Abstract
Small heat shock proteins (sHSP) are a class of ATP-independent protein chaperones found throughout nature. They share a common ability to maintain partly unfolded proteins in soluble states under cellular stress conditions. All sHSPs contain a central domain called the α-crystallin domain (ACD); the domain is found in all sHSPs and in no other proteins and therefore defines the family. Though most sHSPs form large, often polydisperse oligomers from varying numbers of subunits, the ACD is both necessary and sufficient for formation of a dimer, the fundamental building block for oligomers. HSPB1 (also known as Hsp27) is unique among the ten human sHSPs because it contains a Cys residue in its dimer interface. HSPB1 is highly expressed under conditions of oxidative stress and is proposed to serve as a redox-sensitive chaperone. HSPB1 residue Cys137 has been proposed to modulate function by existing in either its oxidized (disulfide) or reduced (thiol) form (Chalova et al 2014). Here we report the solution-state NMR structure of oxidized HSPB1-ACD and compare it to a previously determined crystal structure of the reduced state. Formation of the disulfide-bond across the dimer interface yields a locked dimer structure with increased accessible hydrophobic surface. In the context of full-length HSPB1 oligomers, oxidation of Cys137 is associated with enhanced ability to bind the hydrophobic dye, 8-Anilinonapthalene-1-sulfonic-acid, implying an increased ability to interact with client proteins under oxidative stress.
Keywords: Molecular Chaperone, Small heat shock protein, Redox regulation, HSPB1, crystallin, oxidative stress
Biological context
Cells subject to various stresses such as ischemia, oxidation, and heat shock are at risk of accumulating unfolded or misfolded proteins that can aggregate and negatively affect cellular function. Chaperones are expressed at high levels under stress conditions to 1) maintain proteins in soluble states and 2) help proteins regain their native fold. Small Heat Shock Proteins (sHSPs) are ATP-independent chaperones that perform the former function and HSP90 or HSP70 are ATP-dependent chaperones that perform the latter. Ten sHSPs are encoded by the human genome. Progress towards structural and functional understanding of sHSPs has proven challenging. sHSPs form dynamic supramolecular and oligomeric structures from subunits that are ~ 20 kDa in size. Two pseudo-atomic structural models of HSPB5 24-mers composed of full-length subunits, (also known as αB-crystallin and CRYAB) have been determined using a combination of techniques including solid-state NMR, SAXS (Small Angle X-ray Scattering), and EM (Electron Microscopy) (Braun, Zacharias et al. 2011; Jehle, Vollmar et al. 2011), and a third model has been proposed on the basis of Tandem MS (Mass Spectrometry), IM (Ion Mobility)-MS, and EM (Baldwin, Lioe et al. 2011). These remain the only models for mammalian sHSP oligomers. Much of the current structural information available for sHSPs is for their central regions, which form the so-called α-crystallin domain (ACD; see Figure 1A). The ~100-residue ACD is found in all sHSPs, with sequence identity ranging from 40 to 60% among mammalian sHSPs (Supplemental Figure 1). Inter-subunit interactions that involve the N- and C- terminal regions that flank the ACDs are responsible for formation of oligomers. The ACD, however, is necessary and sufficient for dimer formation and removal of the N- and C-terminal regions of sHSPs yields homodimeric ACDs (~20 kDa) that are amenable to structural studies. Crystal and nmr structures of several mammalian ACDs have been determined and all reveal a topology in which each protomer forms a six- or seven-stranded β-sandwich with a dimer interface formed by two long β-strands aligned antiparallel to each other (Bagneris, Bateman et al. 2009; Laganowsky, Benesch et al. 2010; Clark, Naylor et al. 2011; Hochberg, Ecroyd et al. 2014). A detailed bioinformatics analysis of sHSPs revealed that their sequences are depleted in cysteine relative to the relevant proteome in which they reside (Kriehuber, Rattei et al. 2010). HSPB1 is unique among human sHSPs because it contains a Cys residue in its dimer interface. Notably, HSPB1 is highly expressed under conditions of oxidative stress where increased reactive oxygen species (ROS) can lead to harmful oxidation of proteins and their subsequent aggregation. HSPB1 residue Cys137 has been proposed to modulate function by existing in either its oxidized (disulfide) or reduced (thiol) form (Chalova, Sudnitsyna et al. 2014). Two crystal structures of reduced HSPB1-ACD have been solved. Here we report the solution-state NMR structure of oxidized HSPB1-ACD and present a hypothesis for the ramifications of Cys137 oxidation to HSPB1 function.
