Abstract
Specialized junctional sites that connect the plasma membrane (PM) and endoplasmic reticulum (ER) play critical roles in controlling lipid metabolism and Ca2+ signaling1–4. Store operated Ca2+ entry mediated by dynamic STIM1-ORAI1 coupling represents a classical molecular event occurring at ER-PM junctions, but the protein composition and how previously-unrecognized protein regulators facilitate this process remain ill-defined. Using a combination of spatially-restricted biotin-labelling in situ coupled with mass spectrometry5, 6 and a secondary screen based on bimolecular fluorescence complementation7, we mapped the proteome of intact ER-PM junctions in living cells without disrupting their architectural integrity. Our approaches lead to the discovery of an ER-resident multi-transmembrane protein that we call STIMATE (STIM-activating enhancer, encoded by TMEM110) as a positive regulator of Ca2+ influx in vertebrates. STIMATE physically interacts with STIM1 to promote STIM1 conformational switch. Genetic depletion of STIMATE substantially reduces STIM1 puncta formation at ER-PM junctions and suppresses the Ca2+-NFAT signaling. Our findings enable further genetic studies to elucidate the function of STIMATE in normal physiology and disease, and set the stage to uncover more uncharted functions of hitherto underexplored ER-PM junctions.
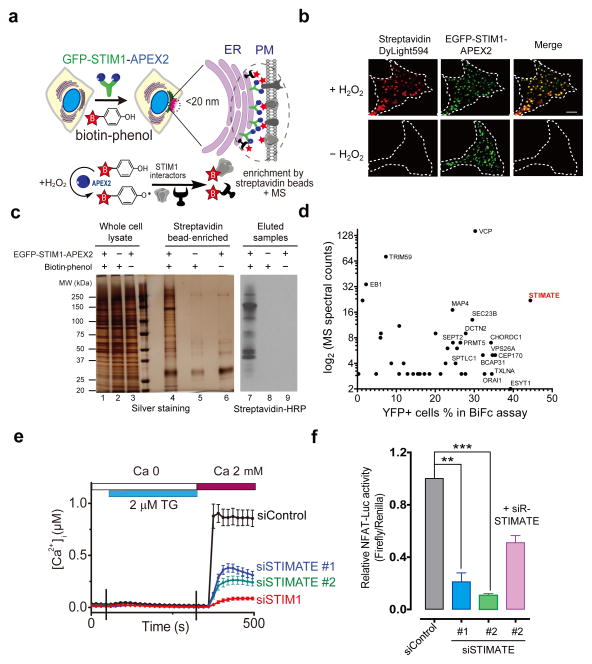
ER-PM junctions are defined as spatially extended or small circular compartments in which the PM and ER membranes are stably separated at a distance of approximately 10–20 nm without direct membrane fusion1. The broad significance of this structure has only recently begun to be appreciated, with emerging roles in lipid metabolism and Ca2+ homeostasis1–4. Although ER-PM junctions were first observed over half a century ago8, systematic dissection of this specialized subcompartment at the molecular level remains challenging due to the lack of appropriate methods and convenient tools, which motivated us to explore a non-biased yet effective approach to obtain an integrated picture of ER-PM junctions. By taking advantage of an in situ protein labeling technique5, 6, we set out to map the proteome of ER-PM junctions that are dedicated to store operated calcium entry (SOCE), a fundamental physiological process that is mediated by two protein families, STIM and ORAI3. The ER-resident Ca2+ sensor protein, STIM1, when fused to an engineered ascorbate peroxidase 2 (APEX2), enables proximity biotin labeling with least perturbation to the architecture of ER-PM junctions, thereby opening new avenues for capturing protein complexes situated at or in close proximity of STIM1 in living cells (Fig. 1a). At the heart of this in situ protein labelling technique is APEX2-catalyzed conversion of biotin-phenol in the presence of H2O2 to phenoxyl radicals, which could attack electron-rich amino acids and covalently attach biotin tags to targeted proteins5, 6. Biotinylated proteins can be subsequently enriched by streptavidin beads and analyzed by mass spectrometry. Notably, these radicals have very short lifetimes (<1 ms) with an estimated labelling radius of <20 nm5, 9, 10, thus matching the distance that separates ER and PM at junctional sites. One additional benefit of this approach is that it enables dynamic sampling of this specialized cellular compartment during the translocation of STIM1 from ER toward PM, thereby allowing us to compare protein complexes surrounding STIM1 before and after store depletion.
Figure 1. STIMATE identified as a positive regulator of the Ca2+-NFAT pathway.
a, Schematic depiction of proteomic mapping of potential STIM1 interactors at ER-PM junctions through APEX2-mediated biotin-labelling in situ. APEX2 fused to STIM1 enables biotin labelling on proteins located <20 nm from STIM1. This reaction requires externally added H2O2. Biotinylated proteins were further affinity-enriched with streptavidin beads, with the eluted products analyzed by mass spectrometry (MS).
b, Fluorescence imaging of biotinylated proteins (stained with streptavidin conjugated with Dylight594) in HEK293 cells expressing GFP-STIM1-APEX2 as illustrated in panel a. Store depletion was induced by 1 μM thapsigargin (TG) in nominally Ca2+ and serum free medium. Scale bar, 5 μm.
c, Silver staining and immunoblotting of biotinylated protein complexes surrounding STIM1 puncta at ER-PM junctions. Shown on the SDS-PAGE were samples before (lanes 1–3) and after (lanes 4–6) streptavidin bead enrichment, as well as the eluted samples immunoblotted by streptavidin conjugated by horseradish peroxidase (streptavidin-HRP, lanes 7–9).
d, Scatter plot of identified potential STIM1 binding partners. Bimolecular fluorescence complementation (BiFc, Supplementary Fig. 1a-c) was used as a secondary assay to confirm strong hits that emerged from the proteomic study. The N-terminal fragment of YFP (YFPN, residues 1–155) is fused to the STIM1 C-terminus as the bait to interact with prey in a customized library containing candidate genes fused with the C-terminal half of YFP (YFPC, residues 156–238). Each candidate was plotted according to its MS spectral counts (Y axis) and the degree of YFP fluorescence restoration (X axis, as shown in Supplementary Fig. 1c).
e, TG-induced Ca2+ influx in HEK293 cells transfected with siControl (n = 30), siSTIMATE oligo 1 (n = 34), siSTIMATE oligo 2 (n = 45) or siSTIM1 (n = 60 cells pooled across three independent experiments). Error bars denote s.e.m.
f, Quantification of NFAT-dependent luciferase activity in HEK293 cells treated with indicated siRNA oligos or co-transfected with an siRNA-resistant STIMATE construct (siR-STIMATE). **P<0.01; ***P < 0.001 (paired Student’s t-test); Error bars denote s.d. for n = 12 wells of cultured cells (0.1–0.5 million cells per well) pooled across three independent experiments.
Following ER Ca2+ depletion elicited by thapsigargin (TG), a blocker of the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA), total internal reflection fluorescence (TIRF) microscope imaging showed that EGFP-tagged STIM1-APEX2 formed puncta and colocalised tightly with biotinylated proteins, the latter of which were labelled by fluorophore-conjugated streptavidin (Fig. 1b). This process was dependent on H2O2 since biotinylation was not observed whilst omitting H2O2 in the reaction medium. The H2O2- and APEX2-dependent biotinylation of protein complexes surrounding STIM1 was further confirmed by silver staining after affinity enrichment and independently by western blotting probed by streptavidin-HRP (Fig. 1c). Because ORAI1 (MW: 33 kDa) is a known binding partner of STIM1, we first analysed a gel slice corresponding to 25–37 kDa on the SDS-PAGE with mass spectrometry in our initial experiment and confirmed the presence of ORAI1 using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). In the same gel band, we repeatedly detected the gene product of TMEM110 (RefSeq ID: NM_198563), hereafter designated as STIMATE (for STIM-activating enhancer, Supplementary Fig. 1). In our subsequent LC-MS/MS analyses on all the eluted products, we further identified a total of 73 potential STIM1 interactors, with 17 detected in both store-full and store-depleted HEK293 cells (Supplementary Tables 1 and 2). The majority of them fell into the categories of ER- or PM-resident proteins, cytoskeletal components, and proteins functioning in intracellular membrane trafficking or posttranslational modifications. Notably, 18% of them also appeared as strong hits identified in an independent genome-wide screen for regulators of nuclear factor of activated T-cells (NFAT)11, a transcriptional factor that is downstream of Ca2+ influx mediated by STIM-ORAI signaling. The prowess of this approach was further attested by the identification of EB1, SERCA2, ORAI1, voltage-dependent Ca2+ channels, extended synaptotagmin 1 and septins as candidate proteins that are in close proximity to STIM1 (Supplementary Tables 1 and 2). These proteins are known to either directly interact with STIM1 or reside in the ER-PM junctions3, 11–21.
