilities of each individual event (see Methods for details). Thus, if co-occurrence is favored, the probability of having both events is higher than expected by chance. We found that the observed co-occurrence of PIK3CA mutation and HER2 amplification in the same cell was higher than expected both in the pre- and post-treatment samples (Fig. 4a), implying higher fitness of cells with both mutations compared to cells with each individual mutation. The co-occurrence of PIK3CA His1047Arg mutation with HER2 amplification within the same tumor has been previously reported12,39. However, whether these two genetic alterations are in the same cancer cell or in different subpopulations has not been defined conclusively. Our results indicate that most cells with PIK3CA His1047Arg mutation do not have HER2 amplification. Yet, the frequency of cells with both genetic events is significantly higher than expected by chance. These results suggest that PIK3CA His1047Arg mutation and HER2 amplification are independent genetic events, and that cells with both events have a fitness advantage over others. Our results are in line with data in animal models, which show that the combination of PIK3CA His1047Arg mutation with HER2 amplification accelerates tumor formation, metastatic progression, and confers resistance to HER2-targeted therapies42.
Figure 4.
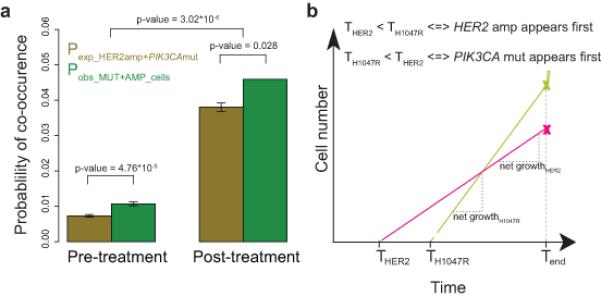
Probable course of tumor evolution based on the co-occurrence of PIK3CA mutation and HER2 amplification. (a) Co-occurrence of PIK3CA His1047Arg mutation and HER2 amplification in the same cell. Expected probability (Pexp) of co-occurrence of the two genetic events is compared with observed probability (Pobs) based on the frequency of MUT+AMP cells. Error bars are defined as the standard deviation of expected or observed values divided by the square root of the total number of samples. (b) Mathematical model of probable tumor evolution. Prediction of the time at which each event arises (THER2 and THis1047Arg) based on STAR-FISH counts for both events (x marks) at observation time (Tend) and the net growth rates attributed to these events based on published data42.
Model of relative order of genetic events
Next, we examined the relative order of PIK3CA mutation and HER2 amplification during tumor progression. To determine the time during tumor progression at which these genetic events arose, a mathematical model was constructed based on STAR-FISH-derived numbers of cells with PIK3CA mutation and with HER2 amplification and cell death and proliferation rates of cells with individual and both genetic alterations in a transgenic model of HER2+ breast cancer42, assuming that the characteristics of human and mouse mammary tumor cells with PIK3CA His1047Arg mutation and HER2 amplification are similar (see Methods for details). This model suggested that HER2 amplification arises first and that PIK3CA mutation is acquired at later stages of tumor progression (Fig. 4b). Although with the current data it is not possible to predict whether the observed diversity for PIK3CA mutation and HER2 amplification is a result of parallel evolution of two independent cell populations or diversification at a later stage, our mathematical modeling of the temporal order of events supports the notion that HER2 amplification is an early event in breast tumor evolution.
Spatial distribution of genetically distinct subpopulations
Our data showing associations between intratumor diversity across different areas of the same tumor and long-term clinical outcome (Fig. 3f) raised the possibility of local microenvironments within primary tumors favoring the expansion of genetically distinct cell populations. To investigate this further, we asked whether different cell types within the tumor are distributed randomly or whether there is any spatial clustering of cell types. While we found no significant correlation of the presence of any two individual cell types in the same sample, there is a trend for the co-occurrence of cells with PIK3CA mutation alone (MUT) and cells with both PIK3CA mutation and HER2 amplification (MUT+AMP) (Fig. 5a).
Figure 5.