Fig. 1.
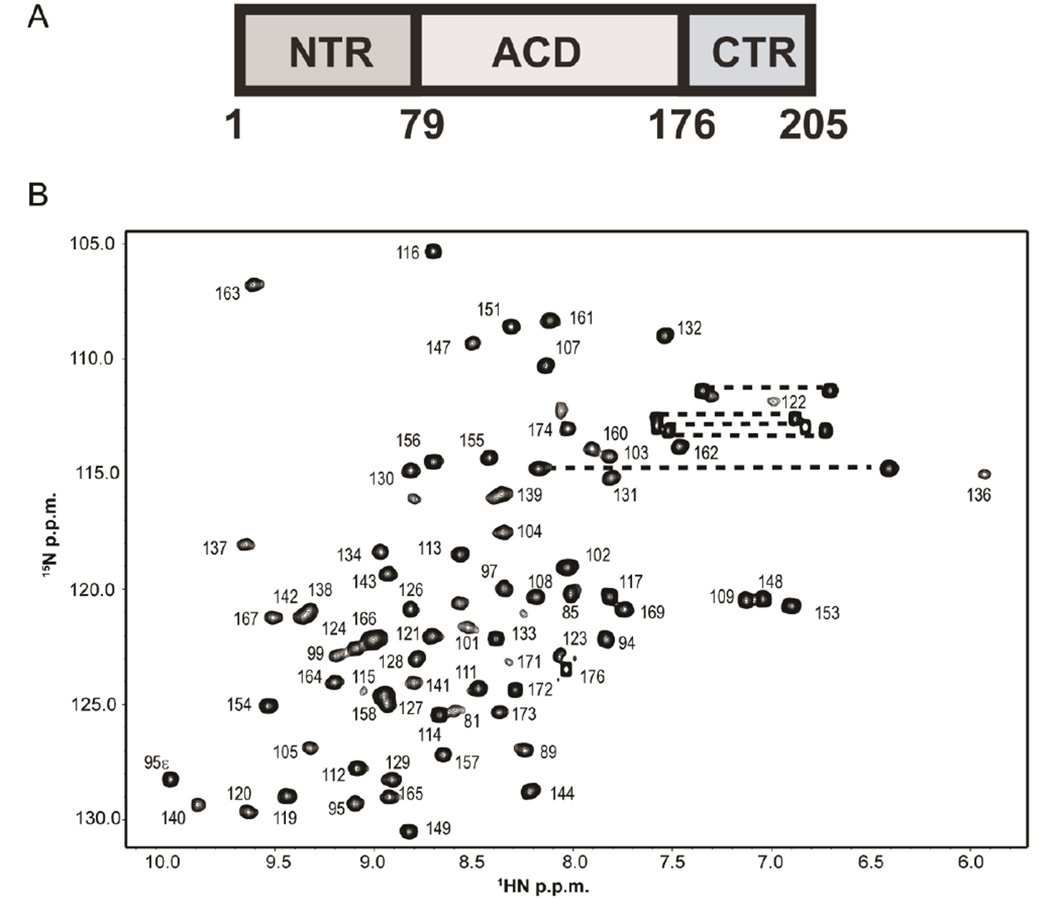
HSPB1 contains a central α-crystallin domain that is necessary and sufficient for dimer formation A) Domain organization of human HSPB1. The 100 α-crystallin domain (ACD) is flanked by a 78-residue N-terminal region (NTR) and a 29-residue C-terminal region (CTR). In the absence of the NTR and CTR, the ACD forms a homodimer in solution. B) 1H-15N HSQC spectrum of HSPB1-ACD with assignments. Dashed lines connect side-chain amide groups of Gln and Asn residues. The spectrum was acquired at 800 MHz at 22 °C on a 0.5 mM sample in sodium phosphate buffer (pH 7.5) containing 100 mM NaCl.