Given the major interest in identifying protein regulators at ER-PM junctions that are directly involved in STIM1-dependent Ca2+ influx, we created a customized prey library of 38 genes representing the majority of gene products (e.g., STIMATE) identified under both store-full (resting) and store depleted conditions, as well as candidate genes that have an annotated function in Ca2+ signaling or harbor predicted transmembrane domains (Supplementary Table 3). A secondary screen based on bimolecular fluorescence complementation (BiFc, Supplementary Fig. 1a–c) was performed to examine their potentials as STIM1 interactors either before or after TG-induced store depletion. Our assay is based on the reconstitution of an intact yellow fluorescent protein (YFP) from two complementary non-fluorescent fragments when they are brought together through STIM1 and STIM1-binding partners. Again, STIMATE stood out as one of the strongest hits in the screen (Fig. 1d and Supplementary Fig. 1b–sc). Collectively, STIMATE, a positive regulator of NFAT activation identified in a previous genome-wide RNAi screening11, emerged early and prominently in both screens, which prompted us to further examine its role in modulating Ca2+ flux at ER-PM junctions, and in parallel, to dissect the molecular mechanism underlying its interplay with STIM1.
To examine the effect of STIMATE on SOCE and its downstream effectors, we monitored the Ca2+ flux and NFAT-dependent activities by treating HEK293 cells with siRNA oligonucleotides designed to specifically target STIMATE. The knockdown efficiency (~60–80%) of two synthesized siSTIMATE oligos was confirmed by qPCR (Supplementary Fig. 2). As shown in Fig. 1e–f, TG-triggered SOCE, as well as NFAT-dependent luciferase activity, was significantly suppressed in HEK293 cells transfected with either of the two siSTIMATE oligos. The inhibitory effect could be rescued in siSTIMATE-treated cells by overexpression of an siRNA-resistant variant of STIMATE (Fig. 1f).
To further demonstrate the role of STIMATE in modulating SOCE, we generated STIMATE knockout (STIMATE-KO) HEK293 stable cell lines by disrupting exon 4 of human STIMATE with the CRISPR-Cas9 genome-editing tool22. Gene disruption was validated using both the Surveyor nuclease assay and Sanger’s sequencing of exon 4 (Supplementary Fig. 2c–d). In two stable STIMATE-KO clones harboring early stop codons, the cytoplasmic Ca2+ increase in response to ionomycin-induced store depletion (Supplementary Fig. 2e) was profoundly reduced, whilst the Ca2+ release from the ER Ca2+ store remained largely unaltered (first peak, Supplementary Fig. 2e). Together, these results suggest that STIMATE is a previously-unrecognized positive modulator of SOCE.
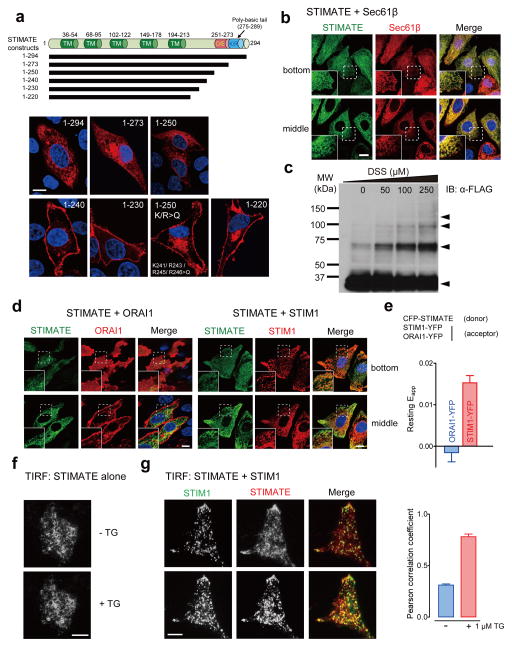
STIMATE is broadly expressed in mouse and human tissues (Supplementary Fig. 2a). STIMATE protein is predicted to contain multiple putative transmembrane (TM) segments with a polybasic C tail (Fig. 2a and Supplementary Fig. 1d), but its subcellular localization and membrane topology remain unknown. When overexpressed in HEK293, HeLa or COS-7 cells, STIMATE localised to ER and overlapped tightly with the ER marker Sec61β23 or DsRed-ER (Fig. 2b and Supplementary Fig. 3). Interestingly, STIMATE exhibited a predominant PM-like localisation with a small pool trapped intracellularly when the last 74 amino acids was truncated (STIMATE1–220, Fig. 2a). Through a serial truncation study combined with site-directed mutagenesis, we further mapped a region (residues 241KXRXRR246) that seems to be critical for keeping STIMATE localised within the ER. Substitution of these positively charged residues with glutamines (K or R>Q mutant) caused a large fraction of the ER-resident STIMATE1–250-mCherry to exhibit a PM-like distribution (Fig. 2a). Next, we performed a chemical cross-linking experiment to examine the oligomeric state of STIMATE. As shown in Fig. 2c, addition of membrane-permeable cross-linker DSS resulted in the appearance of protein bands corresponding to multimers of STIMATE on SDS-PAGE, suggesting that STIMATE could be assembled as oligomers in ER membrane. This result echoes the presence of a GXXXG transmembrane association motif24 in one of its predicted TM segments (Supplementary Fig. 1d). The tendency of STIMATE to multimerize was further attested by the efficient restoration of YFP fluorescence when STIMATE was C-terminally tagged with two complementary non-fluorescent YFP fragments in the BiFc assay (Supplementary Fig. 1b–c). The membrane topology of STIMATE was further determined by using a fluorescence protease protection assay (FPP)25, with EGFP-STIM1 and STIM1-YFP as controls. We found that both termini of STIMATE face toward the cytosolic side of ER (Supplementary Fig. 1e–h).
Figure 2. STIMATE is an ER-resident protein that colocalises with STIM1.
a, Predicted domain architecture of STIMATE and confocal images of indicated STIMATE variants tagged with a C-terminal mCherry. STIMATE is predicted to contain 4–5 transmembrane segments (see Supplementary Fig. 1). TM, predicted transmembrane segment. D/E: a negatively-charged region in the C-terminus of STIMATE (STIMATE-CT). K/R: a ploy-basic domain in STIMATE-CT. Blue, nuclear staining with Hoechst 33342. Scale bar, 10 μm.
b, Confocal sections (at the footprint or the middle plane) of HEK293 cells coexpressing EGFP-STIMATE (green) and the ER marker mCherry-Sec61β (red). Nuclei were labeled in blue by Hoechst 33342. The insets show the regions outlined by dotted lines at higher magnification. Scale bar, 10 μm.
c, Cross-linked FLAG-STIMATE resolved on SDS-PAGE. FLAG-tagged STIMATE was transiently expressed in HEK293 cells and subjected to chemical cross-linking with 0–250 μM DSS (a membrane-permeable cross-linker) on ice for 5 min. The arrows indicate the appearance of FLAG-STIMATE oligomers upon cross-linking.
d, Confocal images of EGFP-STIMATE in HEK293 cells coexpressing mCherry-ORAI1 or mCherry-STIM1 without store depletion. Blue, nuclear staining with Hoechst 33342. Scale bar, 10 μm.
e, Quantification of FRET signals in HEK293 cells coexpressing CFP-STIMATE with ORAI1-YFP (n = 5 cells) or with STIM1-YFP (n = 10 cells) pooled across three independent experiments). Error bars denote s.e.m..
f, Representative TIRF images of an HEK293 cell expressing STIMATE-EYFP before and after store depletion induced by 1 μM TG. Scale bar, 10 μm.
g, Representative TIRF images of an HEK293 cell coexpressing EGFP-STIM1 (green) and mCherry-STIMATE (red) before and after store depletion induced by 1 μM TG. The single channel images are shown in grayscale. The plot on the right shows the quantification of STIM1-STIMATE colocalisation under TIRFM by Pearson’s correlation coefficient. Error bars denote s.e.m. for n = 6 cells pooled across two independent experiments. Scale bar, 10 μm.