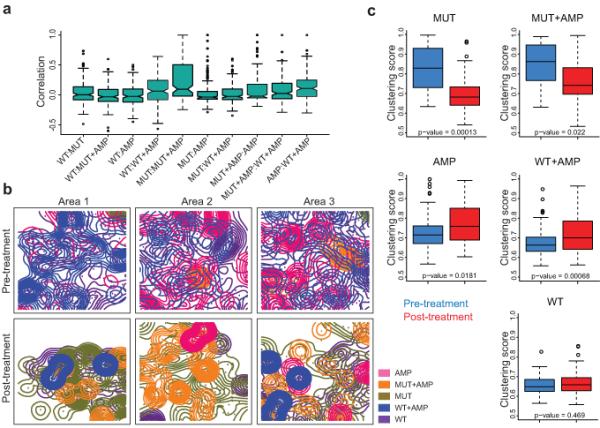
Intratumor topology. (a) Co-occurrence of the indicated cell type combinations. None of the correlations are significant. The median value is the horizontal bar in the middle of each box. IQR (interquartile ranges) is defined by the upper and lower edge of each box. Notches are the 95% confidence interval of the median. The lower whisker is defined as the maximum between 25 percentile - 1.5 × IQR and the minimum value of the data, whereas the upper whisker is defined as the minimum between 75% percentile + 1.5 × IQR and the maximum value of the data. (b) Kernel plots depicting the two-dimensional spatial distribution of the indicated cell types in three different regions of the same case before and after treatment. Scale bar represents 75 μm. (c) Spatial dispersion of different cell types within tumors. Formation of larger clusters in 2D space is indicated by an increase in clustering score, the ratio of between and within-cluster variation of Kernel means – a higher ratio means more clustering, while lower ratio signifies more random distribution.
To further investigate potential relationships between the intratumor localization of different cell types, we performed spatial distribution analyses. Kernel density and counts of cell type densities were calculated for each cell type (Fig. 5b; see Methods for details). These measurements were used to assess if cells of a certain type tend to cluster together or be randomly distributed across the investigated area. K-means clustering43 showed that cells with PIK3CA mutation, irrespectively of their HER2 amplification status, tend to form clusters in situ (Fig. 5b). Cells with wild type PIK3CA and no HER2 amplification (WT) have the lowest clustering score and this remained unchanged after treatment (Fig. 5c). Neoadjuvant therapy seems to have an opposing effect on MUT and MUT+AMP cells compared to WT and WT+AMP cells: for cells with PIK3CA mutation the clustering score after treatment is decreased to the level of WT cells (cells are more dispersed after treatment), whereas for AMP and WT+AMP cells it is increased (cells are less dispersed after treatment). These results suggest that WT cells are most likely non-malignant stromal cells since their distribution within tumors neither show any specific pattern nor is affected by treatment. In contrast, the increased dispersion of cancer cells with PIK3CA mutation implies that these cells might be more migratory. Indeed, PIK3CA mutations were shown to increase cell motility42,44, supporting our hypothesis.
DISCUSSION
The STAR-FISH method we described here is relatively inexpensive and easy to interpret. It is also much more sensitive than sequencing or mass spectrometry-based45 methods, since even a single mutant cell can be detected among thousands of cells analyzed with FDR = 0.001. Other methods, such as padlock-probe rolling-circle amplification, can be used to detect mutations in situ based on mRNA46. However, mRNA levels can be affected by tissue handling, the nature of the mutation (e.g., nonsense mediated decay47), and also by cancer treatment. Thus, methods based on DNA detection such as STAR-FISH are more suited for clinical studies, especially because it utilizes standard FFPE samples which are routinely available. Since intratumor heterogeneity is one of the main drivers of disease progression and therapy resistance, including STAR-FISH analysis in clinical practice could improve patient stratification and the design of optimal individualized therapies. Due to its high sensitivity and specificity for detecting mutant cells, STAR-FISH could especially be useful for guiding the design of targeted therapies with known genetic resistance mechanisms. Limitations of STAR-FISH include the need for a priori knowledge of the mutation to be analyzed and the number of fluorophores that can be applied to detect in situ PCR products and chromosomal regions. The ability to increase multiplexing by combining it with other detection methods such as FISSEQ48 or CyTOF-based approaches49 can transform STAR-FISH into an exceptionally powerful diagnostic pathology assay for single cell mutation detection and genetic heterogeneity analysis.
In summary, we applied a novel in situ technique to assess the clinical impact of changes in intratumor heterogeneity during neoadjuvant therapy. STAR-FISH detected mutant cells with high sensitivity and in intact tissues, enabling the mapping of spatial distribution of subpopulations of cancer cells. Our results show that chemotherapy affects both the frequency and topology of these subpopulations. Changes in diversity across different areas of the same tumor proved to be clinically more significant than overall diversity changes. This suggests that the application of in situ techniques might be key for the design of more effective therapies for heterogeneous tumors.