Methods and results
Protein expression and sample purification
A human HSPB1 ACD construct suitable for NMR studies was optimized by generating constructs of different lengths by truncation of the N- and C-terminal regions from HSPB1 (P04792). Constructs were expressed in Escherichia coli (E.coli) BL21 DE3 cells (Invitrogen), grown and purified using protocols similar to HSPB5-ACD (α-crystallin domain from human HSPB5) (Jehle, van Rossum et al. 2009). An ACD construct spanning residues Gln80 to Ser176 was found to be suitable for solution state studies as judged from the quality of 1H-15N HSQC spectra.
NMR spectroscopy
Spectra were acquired on a 1 mM HSPB1-ACD sample in 50 mM sodium phosphate buffer (pH 7.5) containing 100 mM NaCl at 22 °C. TROSY versions of the standard suite of triple resonance experiments such as HNCA, HNCOCA, HNCACB, HN(CO)CAB, HNCO, HBHA(CO)NH were acquired on a 2H, 13C, 15N-labeled sample to assign backbone resonances. Side chain assignments were obtained from CC(CO)NH- and HCC(CO)NH)-TOCSY experiments on a sample with 50% deuteration level. Side-chain assignments were further augmented from HCCH-TOCSY acquired on a uniformly 13C, 15N-labeled sample. Distance constraints were obtained from 15N-edited and 13C-edited NOESY experiments acquired with a mixing time of 150 ms on a uniformly 13C, 15N-labeled sample. Triple resonance spectra and TOCSY-type spectra were acquired on 800 MHz and 600 MHz Varian INOVA spectrometers, respectively, both equipped with a triple-resonance, z-gradient cold probe. NOESY spectra were acquired on 800 MHz and 900 MHz Varian INOVA spectrometers both equipped with a triple-resonance, z-gradient cold probe. Spectra were processed with NMRPipe (Delaglio, Grzesiek et al. 1995) and analyzed with SPARKY (Goddard and Kneller). Residual dipolar couplings were measured from IPAP (InPhase-AntiPhase) spectra acquired on a Bruker AVANCEIII 800 MHz spectrometer equipped with a triple-resonance, z-gradient cryoprobe. Spectra were processed with NMRPipe and analyzed with NMRView(Johnson 2004). 1H chemical shifts were referenced with respect to external DSS and 13C and 15N dimensions were referenced indirectly(Wishart, Bigam et al. 1995).
The 1H-15N HSQC spectrum of HSPB1-ACD labeled with assignments is shown in Fig.1B. Of 90 non-proline residues, NH resonances for 73 are observed and assigned. Residues at the N-terminal end of the construct account for the majority of missing resonances, indicating an unstructured region with residues in exchange with H2O. Residues that are not observed are: residues 80, 82–84, 86–88, 90–92 in the N-terminus; residues 96, 98, and 100 in the β3 strand; and residues 110, 118, 125, 135, and 175.