We next examined whether STIMATE is located in the vicinity of the two major players of SOCE, STIM1 and ORAI1. When expressed together with ORAI1 or STIM1 in HEK293 cells, STIMATE colocalised tightly with STIM1 to ER but not with ORAI1 at resting conditions (Fig. 2d), a finding that is further corroborated by results from an independent FRET assay (Fig. 2e). Discernible FRET signals were only detected in HEK293 cells cotransfected with CFP-STIMATE and STIM1-YFP but not in cells coexpressing CFP-STIMATE and ORAI1-YFP (Fig. 2e). When expressed alone or coexpressed with an ER marker in HEK293, HeLa or COS-7 cells, STIMATE did not seem to globally translocate to ER-PM junctions as STIM1 did after store depletion (Fig. 2f and Supplementary Fig. 4a). However, when STIMATE was coexpressed with STIM1 in the same cell, we observed the colocalisation of STIMATE with STIM1 at ER-PM junctions in a fraction of cells after store depletion. This phenomenon was less obvious under the confocal microscope but more pronounced when cells were examined by TIRF microscopy, which reports exclusively on fluorescence within 200 nm above the coverslip (Fig. 2g and Supplementary Fig. 4b). The coexpression of ORAI1 did not seem to increase the degree or tendency of STIMATE to colocalise with STIM1 (Supplementary Fig. 4c–d). Thus, a portion of STIMATE might translocate to ER-PM junctions in a STIM1-dependent manner in store-depleted cells. Together, our data imply that STIMATE colocalises with STIM1 within the ER and might also reside in close proximity to STIM1 at ER-PM junctions in response to store depletion in mammalian cells.
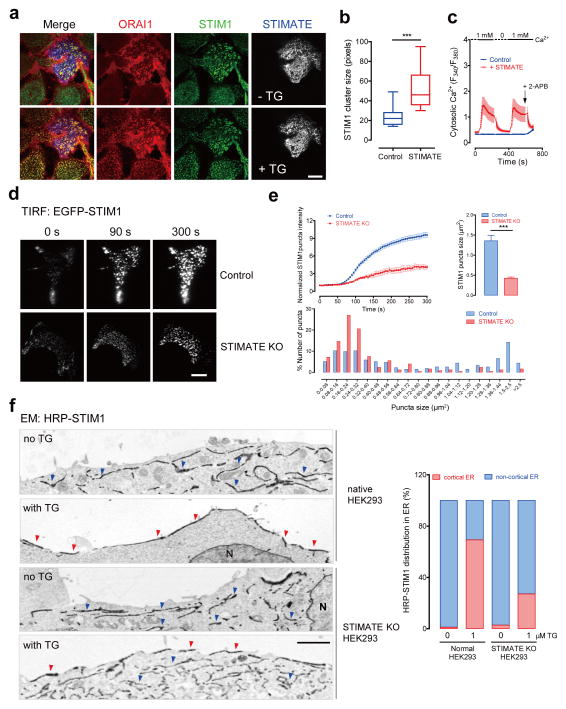
After expression of mCherry-STIMATE in HEK293 cells cotransfected with STIM1, we frequently observed the spontaneous formation of STIM1 puncta even when the ER store was not depleted (Fig. 2g and Supplementary Fig. 4b–c). This scenario was most prominently visualized in cells stably expressing both ORAI1-CFP and STIM1-YFP (Fig. 3a–c). STIM1 puncta could be readily detected in cells transfected with mCherry-STIMATE before store depletion. In contrast, the other cells in the same field without STIMATE overexpression showed an even distribution of STIM1 at the footprint of HEK293 cells (Fig. 3a). Upon store depletion, STIMATE-expressing cells formed significantly larger STIM1 puncta (Fig. 3b). Consistent with the pre-formation of STIM1 puncta before store depletion, overexpression of mCherry-STIMATE also elicited constitutive Ca2+ influx in these stably expressing cells when we switched the extracellular Ca2+ concentration from 0 to 1 mM (Fig. 3c). The Ca2+ flux could be reversed by withdrawing Ca2+ or by adding 2-APB, a pharmacological tool that is widely used to block SOCE at high concentrations (~50–100 μM).
Figure 3. STIMATE facilitates efficient formation of STIM1 puncta at ER-PM junctions.
a, Confocal images at the footprint of STIM1-YFP + ORAI1-CFP HEK293 stable cells transiently transfected with mCherry-STIMATE. STIM1, ORAI1 and STIMATE are artificially colored in green, red and gray, respectively, to aid visualization. Store depletion was triggered by 1 μM TG. Scale bar, 10 μm.
b, Quantification of STIM1 cluster size in TG-stimulated cells treated as in panel a. The rectangle in the box plot shows the distribution of data between the first and third quartiles. The segment inside the rectangle shows the median, whilst the whiskers above and below the box mark the minimum and maximum. ***P<0.001 (n=15 cells pooled across three independent experiments; paired Student’s t-test).
c, Constitutive Ca2+ influx elicited by transient expression of STIMATE in STIM1-YFP + ORAI1-CFP HEK293 stable cells. Ca2+ influx was inhibited by washing away external Ca2+ or by the addition of 50 μM 2-APB. Error bars denote s.e.m. for n=15 (control) or n=20 (STIMATE group) cells pooled across three independent experiments.
d, Representative TIRF images of STIM1 acquired 0, 90 or 300 seconds after depletion of ER Ca2+ stores with 1 μM TG in normal or STIMATE-KO HEK293 stable cells. Scale bar, 10 μm.
e, Time course of EGFP-STIM1 puncta formation (upper left panel) and quantification of STIM1 puncta size (upper right panel) and their distribution profiles (lower panel). Store depletion was induced by 1 μM TG. ***P<0.001 (compared to control; paired Student’s t-test). Error bars denote s.e.m. for n=6 cells pooled across three independent experiments.
f, Electron micrographs of HRP-STIM1 in normal or STIMATE-KO HEK293 cells before and after store depletion triggered by 1 μM TG. Blue arrowheads, HRP-STIM1 staining in non-cortical intracellular ER pools; red arrowheads, HRP-STIM1 distribution at cortical ER that is separated <20 nm from PM. Bar graph on the right represents the quantification of HRP-STIM1 distribution in different pools of the ER (average of 5–10 cells). Scale bar, 2000 nm.
These findings led us to propose that STIMATE could modulate STIM1 puncta formation at ER-PM junctions. Indeed, genetic disruption of STIMATE in HEK293 (STIMATE-KO) cells induced a substantial reduction in the amounts of STIM1 puncta at ER-PM junctions (Fig. 3d and Supplementary Video 1). Compared to TG-stimulated normal HEK293 cells, the fluorescence intensity of EGFP-STIM1 at the TIRF layer of STIMATE-KO cells dropped by almost 70%. STIMATE knockout appreciably delayed the formation of STIM1 puncta, and resulted in a significant reduction in the puncta size at ER-PM junctions (Fig. 3e). An appreciable (~10–15%), but less pronounced, reduction in puncta formation was also observed in STIMATE knockout cells expressing the STIM1 gain-of-function mutants D76A and L258G (Supplementary Fig. 5a–d). Subsequent scrutiny under high-resolution electron microscopy (EM) further revealed that fewer STIM1 proteins migrated into cortical ER (cER, Fig. 3f and Supplementary Fig. 5e), a specialized ER network that lies immediately beneath the PM (<20~30 nm). In aggregate, these results indicate that STIMATE is required to promote efficient STIM1 clustering at ER-PM junctions, which might be achieved through modulating cER accumulation or modulating STIM1 activation as described below.