ONLINE METHODS
Human breast cancer samples
Archival formalin-fixed paraffin embedded HER2+ breast tumor samples were obtained from patients matched biopsies and tumors post-neoadjuvant treatment, from Seoul National University Bundang Hospital. The Institutional Review Boards of Seoul National University Bundang Hospital and DF/HCC approved the protocol and waived the informed consent requirement. Samples were de-identified prior to analysis. Tumor histology and expression of standard biomarkers (ER, PR, and HER2) were evaluated at the time of diagnosis according to ASCO/CAP guidelines50,51. For the comparison of STAR-FISH and Sequenom MassARRAY, IDC and adjacent DCIS formalin-fixed paraffin embedded tissue slides were obtained following approval from Institutional Review Board at Memorial Sloan Kettering Cancer Center, informed consent requirement was waived. Tumor histology and marker analysis was performed as previously described23.
Cell lines and histogel embedding
Breast cancer cell lines MDA-MB-231, T-47D, AU565, and BT-483 were obtained from ATCC and SUM-185PE was obtained from Steve Ethier (University of Michigan). Cells were cultured in media recommended by the provider. The identity of the cell lines was confirmed by short tandem repeats analysis. Tests for mycoplasma contamination were performed regularly. For histogel embedding, BT-483 cells were fixed in 4% paraformaldehyde for 20 minutes, dehydrated in a series of ascending ethanol and resuspended in 2 volumes of Histogel (Thermo Scientific), pre-warmed to 55°C. Once solidified, the histogel was kept in 70% ethanol and processed into paraffin block.
Xenograft assays
All animal procedures were approved by the DFCI ACUC and followed NIH guidelines. Bi-lateral mammary fat pad injection of 1×106 cells re-suspended in 50% Matrigel (BD Biosciences) per transplant were performed on 4–5-weeks old female NCRNU-F mice (Taconic). Tumor volumes were monitored by bi-weekly caliper measurements until 1.2cm3 was reached. Then, tumors were harvested and processed into formalin-fixed paraffin embedded blocks. For mixed cell line xenograft assay, MDA-MB-231 and SUM-185PE cells expressing mCherry and GFP, respectively, were generated by lentiviral infection as previously described52. Cells were collected, mixed at different ratios, total cell number was 2×106, mixed with 50% Matrigel and injected into mammary fat pad of NCRNU-F mice (Taconic). Tumors were removed when they reached 1 cm diameter and cut into two equal pieces – one for FFPE processing and one for FACS analysis.
Fluorescence activated cell sorting for GFP and mCherry labeled cells
Tumor tissue was digested with 2 mg/ml collageanse I (Worthington Biochemical Corporation), 2 mg/ml hyaluronidase (Sigma-Aldrich). Following dissociation, the sample was filtered through 40 μm cell strainer (BD Biosciences), washed in PBS with 0.5mM EDTA and 5% BSA and analyzed on BD LSRFortessa analyzer. The gates were set up based on forward and side scatters to exclude dead cells and doublets.
Immunofluorescence analyses
After deparaffininzation and rehydration, the slides were subjected to antigen retrieval in citrate buffer pH 6 (DAKO), for 20 min. in a steamer. Next, blocking solution (10% FCS in PBST) was applied for 10 min. Incubation with primary antibody in PBS was held overnight at 4°C in a moist chamber. Secondary antibody was applied for 45 min. at room temperature. Samples were mounted with VectaShield HardSet Antifade Mounting Medium with DAPI (Vector). Imaging was performed on Leica SP5 confocal microscope. Antibodies used52: anti-GFP antibody (sc-9996, mouse monoclonal IgG2a, 1:50 dilution, Santa Cruz Biotechnology), goat-anti-rabbit Alexa488-conjugated (1:100 dilution, Life Technologies), anti-SMA (M085101, clone 1A4, mouse monoclonal IgG2a, 1:250 dilution; Dako).