Structure calculation
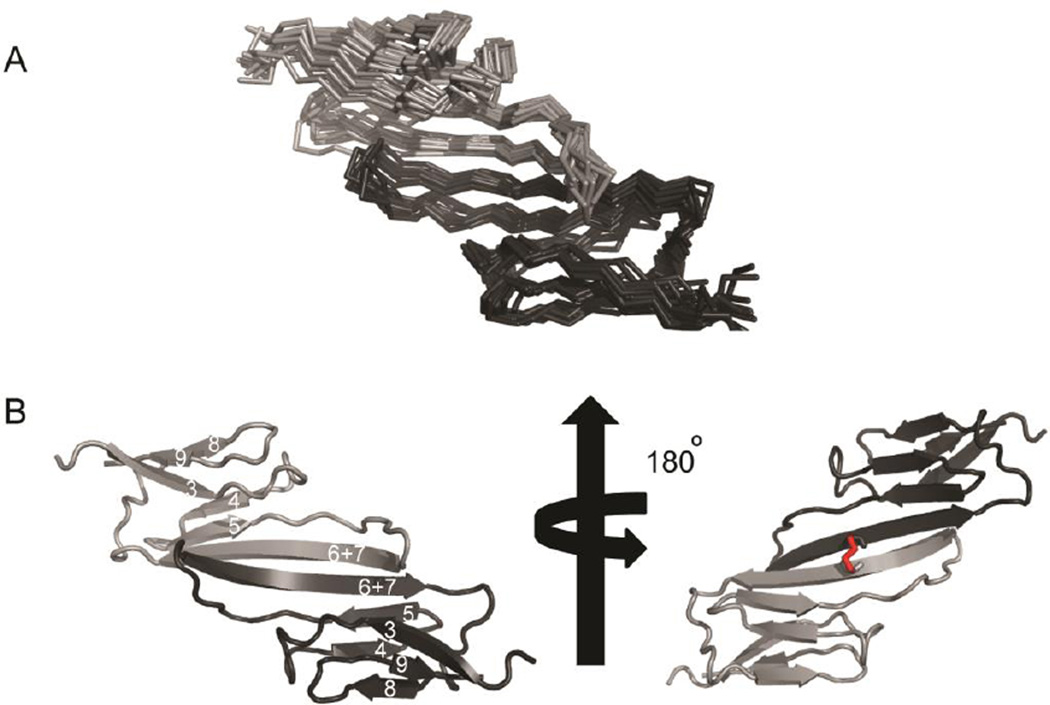
Sedimentation velocity experiments of HSPB1-ACD under reducing conditions confirmed that it exists predominantly as a homodimer at concentrations > 20 µM (Supplemental figure 2). Furthermore, HSPB1-ACD elutes as a dimer on size exclusion chromatography in both the presence and absence of DTT (data not shown). We collected 1H-15N HSQC spectra in the absence and presence of 1 mM H2O2 to see if external oxidizing agent is needed for disulfide bond formation. No differences between the spectra in the presence and absence of H2O2 were detected, indicating that the disulfide bond forms spontaneously under non-reducing conditions. NMR data were collected on disulfide-linked ACD dimer for structure calculation. A standard protocol for obtaining inter-molecular NOEs in a homodimer involves observation of NOEs from 13C-edited/filtered experiments performed on a sample containing equimolar amounts of labeled and unlabeled protein. Low signal-to-noise in these spectra prevented such analysis on HSPB1-ACD, necessitating an alternative procedure. A homology model was built using SWISS-MODEL based on the structure of the closest relative of HSPB1, HSPB5-ACD (56.18% sequence identity). The Z-score for Cβ and All Atom was −3.68 and −3.37, respectively. The model was used only as a rudimentary step for distinguishing between inter-molecular and intra-molecular NOEs. The structure of HSPB1-ACD was determined entirely from experimental restraints obtained from 13C- and 15N-edited NOESY spectra. NOESY spectra were analyzed with CcpNmr and distance restraints were obtained by binning NOEs into weak, medium, and strong categories. 1H-15N and Cα-C′ residual dipolar couplings were measured on a 200 µM sample containing 12% pf1 phage (ASLA Biotech). Structure calculations were performed using chemical shifts, ~1500 NOEs, and 1H-15N RDCs as input into RosettaOligomer, using a protocol described previously (Sgourakis, Lange et al. 2011). An explicit restraint for the inter-subunit disulfide bond was included in the inter-molecular restraint file and the structure was further refined with XPLOR-NIH. The final ensemble and a representative cartoon structure are shown in Fig. 2; structure statistics are reported in Table 1. Each protomer forms a six-stranded β-sandwich and the dimer interface is formed by two ten-residue, antiparallel β strands, centered about Cys137. The numbering of the β-strands follows the convention adopted in the sHSP field.
Fig. 2.
Solution structure of oxidized HSPB1-ACD dimer A) A 10-membered ensemble of the solution structure of HSPB1-ACD is shown (PDB ID 2N3J). Each chain in the dimer is colored in a different shade of gray. B) A cartoon representation of one member of the solution ensemble is shown. Each protomer forms a six-stranded β-sandwich and the ten-residue β6+7 strand from each protomer forms the dimer interface. The β-strands are labeled in the cartoon on the left using the numbering convention adopted in the sHSP field. The disulfide formed by Cys137 and the symmetry-related residue in the other protomer is colored red on the right.