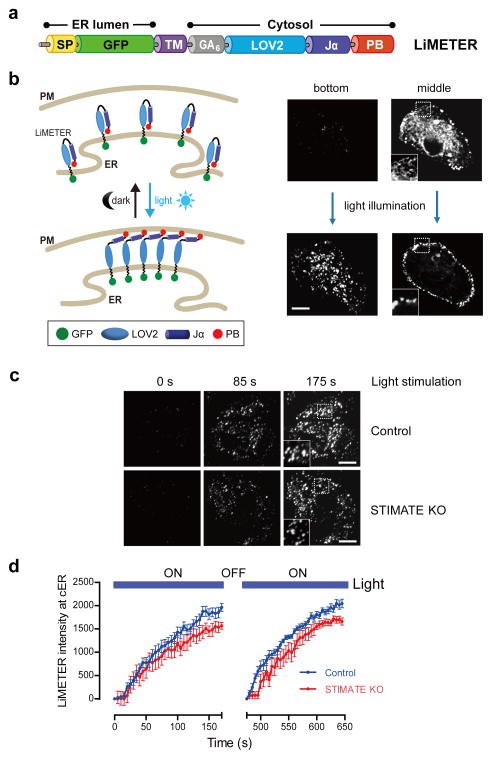
To examine the effect of STIMATE depletion on cER, we compared the cER accumulation in native and STIMATE-KO HEK293 cells. We used the genetically-encoded fluorescent cER marker, MAPPER (membrane-attached peripheral ER), as readout because it has been recently shown to selectively label cER and colocalise tightly with activated STIM1 puncta at ER-PM junctions20. We noted that depletion of STIMATE caused approximately 8–10% decrease in the intensity or density of cER in HEK293 cells (Supplementary Fig. 5f–h). To rule out the possibility that the constitutive localization of MAPPER at ER-PM junctions might cause bias in our analysis on cER, we created an optogenetic tool, LiMETER (light-inducible membrane-tethered peripheral ER, Fig. 4 and Supplementary Video 2), to reversibly label cER. The ER luminal domain of LiMETER is composed of a signal peptide and the single TM domain derived from STIM1, with GFP placed in between as reporter. The cytoplasmic region of LiMETER contains a flexible linker and a genetically-encoded lightswitch LOV2 domain (Light Oxygen Voltage-sensing domain, residues 404–546) derived from Avena sativa phototrophin 1 (refs. 26, 27), followed by a C-terminal PM-targeting polybasic tail isolated from the small G protein Rit28 (Fig. 4a). In the dark, the Jα helix docks to the LOV2 domain and cages the polybasic tail to prevent its interaction with negatively-charged PM-resident phosphoinositides. Upon blue light stimulation, photoexcitation generates a covalent adduct between a cysteine residue (C450) and the flavin cofactor in LOV2, and subsequently promotes the undocking and unwinding of the Jα helix26, 27, thereby exposing the Rit polybasic C-tail to enable translocation of the protein toward PM to form puncta-like structures28. As a result, LiMETER underwent photo-inducible accumulation at ER-PM junctions to specifically label cER (Fig. 4 and Supplementary Video 2). Notably, this process can be reversibly repeated with multiple light-dark cycles without significant loss in the magnitude of response (Fig. 4d). This unique tool enables us to quantitatively examine the effect of STIMATE depletion on the dynamics of cER accumulation at defined spatiotemporal resolution. When compared to native HEK293 cells, we observed 10–12% decrease in the rate and extent of LiMETER accumulation at ER-PM junctions in STIMATE-KO HEK293 cells after blue light illumination (Fig. 4c–d). Thus, by using either MAPPER or LiMETER as cER marker, we found that genetic depletion of STIMATE noticeably affects cER accumulation in HEK293 cells. Nonetheless, the ~10% decrease in the efficiency of cER accumulation could not fully explain the more pronounced effect (~70%) on STIM1 puncta formation in STIMATE-depleted cells (Fig. 3d–e). This prompted us to further explore alternative mechanisms by focusing on the initial activation steps of STIM1.
Figure 4. Effect of STIMATE depletion on cER accumulation reported by LiMETER.
a, Design of LiMETER as an optogenetic tool to photo-manipulate ER-PM junctions and reversibly label cER. The ER luminal region of LiMETER includes a signal peptide (SP) and the single transmembrane domain (TM) derived from human STIM1, with GFP inserted in between as reporter. The cytoplasmic region is composed of a (GA)6 linker, the lightswitch LOV2 domain and a PM-targeting polybasic tail (PB) derived from the small GTPase Rit.
b, In the dark, the LOV2 Jα helix keeps the polybasic C-tail caged to prevent its targeting toward PM. LiMETER appears as a typical ER-resident protein like STIM1 at its resting state. Upon blue light stimulation (450–490 nm), the unwinding of the Jα helix exposes the PB domain to associate with phosphoinositides enriched in the inner leaflet of the plasma membrane, thereby promoting the formation of LiMETER puncta at cER (much as STIM1 forms puncta at ER-PM junctions). The confocal images on the right are representative confocal sections at the footprint or the middle layers of a typical LiMETER-expressing HEK293 cell before (upper) and after photoactivation (lower). A 488-nm laser was used as the light source to activate LiMETER while acquiring the GFP signals. Scale bar, 10 μm.
c-d, Representative TIRF images (c) and time course (d) of LiMETER accumulation at cER in response to light stimulation (488 nm with an estimated power density of 50 mW cm-2) in normal (upper) or STIMATE-KO HEK293 cells (lower). For points falling within the time window of 125–175 s or 600–650 s, the difference of signal between control and STIMATE-KO was statistically significant (P < 0.05). Nonetheless, no significant difference was observed in the rate of LiMETER translocation toward cER (P = 0.07, paired Student’s t-test). The LiMETER intensity was quantified using ImageJ software. Error bars denote s.e.m. for n=10 (control) or n=12 cells (STIMATE-KO) pooled across three independent experiment that emitted comparable total fluorescence signals under epifluorescence microscopy. Scale bar, 10 μm.
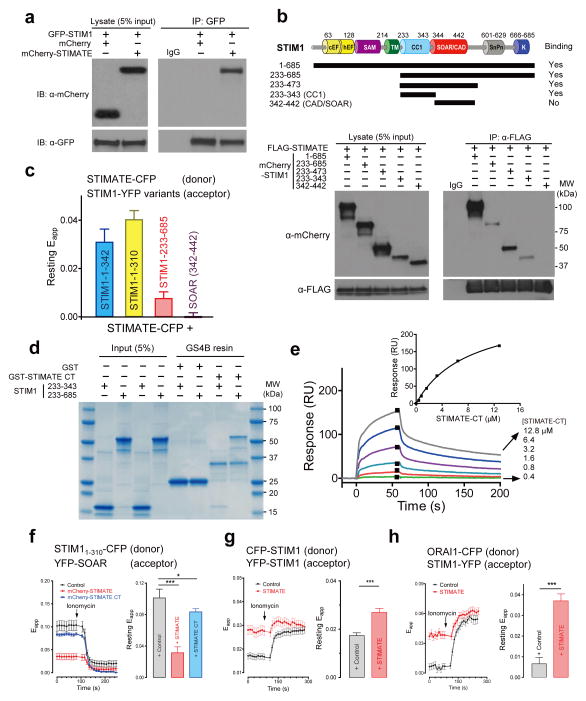
Results from both the BiFc (Supplementary Fig. 1a–c) and FRET (Fig. 2e) assays suggest a possible interaction between STIMATE and STIM1 in HEK293 cells. We further confirmed this by using coimmunoprecipitation (CoIP, Fig. 5a). To investigate how STIMATE modulates STIM1 actions, we applied a combination of CoIP (Fig. 5b) and FRET (Fig. 5c) methods to map the domain(s) within STIM1 that mediate its interaction with STIMATE. We narrowed down the minimal STIMATE-interacting domain to the juxtamembrane coiled coil region of STIM1 (STIM1-CC1, residues 233–343). In both assays, the minimal ORAI-activating domain of STIM1 (CAD/SOAR)29, 30 showed negligible or no discernible interaction with STIMATE. Since the longest cytosolic fragment of STIMATE protein is its C-terminal region (STIMATE-CT, residues 214–294, Fig. 2a and Supplementary Fig. 1d), we further expressed STIMATE-CT in bacteria and purified the recombinant protein to examine whether it would directly interact with STIM1 cytosolic domains. GST-tagged STIMATE-CT immobilized on GS4B resin was able to pull down both the entire STIM1 cytoplasmic domain (233–685) and STIM1-CC1 (Fig. 5d). The dissociation constant of the interaction between STIMATE-CT and STIM1-CC1 was further determined to be 8.9 ± 0.7 μM (Fig. 5e). Overall, these studies clearly demonstrated a direct physical contact between STIMATE and STIM1 in vitro.