STAR-FISH in situ PCR conditions
The 1st round of in situ PCR for PIK3CA H1047H/R was performed with 2 mM MgCl2, 200 μM dNTPs, 250 nM primers (for primer design see Supplementary Note), 0.1 U/μl Platinum Taq Polymerase (Life Technologies) in 10x Platinum Taq Buffer (without MgCl2; Life Technologies). Total of 9 cycles were performed, in 3 steps of 3 cycles with annealing temperature at 45°C, 52°C and 58°C, respectively. All cycling steps were 30s long. The 2nd round of PCR for PIK3CA H1047H/R was performed with 3.5 mM MgCl2, 200 μM dNTPs, 250 nM primers, 0.1 U/μl Platinum Taq Polymerase (Life Technologies) in 10x Platinum Taq Buffer (without MgCl2; Life Technologies). Total of 30 cycles were performed, 5 cycles with annealing temperature at 45°C, 5 cycles with annealing temperature at 52°C, and 20 cycles with annealing temperature at 58°C. All cycling steps were 30s long. The 1st round of PCR for PIK3CA E542E/K was performed with 4 mM MgCl2, 150 μM dNTPs, 250 nM primers, 0.1 U/μl Platinum Taq Polymerase (Life Technologies) in 10x Platinum Taq Buffer (without MgCl2; Life Technologies). Total of 9 cycles were performed, annealing temperature was 56°C, cycling steps were 30s long. Second round of PIK3CA E542E/K PCR was performed as for PIK3CA His1047Arg, with 4 mM MgCl2 and 150 μM dNTPs. For TP53 R175R/H 2 mM MgCl2 and 200 μM dNTPs were used in both PCR rounds. Since the amplicon resides in a GC rich region, dc7GTP was added and slowdown PCR program was used, as described before53. Second round of PCR was performed as for PIK3CA H1047H/R.
STAR-FISH
5μM Formalin fixed paraffin embeded tissue sections on syliated glass slides were baked overnight at 70°C, dewaxed in xylene (Leica Microsystems), washed in 100%, 70%, and 50% ethanol (Leica Microsystems), rinsed with H2O and air dried for 1h. After baking at 105°C for 2 minutes, the samples were treated with 20 μg/ml Proteinase K in PBS (Ambion) at 37°C for 12 minutes (xenografts) or 18-20 minutes (human tumors). To increase signal to noise ratio, the slides were then rinsed in H2 O and treated with 100 μg/ml RNase in 2xSSC buffer (Fisher Scientific) for 1h at 37°C. Next, 5min wash in 5% Chelex beads (Bio-Rad) in H2O with agitation was applied and followed by 2 more washes with H2O. Excess water was drained and frame-seal incubation chambers (Bio-Rad) were applied to the slides, the master-mix for the 1st round of PCR was pipetted on top of the tissue, the chambers were sealed, and placed in the in situ PCR machine (Bio-Rad DNA Engine with Slide Tower). The first round of PCR was run for 9 cycles. Next, the frame-seals were removed, excess of remaining reagents drained, and new frames applied before the addition of master-mix for the 2nd round of PCR. The second round of PCR was run for 30 cycles. After removal of the frame-seals, the slides were washed in H2O and dehydrated in increasing concentrations of ethanol (50%, 70%, 85%, 100%), and air-dried. FISH probe mix, containing custom made probes recognizing WT and MUT PCR product (labeled with dFluorescein and TYE, respectively; Exiqon), HER2 BAC probe (RP11-94L15, kindly provided by Drs. Hege Russness and Inga Rye, Oslo, Norway; labeled with SpectrumOrange dUTP by Nick Translation Kit; Abbott Molecular), CEP17Aqua probe (Abbott Molecular) was then applied to the slides, covered with coverslip, and sealed with rubber cement. Hybridization was performed for 7 minutes at 75°C followed by overnight incubation at 37°C in a humid chamber. Next, the slides were washed in 0.4x SSC with 0.3% NP-40 for 2 minutes at room temperature, in 0.4x SSC with 0.3% NP-40 for 2 minutes at 74°C, then in 2x SSC with 0.1% NP-40, in 2x SSC, and in PBS. Nuclear counterstaining was performed by 10 min. incubation at room temperature with 1 μM To-Pro-3 in PBS (Molecular Probes, Life Technologies). After two additional washes in PBS and one in H2O, the slides were dried and mounted with Vectashield Mounting Medium (Vector Laboratories), covered with coverslips and stored overnight to 3 days at −20°C.
Image acquisition and quantification of STAR-FISH signal
STAR-FISH images were acquired in Z-stacks (0.25 μM per slice, average 7 slices per image) with Leica SP5X laser scanning confocal microscope with white light laser, 63x Plan Apo objective. Image resolution was set at 1,024×1,024, scanning speed was set at 400 Hz. All images were acquired in sequential mode to avoid fluorophore crosstalk. Per each sample 3 to 5 images of different regions were collected to account for intratumor heterogeneity. Analysis of the images was conducted using ImageJ software. Maximum projection from the z-stack was used to facilitate quantification of the FISH signal. Automated counting of STAR-FISH signal in each individual nucleus was performed with custom-made macro in ImageJ (code available upon request). The image of the nuclei was used to create an outline of each nucleus, which was then applied as region of interest for particle analysis for all STAR-FISH channels. As a result for each image a table was generated, containing cell index and measurements of STAR-FISH particles found in each nucleus. Size restriction for the nuclei has been used to filter out stromal cells, which usually have smaller nuclei than tumor cells.