TABLE 1.
NMR data and refinement statistics for HSPB1-ACD solution structure
| NMR distance and dihedral constraints | HSPB1-ACD |
|---|---|
| Distance constraints | |
| Total NOE | 2021 |
| Intraresidue | 228 |
| Inter-residue | |
| sequential (|i-j| = 1) | 635 |
| Medium-range (|i-j| < 4) | 279 |
| Long-range (|i-j| > 5) | 879 |
| Inter-molecular | 266 |
| Total dihedral angle restraints | 230 |
| φ (TALOS) | 115 |
| ψ (TALOS) | 115 |
| Residual Dipolar Couplings (RDCs) | |
| 1H-15N RDCs | 59 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.16±0.21 |
| Dihedral angle constraints (°) | 1.07 ± 1.29 |
| Average pairwise r.m.s deviation (Å)a | |
| Heavy | 1.63 ± 0.56 |
| Backbone | 0.79 ± 0.37 |
Average pairwise r.m.s.d. was calculated among ten refined structures.
ANS Fluorescence
1 mM ANS (8-Anilinonapthalene-1-sulfonic-acid; 10417 SIGMA) was mixed with either oxidized or reduced (full-length) HSPB1 (5 µM). The reduced form of HSPB1 was prepared by incubating the sample with 1 mM DTT overnight at room temperature. Fluorescence was monitored on an SLM/Aminco steady state fluorimeter AB-2. The emission spectrum was scanned from 400 to 520 nm using an excitation wavelength of 372 nm. Fluorescence enhancement was determined from the ratio of fluorescence in the presence of protein to the fluorescence in the absence of protein.
Discussion and conclusions
Cellular oxidative stress leads to an increase in the levels of ROS (Reactive Oxygen Species) that in turn may catalyze the oxidation of the single HSPB1 cysteine, Cys137 (colored red in Figure 2b). This raises the question of whether oxidized HSPB1 is structurally different from the reduced species. We determined the structure of oxidized HSPB1-ACD (Fig. 2), as it likely represents the prevalent species under oxidative stress conditions. Comparison of the solution structure of oxidized HSPB1-ACD with a recent crystal structure of reduced HSPB1-ACD (4MJH, residues V85-P170; (Hochberg, Ecroyd et al. 2014)) is shown in Figure 3. Protomers align with Cα atom r.m.s.d. of 2.04 Å. The dimer interface is formed by the antiparallel arrangement of two identical ten-residue β-strands, a signature characteristic of mammalian ACD structures. In the reduced ACD dimer, Cys137 is directly across from Cys137′ at the center of the dimer interface. The placement of two Cys137 residues across from each other allows for the rapid formation of the disulfide-linked form under non-reducing conditions. Furthermore, the Cys137 sidechains are surface-exposed in both the reduced and oxidized structures, suggesting that HSPB1 could be act as a scavenger for ROS, especially in light of the very high HSPB1 concentrations in cells responding to stress (Gorman, Szegezdi et al. 2005).
Fig. 3.
Comparison of the oxidized and reduced HSPB1-ACD dimer A) Solution structure of oxidized HSPB1-ACD (left) and crystal structure of reduced dimer (4MJH,right) are shown. The crystal structure was obtained on a sample co-crystallized at pH 8.5 in the presence of DTT with an 8-residue peptide that binds at the distal edge of each protomer, away from the dimer interface. The two chains of 4MJH are colored cyan and green. B) The two structures are shown edge on, with a β4-β8 groove facing the viewer. A difference in shape between the oxidized (solution) and reduced (4MJH) can be seen.