Figure 5. STIMATE interacts with STIM1 cytosolic fragments and promotes STIM1 conformational switch.
a, GFP-STIM1 coimmunoprecipitated with mCherry-tagged STIMATE but not mCherry (control).
b, FLAG-STIMATE coimmunoprecipitated with mCherry-STIM1 and three indicated STIM1-CT fragments, but not with the minimal ORAI-activating domain CAD or SOAR. The domain architecture of STIM1 and design of truncated variants are shown at the top.
c, FRET signals measured in HEK293 cells co-transfected with indicated donor-acceptor pairs without store depletion. YFP was C-terminally tagged to STIM11–342 and STIM11–310, and N-terminally fused with STIM1-CT (233–685) and SOAR or CAD. Error bars denote s.e.m. for n= 15, 18, 20 or 17 cells, respectively (from left to right) pooled across three independent experiments.
d, In vitro binding of recombinant STIM1-CT and STIM1-CC1 to recombinant GST-STIMATE-CT proteins immobilized on glutathione (GS4B) resin. GST was used as negative control.
e, Surface plasmon resonance (SPR) measurements of the interaction between STIM1-CC1 and STIMATE-CT in vitro. Sensorgrams monitor the binding of STIMATE-CT (with concentrations labeled on the right) to immobilized STIM1-CC1 (with an N-terminal CGG linker to aid thiol coupling to the CM3 sensor chip). Inset, The peak values for each sensorgram were collected and a dose-response binding isotherm was created to obtain an apparent dissociation constant of 8.9 μM. One set of data representing three reproducible, independent experiments is shown.
f, FRET signals between STIM11–310-CFP (donor) and YFP-SOAR (acceptor) in HEK293 cells coexpressing mCherry (control, black, n=20), mCherry-STIMATE (red, n=24) or mCherry-STIMATE-CT (blue, n=30 cells pooled across three independent experiments). Ionomycin (2.5 μM) was added to deplete ER Ca2+ stores. Left, time course. Right, bar graph of resting FRET signals. *P<0.05; ***P < 0.001 (paired Student’s t-test); Error bars denote s.e.m.
g-h, FRET signals in HEK293 cells stably expressing indicated donor-acceptor pairs and transiently transfected with mCherry (control) or mCherry-STIMATE (red). The time course was shown on the left whilst the quantification of resting FRET signals was plotted to the right. n = 12 cells for panel g; n = 15 (control) or n = 20 (STIMATE) cells pooled across three independent experiments for panel h. ***P < 0.001 (paired Student’s t-test); Error bars denote s.e.m..
After store depletion, STIM1 is activated through a conformational switch mechanism by undocking CAD/ SOAR from CC1, thus projecting CAD/ SOAR toward the PM to engage and gate ORAI1 channels31. The observation of a direct interaction between STIMATE-CT and STIM1-CC1 raised the possibility that STIMATE might promote STIM1 conformational switch through disrupting the intramolecular trapping mediated by CC1-SOAR association32. To test this, we developed a two-component FRET assay by fusing CFP or YFP to STIM11–310 and SOAR, respectively. This assay enabled us to monitor CC1-SOAR interaction in real time, and more importantly, to examine the effect of STIMATE on the STIM1 conformational switch in living cells32. Under resting conditions, YFP-SOAR was tightly docked toward the ER-resident STIM11–310 and showed a high resting FRET signal (Fig. 5f). Upon ionomycin stimulation, YFP-SOAR undocked from CC1 and thus caused a profound decrease of the FRET signal. The coexpression of STIMATE in this assay significantly reduced the resting FRET signal (Fig. 5f), implying that STIMATE could at least partially release STIM1 autoinhibition possibly by weakening CC1-SOAR association at rest. STIMATE-CT exhibited a similar effect but was much less potent, likely because this process requires anchoring of the STIMATE C-tail in the vicinity of the ER membrane or it involves coordinated actions of the C-tail with other regions of STIMATE. The consequence of STIMATE-mediated action on STIM1 was further reflected in enhanced FRET signals between CFP-STIM1 and YFP-STIM1 (Fig. 5g) or STIM1-YFP and ORAI1-CFP at rest (Fig. 5h), and culminated in constitutive Ca2+ influx (Fig. 3c). Clearly, our data provide compelling evidence to support a model in which STIMATE interacts with the juxtamembrane CC1 region of STIM1 to perturb the CC1-SOAR association that is the basis for STIM1 autoinhibition32, thereby shifting the equilibrium toward activated states to promote the conformational switch and subsequent translocation of STIM1 toward ER-PM junctions.
In summary, we have applied a non-disruptive biotin labeling approach to capture protein complexes in close proximity to STIM1 at ER-PM junctions in living cells. Our findings not only afford an initial view on the protein composition of intact ER-PM junctions under physiological conditions, but also provide a framework for future dissection of their roles in cellular signaling, human health and disease. In particular, our study has established an irreplaceable role of the previously-unrecognized ER-resident protein STIMATE in modulating STIM1-depedent Ca2+ signaling at ER-PM junctions. Major questions that warrant further investigation include i) how STIMATE choreographs with other components at ER-PM junctions to coordinate Ca2+ signaling and other cellular events; and ii) how STIMATE itself contributes to the formation and maintenance of ER-PM junctions.
METHODS
Chemicals
Tris(2-carboxyethyl)phosphine and disuccinimidyl suberate were obtained from Pierce. All other reagents were from Sigma Aldrich unless otherwise indicated. Biotin-phenol was initially obtained as a gift from Dr. Alice Ting at Massachusetts Institute of Technology and later synthesized by following the reported synthesis scheme5.
Antibodies
Streptavidin-DyLight594 (catalogue no. 21842, 1:2,000 for immunofluorescence staining) and streptavidin-HRP (N100, 1:4,000 for Western blotting [WB]) were purchased from Thermo scientific. Mouse monoclonal anti-FLAG antibody (clone number M2, catalogue no. F3165, 1:3,000 WB, 1.25 μg per 100 μg of total protein for immunoprecipitation or IP) was purchased from Sigma. The rabbit polyclonal anti-GFP antibody (sc-8334, 1:1,000 WB, 0.5 μg per 100 μg of total protein for IP) was obtained from Santa Cruz Biotechnology. The mouse monoclonal and rabbit polyclonal anti-mCherry antibodies were obtained from Clontech (clone number 1C51, catalogue no. 632543, 1:1,000 WB) and Novus Biologics (NBP2–25157, 1:2,000 WB), respectively. Secondary antibodies, including goat anti-mouse IgG-HRP (sc-2005, 1:2,000) and goat anti-rabbit IgG-HRP (sc-2004, 1:2,000), were purchased from Santa Cruz Biotechnology.
Cell lines
Cell lines including HEK293 (CRL-1573), HeLa (CCL-2) and COS-7 (CRL-1651) were purchased from American Type Culture Collection (ATCC). The cell lines were validated by short tandem repeat profiling analysis by ATCC. Cells stably expressing STIM1, ORAI1 or STIM1-ORAI1 were subsequently generated on the basis of the aforementioned cell lines after geneticin or puromycin selection. All the cell lines were free of mycoplasma contamination.