Droplet digital PCR
Tissue from three deparaffinized FFPE slides was scraped into an eppendorf tube and resuspended in LowTE buffer with SDS and 10 μg/ml Proteinase K (Ambion) and 10 μg/ml RNase (Life Technologies). Samples were incubated at 56°C for 48 h, 10 μg/ml Proteinase K and 10 μg/ml RNase were added every 12 h. DNA was extracted using TE-buffered phenol/chloroform pH = 8 (Fisher Scientific) according to standard protocol. For control cell lines, MDA-MB-231 (PIK3CA wild-type), SUM-185PE (PIK3CA His1047Arg homozygous), DNA was prepared in the same manner, except for HCT116 (PIK3CA His1047Arg heterozygous), for which QIAamp DNA Mini kit (Qiagen) was used. PCR reactions (25 ul) that comprised ddPCR™ Supermix for Probes, custom-made Taqman primer/probe mix (for primers and probes sequences see Supplementary Table 1) and appropriate DNA template were prepared in a 96-well PCR plate and subsequently transferred to the sample wells of a ddPCR cartridge. After Droplet Generator Oil (70 ul) was pipetted into the oil wells, the cartridge was loaded into the Droplet Generator. The emulsified droplets were then transferred to a new 96-well PCR plate and amplified by PCR. Cycling conditions were: 10 min at 95°C followed by 40 cycles of a two step thermal profile of 15 s at 94°C denaturation and 60 s at 59°C. Lastly, the 96-well plate was loaded on to and read by a QX100 Droplet Reader (Bio-Rad). Data analysis was performed using QuantaSoft software (v.1.6.6). Genome copies of PIK3CA H1047 (wt) and His1047Arg (mut) populations in the templates were determined by Poisson distribution fit. The assay was validated with cell line DNA (data not shown).
Sequenom MassArray
DCIS samples and their adjacent invasive breast carcinomas, were microdissected with a sterile needle under a stereomicroscope, and subjected to DNA extraction. DNA samples from DCIS and invasive breast carcinoma, when present, were subjected to Sequenom MassARRAY (Sequenom) analysis to detect PIK3CA hotspot mutations as previously described23,24. The multiplexed assays were designed using the Assay Design 3.1 Sequenom software, as previously reported23. In brief, pre-PCR amplification was performed using the same primers employed for Sequenom, prior to the iPLEX Gold genotyping assay. Purified primer extension reaction (7 nl) was loaded on a matrix pad of a SpectroCHIP (Sequenom) and measured by laser desorption/ionization of time-of-flight mass spectrometry. The allelic fractions were estimated by calculating the ratio of the area of the raw spectra of the mutant allele to its wild-type23,54.
Statistical and mathematical analyses
Diversity index calculation
For patient i, where i = 1, 2, …, 22, denote the number of samples at diagnosis by ni1 and the number of samples obtained after the completion of treatment by ni2. We have a total of n1 = 101 samples at diagnosis, n2 = 82 samples after treatment, and a combined number of n = n1 + n2= 183 samples. From the raw data, for each sample, we obtained the following numbers of cells identified by a particular mutation pattern: ma cells with wild type PIK3CA with neither PIK3CA mutation nor HER2 amplification (WT), mb cells with PIK3CA mutation without HER2 amplification (MUT), mc cells with PIK3CA mutation and HER2 amplification (MUT+AMP), md cells with HER2 amplification with neither HER2 wild type signal nor PIK3CA mutation (AMP), and me cells with PIK3CA wild type signal without PIK3CA mutation but with HER2 amplification (WT+AMP). We have a total of ma + mb + mc + md + me = m cells, for each m from the 183 samples at two time points. We also obtained ps = ms / m for each cell type s from types {a, b, c, d, e}. Using this data, we then calculated an integrative Shannon index for each patient as follows:
Analysis of changes in diversity
To make an inference on whether the diversity of cells within each patient changes significantly during treatment, we exploited a nonparametric bootstrap approach55: For sample j of patient i at time t, suppose there are mijt cells. Then we sampled a new set of mijt cells with replacement from those mijt cells and recalculate Hitb for b = 1, 2, …, B. In our analyses, we chose B = 1,000. Subsequently, the standard deviations from the bootstrapped integrative Shannon indices were calculated to form 95% confidence intervals. If the confidence intervals of pre-treatment and post-treatment Shannon indices did not overlap, then we deduced that the diversity of cell types within this patient changed significantly during treatment. The direction of change was subsequently determined by the sign of Hi2 - Hi1.