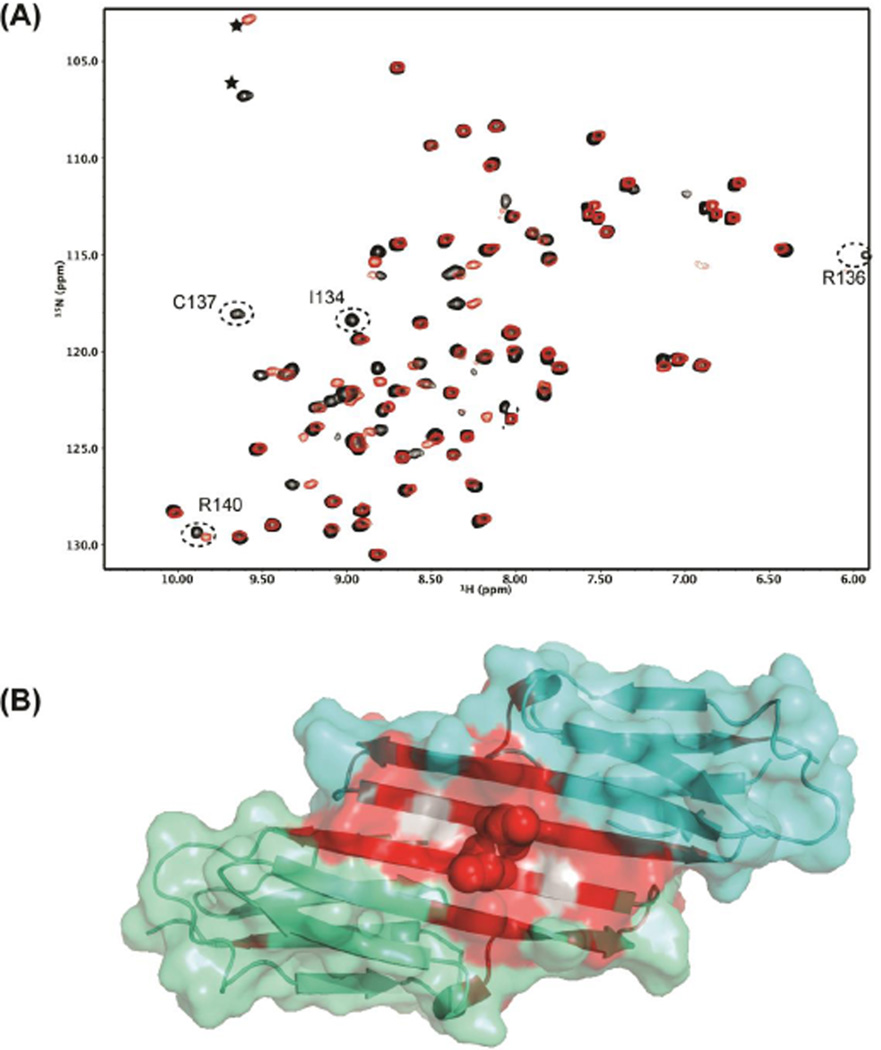
Despite the overall similarities between the NMR structure of oxidized HSPB1 and the crystal structure of the reduced dimer, there are some differences, most notably in the curvature of the dimer (Figure 4). Also, there is an additional N-terminal strand in the crystal structure, but this region is disordered in solution. Finally, the C-terminal β9-strand is shorter in solution compared to the crystal structure. To further characterize which of the observed perturbations in the structure are due to oxidation of Cys137, we collected 1H-15N HSQC spectra in the presence and absence of 5 mM DTT (black and red, respectively in Figure 4A). Small chemical shift perturbations are observed for a majority of residues in the dimer interface as well as for some residues in the neighboring β5-strand. Amide groups of residues I134, R136, and C137 disappear from the spectrum on addition of DTT (S135 is not observed in either spectrum). Exchange with the solvent, H2O, likely accounts for the disappearance of amide groups.
Fig. 4.
Oxidation/reduction of Cys137 causes chemical shift perturbations on the dimer interface A) 1H-15N HSQC spectra of the oxidized (black) and reduced (red) HSPB1-ACD. Reduced HSPB1-ACD was obtained by incubation in 5 mM DTT for several hours. Resonances for I134, R136, and R140 at the dimer interface disappear in the reduced form. Peaks labeled with asterisks are aliased due to different spectral widths in the two spectra. B) Chemical shift perturbations are colored red on the structure of reduced HSPB1-ACD (4MJH). Grey indicates unassigned residues. Perturbations are localized to the dimer interface.