Plasmids
The cDNA clone encoding human STIMATE was purchased from DNASU Plasmid Repository. GFP-STIMATE, STIMATE-GFP and FLAG-STIMATE were generated through gateway cloning by using the vectors pCDNA-DEST54, pCDNA-DEST47 (Life Technologies), and a customized FLAG-tagged destination vector. STIMATE-YFP and STIMATE-CFP was made by inserting STIMATE between NheI-BamHI sites of pEYPF-N1 or pECFP-N1 (ClonTech). mCherry-STIMATE or CFP-STIMATE was made by inserting amplified fragments into the pCDNA3.1(+) vector. mCherry-tagged STIMATE truncation variants were made by ligation into pmCherry-N1 between NheI-BamHI sites. STIMATE mutant constructs were subsequently made by using the QuikChange Lightning Multi site-directed mutagenesis kit (Agilent). Full-length cDNA of human STIM1 was subcloned into pCMV6-XL5 (Origene)33 with the insertion of EGFP or YFP between two additional NarI sites introduced immediately after residue N38. For STIM1-YFP constructs, human STIM1 was inserted into pEYFP-N1 between XhoI and BamHI. pCDNA3.1(+)-mCherry-ORAI1 was made by sequential insertion of mCherry and human ORAI1. The ER labelling plasmid pDsRed2-ER was obatined from Clontech. mCherry-Sec61β, mCherry-STIM1 (ref. 29), STIM1-mOrange, YFP-SOAR30, mCherry-CAD29, ORAI1-YFP and HRP-STIM1 (ref. 33) were obtained from Addgene. The luciferase reporter pGL4.30[luc2P/NFAT-RE/Hygro] and the pRL-TK plasmid encoding Renilla luciferase were purchased from Promega. To generate EGFP-STIM1-APEX2 for in situ biotinylation, we replaced the mitochondria-targeting sequence with EGFP-STIM1 between the AflII-BamHI sites of pCDNA3-mito-APEX (Addgene: 42607) and further introduced the mutation A134P into the soybean APEX to enhance its enzymatic activity5, 6. All the constructs were confirmed by both DNA sequencing and diagnostic digestion.
Human STIM1-YFPN was generated from pDONR221-STIM1 (DNASU) and pBabe-CMV-YFPN-DEST-neo34 through Gateway cloning (Life Technologies). The prey library was generated by mixing pBabe-CMV-YFPC-DEST-puro with Entry clones encoding selected candidate genes (obtained from DNASU, Supplementary Table 3) through Gateway cloning. To generate LiMETER, a human codon-optimized gBlock gene fragment was synthesized (Integrated DNA Technologies, Inc.) based on the scaffold of MAPPERs20 and cloned into pcDNA3.1(+) between NheI and XbaI sites, followed by replacing FRB with LOV2.
The sequences of STIM1 cytoplasmic fragments were amplified via PCR and cloned into the pProEX HTb vector (Life Technologies) between the BamHI and XhoI sites for expression as (His)6-tagged proteins31, 35. STIMATE C-terminal domain (STIMATE-CT residues 214–294) was constructed as a fusion protein with the B1 domain of streptococcal protein G (GB1) for increased solubility or with glutathione S-transferase (GST) for pulldown assays. For the construct GB1-STIMATE-CT, we sequentially inserted GB1 at NcoI-BamHI sites and STIMATE-CT at BamHI-XhoI sites of the pProEx HTb vector. For the construct GST- STIMATE-CT, the gene was inserted into pGEX-6P-1 vector at BamHI-EcoRI sites.
sgRNA-directed knockout of STIMATE using the CRISPR-Cas9 genome editing tool
sgRNA targeting the sequence GGCATGCTGCTCATCTACGTGGGG (underscored, protospacer adjacent motif or PAM) on exon 4 of human TMEM110 was designed with the online tool (http://crispr.mit.edu/)22 and inserted into the BbsI site of the pSpCas9(BB)-2A-Puro vector (Addgene catalogue no. 48139) to generate px459-sgTMEM110. The sgRNA-containing plasmids were transfected into HEK293 cells and subjected to puromycin selection (2 μg ml−1) for 2–4 days. Survival clones were then seeded into 96-well plates and later expanded to 24-well plates for maintenance in the presence of 1 μg ml−1 puromycin. The gene disruption was first confirmed by Surveyor nuclease assay (see below, Supplementary Fig. 2c) and ultimately validated by Sanger’s sequencing (Supplementary Fig. 2d).
Surveyor nuclease assay for detection of genome modifications
Single colonies stably expressing px459-sgTMEM110 were picked up by using limiting-dilution assay. Genomic DNA from control (transfected with empty pX459 vector) or STIMATE-KO stable cell clones was extracted using the Quick-gDNA Kit (Zymol Research). The genomic region flanking the CRISPR/Cas9 targeting site for TMEM110 was PCR amplified by using primers listed in Supplementary Table 4. The PCR products were purified using the Gel DNA Recovery Kit (Zymol Research) according to the manufacturer’s instructions. Purified PCR products from control and stable cells were mixed and subjected to a reannealing process to produce heteroduplex formation by following procedures described in the SURVEYOR Mutation Detection Kit (Transgenomic). After reannealing, products were treated with SURVEYOR nuclease and enhancer S (Transgenomic). The digested products were then subjected to electrophoresis on a 2% agarose gel and visualized with SYBR gold (Life Technologies) staining.
Quantitative real-time polymerase chain reaction (qPCR) analysis on STIMATE distribution
Total RNA was isolated from cells or tissues, and first-strand cDNA was generated from total RNA using oligo-dT primers and reverse transcriptase II (Life Technologies). Real-time qPCR was performed using specific primers and the ABI Prism 7000 analyzer (Applied Biosystems) with the SYBR GreenER qPCR Super Mix Universal kit (Life Technologies). Target gene expression values were normalized to human GAPDH or mouse GAPDH. The primers used for qPCR were summarized in Supplementary Table 4.
Proteomic mapping of ER-PM junction in HEK293 cells
A step-by-step protocol describing GFP-STIM1-APEX2 mediated in situ biotinylation and further proteomic studies on ER-PM junctions can be found at Nature Protocol Exchange (doi:10.1038/protex.2015.072)
Bimolecular fluorescence complementation (BiFc) assay and flow cytometry34
1×104 HEK293 cells seeded in 96-well plate were cotransfected with 50 ng μl−1 STIM1-YFPN and prey-YFPc using Lipofectamine 3000. Each condition was replicated in at least three wells. 24 h post-transfection, cells were harvested for high-throughput flow cytometric analysis on a LSRII flow cytometer equipped with a HTS sampler (BD Biosciences) through the Cell-Based Assay Screening Service core facility at Baylor College of Medicine. Percent of YFP positive cells was calculated by the software FlowJo.
Single-cell intracellular Ca2+ measurements
Intracellular [Ca2+] were measured with Fura-2 AM as previously described35–38. In brief, HEK293 WT or STIM1-YFP/ORAI1-CFP stable cells were seeded and cultured on cover slips at least one day before imaging. To load Fura-2 AM, HEK293 cells were first kept in the imaging solution with 1 mM CaCl2 and 2 μM Fura-2 AM for 30 min. Next, cells were kept in Fura-2 AM free imaging solution with 1 mM CaCl2 for another 30 min. For cells transfected with STIMATE that has constitutive Ca2+ influx, 300 μM Ca2+ or nominally Ca2+ free solution were used to keep cells healthy. Emission fluorescence at 509 nm generated by 340-nm excitation light (F340) and 380-nm light (F380) was collected every two seconds, and intracellular Ca2+ levels are shown as F340/F380 ratio. After careful calibration, the Fura-2 intensities were further converted to Ca2+ concentrations as we routinely did in previous studies35, 37, 39. All experiments were carried out at room temperature. Traces shown are representative of at least three independent repeats with each including 15–60 single cells.