Co-occurrence analyses of PIK3CA mutations and HER2 amplifications
We then sought to investigate whether cells with PIK3CA mutations are more or less likely to contain HER2 amplifications as compared to cells with wild type PIK3CA. To this end, we determined the following quantities: for each sample, (1) the total number of cells in this sample, n; (2) the number of cells with PIK3CA mutation, np; (3) the number of cells with HER2 amplification, nh; and (4) the number of cells with both HER2 amplification and PIK3CA mutation, nhp. If the two events are independent, then we should have Proportionhp ~ Proportionh Proportionp. Thus the binomial test was adopted to test the null hypothesis that PIK3CA mutation and HER2 amplification emerge independently, where np / n is the background probability of HER2 amplifications, which is compared to a binomial distribution with nhp realizations and nh totals.
Inferring temporal ordering between PIK3CA mutation and HER2 amplification
We then sought to determine whether PIK3CA mutation or HER2 amplification arises first within a cell. We considered that cells grow according to an exponential growth model. The numbers of cells with PIK3CA mutation or HER2 amplification (assuming independence) over time was then modeled as follows:
The parameters λ* and μ* denote the birth and death rates of each cell type, respectively, with the subscript indicating either of the two cell types. We used the subscript p to denote parameters for cells carrying a PIK3CA point mutation, and the subscript h to denote parameters for cells carrying a HER2 amplification. To estimate λ* and μ*, we used previously published measurements obtained from a mouse model42. Using this data (Fig. S2B from Hanker et al. PNAS 201342), we found that the death rates for either cell type are lower than 0.1 while the estimate for λp is around 0.9, and λh is around 0.75. Then, assuming that initially there is one cell of each type, the time it takes to reach np or nh cells are given by
The indicator z = I(th < tp) then provides us with the temporal order of the two types of mutations: if z = 1, then HER2 amplification is the first event, otherwise it is the second event. For each patient, we used the number of cells carrying HER2 amplification at the pre-treatment time point as nh(th) and the number of cells carrying a PIK3CA point mutation as np(tp). Since the birth rates and death rates were measured in the mouse model as outlined above, and these parameters might differ from the rates of human cells, sensitivity analyses were performed to investigate the robustness of the identified temporal order against perturbations in the parameter values: first, we changed the birth rate for HER2-amplified cells to the following values: 0.5, 0.55, …, 0.9. We found that the model using any of these values predicted that HER2 amplification was the first event. Second, we changed the death rate for HER2 amplified cells to the following values: 0.06, 0.10, 0.14, 0.18, 0.22. Similarly, we found that HER2 amplification was the first event in all these scenarios. Thus we concluded that HER2 amplification is very likely to be the first event, followed by PIK3CA mutation, even taking into account the uncertainty of the parameter estimation and differences in estimates between human and mouse cells. To perform a more detailed analysis of the evolutionary dynamics of the acquisition of these mutations, more data obtained from human cells is necessary.
Spatial analyses
We then sought to determine whether there was any spatial clustering in the cell types within samples. For each sample, we obtained a 1,024 × 1,024 pixel square image to display the spatial distribution of each measured cell. Then, for each sample and each mutation type, we performed K-means clustering43 with K = 3. Next we calculated the ratio of between-cluster sums of squares versus the total sums of squares to quantify the randomness in localization of each mutation species. When this ratio is larger, then K = 3 is sufficient to cluster the spatial data, whereas when this ratio is lower, then the spatial data might form smaller clusters and K = 3 is insufficient, i.e. cells are more sporadically distributed. Finally, we used the Wilcox rank test to determine whether there was any significant difference in the distribution of this ratio between pre-treatment and post-treatment samples.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Eric Winer, Ian Krop, Bert Vogelstein, and members of the Polyak and Michor laboratories for their critical reading of the manuscript and useful discussions. We thank Dr. Andriy Marusyk and Doris Tabassum for their help with the xenograft assays, Dr. Robert Witwicki for help with data processing, Lisa Cameron in the DFCI Confocal Microscopy for her technical support, Dr. Andrea Richardson (DFCI) for providing slides from a human breast tumor with known PIK3CA His1047Arg mutation status, and Drs. Hege Russness and Inga Rye (Oslo University Hospital, Oslo, Norway) for providing the BAC probe for HER2. This work was supported by the Dana-Farber Cancer Institute Physical Sciences-Oncology Center (U54CA143798 to F.M.), EMBO (M.J.), Swiss National Science Foundation (M.J.), American Cancer Society (CRP-07-234-06-COUN to C.L.A.), and the Breast Cancer Research Foundation (K.P.).