A crystal structure of a variant HSPB1-ACD in which two glutamic acid residues in a loop region were changed to alanine to promote crystal formation has also been solved (Baranova, Weeks et al. 2011). The mutated residues were chosen to reduce disorder observed in the wild type construct. E125A/E126A-HSPB1-ACD crystallized at pH 5.5 and formed a hexamer in the crystal with a crystallographic six-fold axis (i.e., the canonical dimer interface is not present in this crystal). Nevertheless, the protomer structure is similar to the ACD structures described here even though the observed quaternary structure is markedly different.
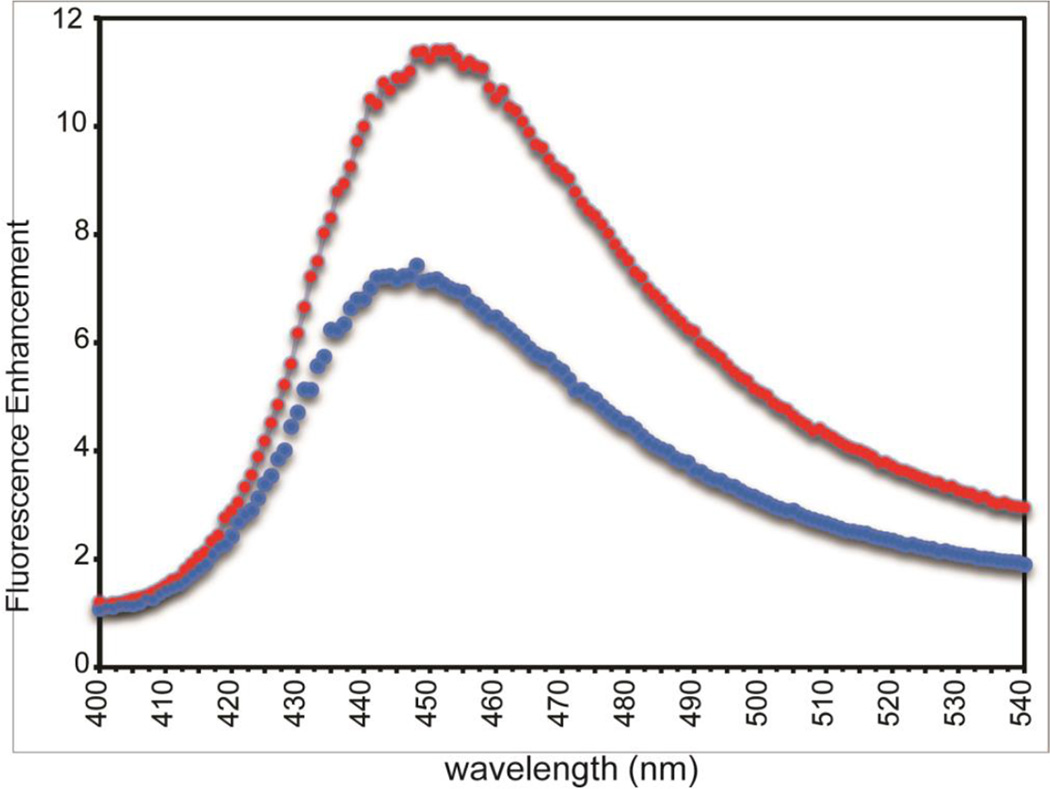
sHSPs are thought to bind hydrophobic surfaces of misfolded proteins, so readily accessible hydrophobic surfaces can potentially allow for increased client binding capacity. To ask whether the oxidation of Cys137 within the ACD has an effect in the context of the very large oligomers formed by full-length HSPB1, we compared the fluorescence of ANS (8-Anilinonapthalene-1-sulfonic-acid) in the presence of reduced and oxidized oligomers (Figure 5). Fluorescence of ANS is enhanced upon binding to proteins with exposed hydrophobic areas. The enhancement is substantially greater for oxidized HSPB1 than for reduced oligomer (red versus blue, respectively). Thus, under oxidative stress conditions, HSPB1 oligomers will have a greater exposed hydrophobic area. A correlation between chaperone activity and ANS binding has been observed for a phospho-mimic of HSPB5 which has increased ANS fluorescence indicating greater accessible hydrophobicity and is a better chaperone than wt-HSPB5 (Peschek, Braun et al. 2013). Formation of a disulfide bond leading to a conformational change is a mechanism observed in several redox-sensitive chaperones, such as Hsp33 (Graumann, Lilie et al. 2001). The unique Cys137 is conserved in all mammalian HSPB1 proteins, so it is likely that a similar mechanism is in play here. Our results indicate that oxidation can cause small but functionally significant alterations in the properties of HSPB1 that can serve to modulate its function in cells undergoing stress.