Fluorescence resonance energy transfer measurements
The same system used in Ca2+ measurements plus an Optosplit II Image Splitter (Cairn Research Limited) was used for FRET measurements. CFP (428.9±5.5Ex/ 465±32Em), YFP (502.6±11.2Ex/ 549±21Em), FRETraw (428.9±5.5Ex/ 549±21Em) filters were used to capture raw images (FCFP, FYFP and Fraw, respectively) every 10 seconds at room temperature. To generate apparent FRET efficiency, Eapp, the following calculations on raw images were performed: First, three channel corrected FRET was calculated using the following formula: , where FRETc represents the corrected total amount of energy transfer, and Fd/Dd represents measured bleed-through of CFP into the FRET filter (0.84), Fa/Da represents measured bleed-through of YFP through the FRET filter (0.13). Second, to reduce variations caused by differences in expression levels, FRETc values were further normalized against donor fluorescence (FCFP) to generate N-FRET (normalized FRET) signal. Third, Eapp, was calculated using the following equation: 40, where G (4.59) is the system-dependent factor40. A fluorescence probe, YFP-STIM1-D76G-CFP was expressed in HEK293 cells to examine the relative expression level of CFP tagged protein to YFP tagged ones. YFP and CFP of this probe is separated by ER membrane with a distance more than 10 nm, thus used as a negative control in our experiments1. To minimize fluctuations in Eapp caused by variations of relative expression levels of donor protein to acceptor protein, only cells with FCFP/FYFP ratio falling between 0.7~1.4 were used for data analysis. All fluorescence images were collected and processed with MetaFluor software (Molecular Devices), and the resulting data were further analyzed with Matlab R2012b software and plotted with Prism5 software. Representative traces of at least three independent experiments are shown as mean ± s.e.m.
NFAT-dependent luciferase assay
HEK293 cells were cultured in DMEM as described above in 24-well plates. After reaching 40–50% confluence, siRNA oligos were transfected using DharmaFECT (GE Dharmacon) for 48h before further transfection of NFAT-luciferase reporter gene pGL4.30[luc2P/NFAT-RE/Hygro] (Promega). The Renilla luciferase gene (pRL-TK) was cotransfected as a control for counting transfected cells and normalizing the luminescence signals. HEK293 cells were treated with phorbol 12-myristate 13-acetate (PMA; 1 μM) and thapsigargin (1 μM) for 8 hours. Three duplicates were used for each treatment. Cells were then harvested and lysed by following the manufacturer’s protocol. Luciferase activity was assayed using the Dual Luciferase Reporter Assay System (Promega) on a Biotek Synergy2 luminescence microplate reader. The ratio of firefly to Renilla luciferase activity was plotted for HEK293 cells. All the data were normalized against the control group. For the rescue group, after 48 h treatment with siRNA, HEK293 cells were further transfected with the siSTIMATE-resistant variant of mCherry-STIMATE, along with the NFAT-luciferase reporter genes and pRL-TK.
DSS-mediated cross-linking in HEK293 cells
HEK293 cells were plated in 6-well plates and maintained at 37 °C and 5% CO2 for 24 hour before transfection. After transient transfection with FLAG-tagged STIMATE, cells were washed three times with ice-cold PBS (pH 8.0) and treated with 0, 50, 100 and 250 μM of freshly prepared DSS. The mixture was incubated on ice for 1–5 minutes and stopped by incubating cells with a quenching solution containing 50 mM Tris-HCl at pH 8.0 for 15 min at room temperature. Cells were then lysed by lysis buffer (20mM HEPES pH 7.4, 150 NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate and 1 mM β-glycerophosphate), mixed with equal volume of 2X sample loading buffer, heated at 42°C for 10 minutes (to avoid aggregation), and subjected to electrophoresis on 4–12% precast gel (Bio-Rad). After transferring, the polyvinylidene fluoride (PVDF) membrane was immunoblotted with a monoclonal mouse anti-FLAG M2 antibody (1:3000, Sigma).
Confocal imaging and total internal reflection fluorescence (TIRF) microscopy
Cell lines used for imaging include HEK293, HeLa and COS-7 cells cultured on 35-mm glass bottom dish (MatTek) at 37 °C with 5% CO2. All cells were grown in Dulbecco's modified Eagle's medium (DMEM, Sigma) supplemented with 10 mM HEPES, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin and 10% heat-inactivated fetal bovine serum, unless otherwise noted. Confocal imaging was performed as previously described32. TIRF images were acquired on a NIKON Eclipse Ti-E microscope customized with Nikon A1R+ confocal laser sources with a 60X, NA 1.49 oil-immersion TIRF objective (Nikon). A 488-nm laser was used to excite GFP, and a 561-nm laser to excite mCherry. For two-color imaging, 100-nm fluorescent beads (TetraSpecK Microspheres, Life Technologies) were deposited onto a coverslip and imaged as markers for later alignment.
To monitor LiMETER accumulation at ER-PM junctions, 50 ng LiMETER was transfected into normal or STIMATE-KO HEK293 cells using Lipofectamine 3000 (Life Technologies). Confocal images were acquired by using the Nikon A1R+ confocal laser sources. Images representative of the dark state of LiMETER (Fig. 4b with relatively low resolution) were acquired within 1 sec using fast scanning mode to minimize photoactivation of LiMETER by the 488-nm light source. TIRF time-lapse imaging of LiMETER was performed 24–36 h posttransfection. The 488-nm laser source used to acquire the GFP signal of LiMETER was applied to activate the photoswitch LOV2. The images were acquired every 5 seconds for 5 minutes as one cycle. The cells were then kept in the dark for 5–10 minutes, followed by subsequent cycles of photostimulation.
Electron microscopy
Normal or STIMATE-KO HEK293 cells were plated onto poly-L-lysine coated chamber slides for electron microscopy. After transient transfection with HRP-STIM1, the cells were treated with or without 1–2 μM TG for 5 minutes in Ca2+- and serum-free medium. The samples were washed with PBS twice and fixed using 2% glutaraldehyde in 0.1 M sodium cacodylate at room temperature for 1 hour, then quickly moved to ice and washed with 0.1 M sodium cacodylate buffer for 10 min. HRP was visualized with 0.5 mg ml−1 diaminobenzidien and 0.03% hydrogen peroxide in 0.1 M sodium cacodylate buffer. Development of HRP took between 5 min to 20 min and was stopped by extensive washes with cold with 0.1 M sodium cacodylate buffer. Cells were postfixed in 2% OsO4 in 0.1 M cacodylate buffer (Electron Microscopy Sciences) at 4 °C for 1 hour, washed in 0.1 M sodium cacodylate buffer and then contrasted in 2% aqueous uranyl acetate (Electron Microscopy Sciences) overnight at 4 °C, dehyrated in ethanol and embedded in epon for conventional EM. Images were acquired on a Morgagni transmission electron microscope (FEI) from Electron Microscopy Services provided by the Image Analysis Lab at Texas A&M University.
Image analyses
All acquired images were analyzed using the NIKON NIS-Elements AR package or the ImageJ (NIH) software. The size of GFP-STIM1 puncta shown in Fig. 3b were quantified using Image J. Regions of interest (ROIs) within the images were determined using a manually selected pixel intensity threshold and the total pixel number per ROI was determined as a measure of puncta size. Using the same image analysis strategy, HRP-STIM1 relative distribution in ER was determined by measuring the total length of HRP-positive STIM1 segments and the length of multiple HRP-positive STIM1 segments migrated into cortical ER (separated within 20 nm from the PM). Pearson correlation coefficient between EGFP-STIM1 and mCherry-STIMATE was calculated using the built-in colocalization analysis module of the NIS-Elements AR software. To determine the intensity, size and number of MAPPER or LiMETER accumulation at cER and STIM1 puncta at ER-PM junctions, the TIRF images were background subtracted and converted to binary images to identify dim puncta and define the edges based on optimized thresholds using the NIS-Elements AR software (Nikon).