Footnotes
AUTHOR CONTRIBUTIONS
M.J. developed the STAR-FISH method, performed the experiments and data analyses. V.A. assisted with image acquisition and analyses. L.L. performed mathematical modeling and data analysis. S.Y.P. provided tumor samples. Y.K. and C.P. performed the digital PCR experiment and data analysis. R.S., B.W., T.K., S.C., and J.R.-F. provided patient samples and performed Sequenome MassARRAY experiment. A.H. and C.A. provided data and tissues from transgenic models of HER2+ breast cancer. K.P. and F.M. supervised the study. All authors helped to design the study and write the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 2.Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps10. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 3.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452–464. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagasra O. Protocols for the in situ PCR-amplification and detection of mRNA and DNA sequences. Nat Protoc. 2007;2:2782–2795. doi: 10.1038/nprot.2007.395. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda S, Takabe K, Inagaki M, Funakoshi N, Suzuki K. Detection of gene point mutation in paraffin sections using in situ loop-mediated isothermal amplification. Pathology International. 2007;57:594–599. doi: 10.1111/j.1440-1827.2007.02144.x. [DOI] [PubMed] [Google Scholar]
- 7.Ebina M, Martínez M, Birrer MJ, Linnoila RI. In situ detection of unexpected patterns of mutant p53 gene expression in non-small cell lung cancers. Oncogene. 2001;20:2579–2586. doi: 10.1038/sj.onc.1204351. [DOI] [PubMed] [Google Scholar]
- 8.Almendro V, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6:514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almendro V, et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res. 2014;74:1338–1348. doi: 10.1158/0008-5472.CAN-13-2357-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SY, Gönen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. The Journal of Clinical Investigation. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachman KE, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 12.TCGA Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Nagata Y, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Liu Y, Du Y, Yin W, Lu J. The predictive role of phosphatase and tensin homolog (PTEN) loss, phosphoinositol-3 (PI3) kinase (PIK3CA) mutation, and PI3K pathway activation in sensitivity to trastuzumab in HER2-positive breast cancer: a meta-analysis. Curr Med Res Opin. 2013;29:633–642. doi: 10.1185/03007995.2013.794775. [DOI] [PubMed] [Google Scholar]
- 16.Rexer BN, Chanthaphaychith S, Dahlman KB, Arteaga CL. Direct inhibition of PI3K in combination with dual HER2 inhibitors is required for optimal antitumor activity in HER2+ breast cancer cells. Breast Cancer Res. 2014;16:R9. doi: 10.1186/bcr3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rexer BN, Arteaga CL. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Crit Rev Oncog. 2012;17:1–16. doi: 10.1615/critrevoncog.v17.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cizkova M, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28. doi: 10.1186/bcr3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cizkova M, et al. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108:1807–1809. doi: 10.1038/bjc.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng G, et al. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer. 2014;14:456–467. doi: 10.1186/1471-2407-14-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupont Jensen J, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.CCR-10-1133. [DOI] [PubMed] [Google Scholar]
- 22.Jurinke C, van den Boom D, Cantor CR, Koster H. Automated genotyping using the DNA MassArray technology. Methods Mol Biol. 2001;170:103–116. doi: 10.1385/1-59259-234-1:103. [DOI] [PubMed] [Google Scholar]
- 23.Sakr RA, et al. PI3K pathway activation in high-grade ductal carcinoma in situ--implications for progression to invasive breast carcinoma. Clin Cancer Res. 2014;20:2326–2337. doi: 10.1158/1078-0432.CCR-13-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandarlapaty S, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18:6784–6791. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunelli M, et al. Genotypic intratumoral heterogeneity in breast carcinoma with HER2/neu amplification: evaluation according to ASCO/CAP criteria. Am J Clin Pathol. 2009;131:678–682. doi: 10.1309/AJCP09VUTZWZXBMJ. [DOI] [PubMed] [Google Scholar]