Fig. 5.
Enhancement of ANS fluorescence by oxidized (red) and reduced (blue) full-length HSPB1 oligomers. The larger enhancement of ANS fluorescence by oxidized HSPB1 is consistent with more extensive exposed hydrophobic surface.
Supplementary Material
Acknowledgements
We thank David Baker for access to RosettaOligomer software and computing facilities, William Atkins for use of his fluorimeter, and Hannah Braughman for help with fluorescence experiments. The work was supported by NIH grant 1R01 EY017370 (to REK).
References
- Bagneris C, Bateman OA, et al. Crystal structures of alpha-crystallin domain dimers of alphaB-crystallin and Hsp20. J Mol Biol. 2009;392(5):1242–1252. doi: 10.1016/j.jmb.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Baldwin AJ, Lioe H, et al. The polydispersity of alphaB-crystallin is rationalized by an interconverting polyhedral architecture. Structure. 2011;19(12):1855–1863. doi: 10.1016/j.str.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Baranova EV, Weeks SD, et al. Three-dimensional structure of alpha-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J Mol Biol. 2011;411(1):110–122. doi: 10.1016/j.jmb.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Braun N, Zacharias M, et al. Multiple molecular architectures of the eye lens chaperone alphaB-crystallin elucidated by a triple hybrid approach. Proc Natl Acad Sci U S A. 2011;108(51):20491–20496. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalova AS, Sudnitsyna MV, et al. Effect of disulfide crosslinking on thermal transitions and chaperone-like activity of human small heat shock protein HspB1. Cell Stress Chaperones. 2014;19(6):963–972. doi: 10.1007/s12192-014-0520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AR, Naylor CE, et al. Crystal structure of R120G disease mutant of human alphaB-crystallin domain dimer shows closure of a groove. J Mol Biol. 2011;408(1):118–134. doi: 10.1016/j.jmb.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Gorman AM, Szegezdi E, et al. Hsp27 inhibits 6-hydroxydopamine-induced cytochrome c release and apoptosis in PC12 cells. Biochem Biophys Res Commun. 2005;327(3):801–810. doi: 10.1016/j.bbrc.2004.12.066. [DOI] [PubMed] [Google Scholar]
- Graumann J, Lilie H, et al. Activation of the redox-regulated molecular chaperone Hsp33--a two-step mechanism. Structure. 2001;9(5):377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- Hochberg GK, Ecroyd H, et al. The structured core domain of alphaB-crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci U S A. 2014;111(16):E1562–E1570. doi: 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, van Rossum B, et al. alphaB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer. J Mol Biol. 2009;385(5):1481–1497. doi: 10.1016/j.jmb.2008.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle S, Vollmar BS, et al. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc Natl Acad Sci U S A. 2011;108(16):6409–6414. doi: 10.1073/pnas.1014656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- Kriehuber T, Rattei T, et al. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010;24(10):3633–3642. doi: 10.1096/fj.10-156992. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Benesch JL, et al. Crystal structures of truncated alphaA and alphaB crystallins reveal structural mechanisms of polydispersity important for eye lens function. Protein Sci. 2010;19(5):1031–1043. doi: 10.1002/pro.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J, Braun N, et al. Regulated structural transitions unleash the chaperone activity of alphaB-crystallin. Proc Natl Acad Sci U S A. 2013;110(40):E3780–E3789. doi: 10.1073/pnas.1308898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgourakis NG, Lange OF, et al. Determination of the structures of symmetric protein oligomers from NMR chemical shifts and residual dipolar couplings. J Am Chem Soc. 2011;133(16):6288–6298. doi: 10.1021/ja111318m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.