Pulldown and coimmunoprecipitation (CoIP) experiments
Expression and purification of recombinant proteins, as well as GST pulldown assay, were carried out as we described previously31, 32, 35. Bound proteins were visualized on SDS-PAGE after Coomassie Brilliant Blue R-250 staining. For CoIP experiments, transfected HEK293 cells were washed in cold PBS for 3 times and lysed directly using lysis buffer including 20 mM Tris-HCl (PH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate and 1 mM β-glyceropphosphate, supplemented with protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma), for 30 minutes at 4°C. The lysates were clarified by centrifugation at 20,000 x g at 4°C for 15 minutes. Equivalent sample amounts were subjected to SDS-PAGE, followed by transferring to PVDF membranes and probing with the appropriate primary and HRP-conjugated secondary antibodies. For immunoprecipitation, the cell lysates were incubated with the indicated antibodies and magnetic A or G beads (Thermo Scientific) overnight at 4 °C. The beads were pelleted and washed with lysis buffer, in the presence of protease inhibitors, for 10 times and were then heated in SDS loading buffer for 10 minutes at 42 °C before resolving on SDS-PAGE. The cell lysates were separated by 4–15% gels (Bio-Rad), transferred to PVDF membranes and probed with indicated antibodies.
BIAcore surface plasmon resonance (SPR) measurements
SPR-based Biacore experiments were performed at 25 °C on a Biacore 3000 instrument (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Immobilized STIM1-CC1 sensor surface was prepared by ligand thiol coupling through the single cysteine residue at the N-terminus of CGG-CC1 (ref. 31). The sensor chip CM3 and reagents for thiol coupling were purchased from GE Healthcare. The immobilization was performed at 5 μl min−1 flow rate with PBS (10 mM sodium phosphate pH 7.4, 150 mM NaCl). Flow cell was activated by 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and 0.1 M N-hyroxysuccinimide followed by washing with 80 mM of 2-(2-pyridinyldithio)ethaneamine hydrochloride (GE Healthcare) in 0.1 M sodium borate (pH 8.5) to introduce reactive disulfide groups on the surface. 30 μl of STIM1-CC1 (10 μg ml−1 in 10 mM sodium acetate pH 5.5) was injected to the activated surface and then blocked with 20 μl of Cysteine-NaCl. Approximately 550 RU (response unit) of STIM1-CC1 was immobilized. A reference flow cell was prepared with the same activation and blocking steps but without any protein coupled. Binding study was performed at 50 μl min−1 flow rate using TBS (20 mM Tris, 150 mM NaCl, pH7.5) as running buffer. After each injection of STIMATE-CT (2 fold serial dilution in running buffer), the CC1 surface was regenerated by injecting 10 mM glycine (pH 1.7) for 30 s to remove bound STIMATE-CT. Background corrected sensorgrams were collected for data analysis.
Statistical analyses
Unless otherwise noted, quantitative data are expressed as the mean and standard deviation of the mean (s.e.m.). Statistical significance was determined with paired Student’s t-test. *P<0.05; **P<0.01; ***P<0.001, when compared to control or WT.
Supplementary Material
Acknowledgments
We are grateful to Dr. Richard Lewis at Stanford University for the HRP-STIM1, mCherry-STIM1, and mCherry-CAD constructs. We thank Dr. Jen Liou at University of Texas Southwestern Medical Center for sharing with us the MAPPERs construct, Dr. Zhou Songyang at Baylor College of Medicine for the BiFc-related constructs. We thank Dr. Magnus Höök at Texas A&M University for access to the Biacore 3000, Dr. Dan Liu at Baylor College of Medicine for access to the Cell Based Assay Screening Facility and advice on BiFc, and Ross Payne at Texas A&M University for technical support in EM studies. This work was supported by the National Institutes of Health grant (R01 GM112003 to Y.Z., R01 AI084167 and R01 GM110397 to P.H.), the Special Fellow Award from the Leukemia & Lymphoma Society (LLS 3013–12 to Y.Z.), the China Scholarship Council (to J.J.), the National Natural Science foundation of China (NSFC-31471279 to Y.W. and NSFC-81222020 to L.C), the Recruitment Program for Young Professionals of China (to Y.W.), the Program for New Century Excellent Talents in University (NCET-13–0061 to Y.W.), the American Heart Association SDG grant (13SDG17200006 to S.L.Z.), the Cancer Prevention Research Institute of Texas grant (to Y.H.), and by an allocation from the Texas A&M University Health Science Center Startup Fund (to Y.Z.).
Footnotes
AUTHOR CONTRIBUTIONS
YZ and YW supervised and coordinated the study. LH, JJ, AQ and YZ designed and generated all the plasmid constructs. LH performed the BiFc assays. JJ, PT and LH generated the knockout cell lines. LH, YD, and MQD prepared the proteomic samples and performed the mass spectrometry analyses. GM, JJ, XL and YZ developed the in vitro assays, and carried out the experiments with assistance from LH, PT and YH. AS, JJ, YW and SLZ performed the Ca2+ influx assay. JJ, AS, AQ, LH, XZ, LC and YW performed all the fluorescence imaging and other cell-based experiments. XS contributed to the synthesis of biotin-phenol. YZ, JJ, YD, LH, AS, GM, YW and MQD analyzed data, with input from the other authors. PGH, YH and CLW provided intellectual inputs to the manuscript. JJ, YW and YZ wrote the manuscript.
AUTHOR INFORMATION
The authors state that they have no competing interests.
Supplementary Information is available in the online version of the paper.
References
- 1.Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends in biochemical sciences. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Hogan PG, Lewis RS, Rao A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol. 2010;28:491–533. doi: 10.1146/annurev.immunol.021908.132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Current opinion in cell biology. 2013;25:434–442. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee HW, et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annual review of biophysics. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. The Journal of biophysical and biochemical cytology. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer G, Bendayan M. Biotinyl-tyramide: a novel approach for electron microscopic immunocytochemistry. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1997;45:1449–1454. doi: 10.1177/002215549704501101. [DOI] [PubMed] [Google Scholar]
- 10.Bendayan M. Tech.Sight. Worth its weight in gold. Science. 2001;291:1363–1365. doi: 10.1126/science.291.5507.1363. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, et al. An siRNA screen for NFAT activation identifies septins as coordinators of store-operated Ca2+ entry. Nature. 2013;499:238–242. doi: 10.1038/nature12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min SW, Chang WP, Sudhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manjarres IM, Rodriguez-Garcia A, Alonso MT, Garcia-Sancho J. The sarco/endoplasmic reticulum Ca(2+) ATPase (SERCA) is the third element in capacitative calcium entry. Cell calcium. 2010;47:412–418. doi: 10.1016/j.ceca.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science. 2010;330:105–109. doi: 10.1126/science.1191086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CY, Shcheglovitov A, Dolmetsch R. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science. 2010;330:101–105. doi: 10.1126/science.1191027. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriev I, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Current biology : CB. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giordano F, et al. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Journal of proteome research. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang CL, et al. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013;5:813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 21.Maleth J, Choi S, Muallem S, Ahuja M. Translocation between PI(4,5)P2-poor and PI(4,5)P2-rich microdomains during store depletion determines STIM1 conformation and Orai1 gating. Nat Commun. 2014;5:5843. doi: 10.1038/ncomms6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. Journal of molecular biology. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz H, Hailey DW, Wunder C, Lippincott-Schwartz J. The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nature protocols. 2006;1:276–279. doi: 10.1038/nprot.2006.42. [DOI] [PubMed] [Google Scholar]
- 26.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 28.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, et al. Initial activation of STIM1, the regulator of store-operated calcium entry. Nat Struct Mol Biol. 2013;20:973–981. doi: 10.1038/nsmb.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma G, et al. Inside-out Ca2+ signaling prompted by STIM1 conformational switch. Nature Communications. 2015 doi: 10.1038/ncomms8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee OH, et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Molecular & cellular proteomics : MCP. 2011;10:M110 001628. doi: 10.1074/mcp.M110.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, et al. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, et al. STIM protein coupling in the activation of Orai channels. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7391–7396. doi: 10.1073/pnas.0900293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, Ramachandran S, Oh-Hora M, Rao A, Hogan PG. Pore architecture of the ORAI1 store-operated calcium channel. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4896–4901. doi: 10.1073/pnas.1001169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, et al. Distinct Orai-coupling domains in STIM1 and STIM2 define the Orai-activating site. Nature communications. 2014;5:3183. doi: 10.1038/ncomms4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang SL, et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zal T, Gascoigne NR. Photobleaching-corrected FRET efficiency imaging of live cells. Biophysical journal. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.