- 29.Glockner S, Buurman H, Kleeberger W, Lehmann U, Kreipe H. Marked intratumoral heterogeneity of c-myc and cyclinD1 but not of c-erbB2 amplification in breast cancer. Lab Invest. 2002;82:1419–1426. doi: 10.1097/01.lab.0000032371.16521.40. [DOI] [PubMed] [Google Scholar]
- 30.Hanna W, Nofech-Mozes S, Kahn HJ. Intratumoral heterogeneity of HER2/neu in breast cancer--a rare event. Breast J. 2007;13:122–129. doi: 10.1111/j.1524-4741.2007.00396.x. [DOI] [PubMed] [Google Scholar]
- 31.Vance GH, et al. Genetic heterogeneity in HER2 testing in breast cancer: panel summary and guidelines. Arch Pathol Lab Med. 2009;133:611–612. doi: 10.5858/133.4.611. [DOI] [PubMed] [Google Scholar]
- 32.Cottu PH, et al. Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. Ann Oncol. 2008;19:595–597. doi: 10.1093/annonc/mdn021. [DOI] [PubMed] [Google Scholar]
- 33.Lewis JT, et al. Analysis of intratumoral heterogeneity and amplification status in breast carcinomas with equivocal (2+) HER-2 immunostaining. Am J Clin Pathol. 2005;124:273–281. doi: 10.1309/J9VX-ABUG-KC4Y-07DL. [DOI] [PubMed] [Google Scholar]
- 34.Striebel JM, Bhargava R, Horbinski C, Surti U, Dabbs DJ. The equivocally amplified HER2 FISH result on breast core biopsy: indications for further sampling do affect patient management. Am J Clin Pathol. 2008;129:383–390. doi: 10.1309/KFKDNK8CENVN24VU. [DOI] [PubMed] [Google Scholar]
- 35.Shin SJ, Hyjek E, Early E, Knowles DM. Intratumoral heterogeneity of her-2/neu in invasive mammary carcinomas using fluorescence in-situ hybridization and tissue microarray. Int J Surg Pathol. 2006;14:279–284. doi: 10.1177/1066896906293055. [DOI] [PubMed] [Google Scholar]
- 36.Seol H, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 37.Bartlett AI, et al. Heterogeneous HER2 gene amplification: impact on patient outcome and a clinically relevant definition. Am J Clin Pathol. 2012;136:266–274. doi: 10.1309/AJCP0EN6AQMWETZZ. [DOI] [PubMed] [Google Scholar]
- 38.Andersson J, Linderholm B, Bergh J, Elmberger G. HER-2/neu (c-erbB-2) evaluation in primary breast carcinoma by fluorescent in situ hybridization and immunohistochemistry with special focus on intratumor heterogeneity and comparison of invasive and in situ components. Appl Immunohistochem Mol Morphol. 2004;12:14–20. doi: 10.1097/00129039-200403000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Ng CK, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol. 2015;16:107–127. doi: 10.1186/s13059-015-0657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magurran AE. Measuring biological diversity. Blackwell; Malden: 2004. p. 256. [DOI] [PubMed] [Google Scholar]
- 41.Martins FC, et al. Evolutionary Pathways in BRCA1-Associated Breast Tumors. Cancer Discov. 2012;2:503–511. doi: 10.1158/2159-8290.CD-11-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanker AB, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci U S A. 2013;110:14372–14377. doi: 10.1073/pnas.1303204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartigan JA, Wong M. A K-means clustering algorithm. Applied Statistics. 1979;28:100–108. [Google Scholar]
- 44.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 46.Grundberg I, et al. In situ mutation detection and visualization of intratumor heterogeneity for cancer research and diagnostics. Oncotarget. 2013;4:2407–2418. doi: 10.18632/oncotarget.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 48.Lee JH, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 50.Hammond ME, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff AC, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marusyk A, et al. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature. 2014;514:54–58. doi: 10.1038/nature13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frey UH, Bachmann HS, Peters J, Siffert W. PCR-amplification of GC-rich regions: 'slowdown PCR'. Nat Protoc. 2008;3:1312–1317. doi: 10.1038/nprot.2008.112. [DOI] [PubMed] [Google Scholar]
- 54.Duprez R, et al. Immunophenotypic and genomic characterization of papillary carcinomas of the breast. J Pathol. 2012;226:427–441. doi: 10.1002/path.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efron B, Tibshirani RJ. An introduction to the bootstrap. Chapman & Hall; London: 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


