Abstract
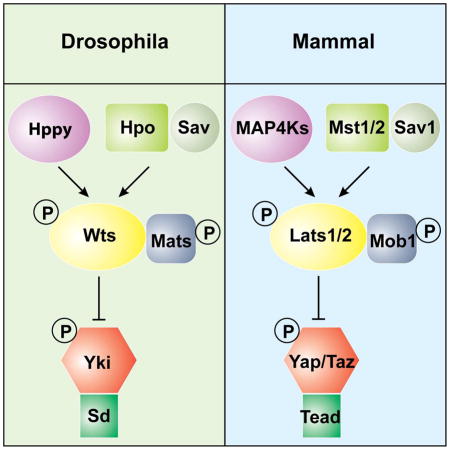
In Drosophila and mammals, the canonical Hippo kinase cascade is mediated by Hpo/Mst acting through the intermediary kinase Wts/Lats to phosphorylate the transcriptional coactivator Yki/YAP/TAZ. Despite recent reports linking Yki/YAP/TAZ activity to the actin cytoskeleton, the underlying mechanisms are poorly understood and/or controversial. Using Drosophila imaginal discs as an in vivo model, we show that Wts, but not Hpo, is genetically indispensable for cytoskeleton-mediated subcellular localization of Yki. Through a systematic screen, we identify the Ste-20 kinase Happyhour (Hppy) and its mammalian counterpart MAP4K1/2/3/5 as an alternative kinase that phosphorylates the hydrophobic motif of Wts/Lats in a similar manner as Hpo/Mst. Consistent with their redundant function as activating kinases of Wts/Lats, combined loss of Hpo/Mst and Hppy/MAP4K abolishes cytoskeleton-mediated regulation of Yki/YAP subcellular localization, as well as YAP cytoplasmic translocation induced by contact inhibition. These Hpo/Mst-like kinases provide an expanded view of the Hippo kinase cascade in development and physiology.
Keywords: Hpo, Yki, YAP, Happyhour, MAP4K1/2/3/5, F-actin, contact inhibition
Graphical Abstract
Introduction
The Hippo signaling pathway was initially delineated in the model organism Drosophila as a critical mechanism that restricts the growth of imaginal discs (Halder and Johnson, 2011; Harvey and Tapon, 2007; Pan, 2007). This pathway is conserved in mammals and has been implicated in diverse physiological processes such as organ size control, cell fate determination, tissue regeneration and stem cell renewal (Barry and Camargo, 2013; Harvey et al., 2013; Johnson and Halder, 2014; Pan, 2010; Zhao et al., 2008). Central to the Hippo signaling pathway is a core kinase cascade wherein the Ste-20 family kinase Hippo (Hpo; Mst1/2 in mammals), in conjunction with the scaffold protein Salvador (Sav, Sav1 in mammals), phosphorylates and activates the NDR family kinase Warts (Wts, Lats1/2 in mammals) and its cofactor Mob as tumor suppressor (Mats, Mob1A/B in mammals). Activated Wts/Lats then phosphorylates the transcriptional coactivator Yorkie (Yki, YAP/TAZ in mammals), leading to the phosphorylation-dependent nuclear exclusion/cytoplasmic sequestration and inactivation Yki/YAP(TAZ). In the nucleus, the Yki/YAP(TAZ) coactivator normally forms a complex with its major DNA-binding partner Sd (TEAD1/2/3/4 in mammals) to promote normal tissue growth. At least in Drosophila, a major function of the Sd-Yki complex is to antagonize default transcriptional repression mediated by Sd and its co-repressor Tgi (Koontz et al., 2013).
There has been longstanding interest in understanding how the Hippo kinase cascade is regulated in development and physiology (Boggiano and Fehon, 2012; Enderle and McNeill, 2013; Yu and Guan, 2013). An emerging paradigm from studies in multiple model systems indicates that the Hippo kinase cascade is a signaling module that integrates multiple biological inputs such as cell polarity, adhesion, mechanical forces and secreted ligands. As the kinase that directly phosphorylates Yki/YAP/TAZ, Wts/Lats represents a converging point of many (if not all) these upstream inputs. For example, multiple upstream tumor suppressors of the Hippo pathway, such as Hpo/Mst, Merlin/NF2, and Fat, all act on Wts/Lats to control the activity of Yki/YAP. Hpo/Mst directly phosphorylates the hydrophobic motif of Wts/Lats (Chan et al., 2005; Wu et al., 2003; Yin et al., 2013; Yu et al., 2010). Merlin/NF2 targets Wts/Lats to cell membrane to facilitate its phosphorylation by Hpo/Mst (Yin et al., 2013). NF2 and Fat stabilize Lats protein in mammals and Wts protein in Drosophila, respectively (Cho et al., 2006; Li et al., 2014b).
Among the multilayers of regulation on Wts/Lats, phosphorylation represents an essential step for Wts/Lats activation. As a NDR family kinase, full activation of Wts/Lats requires two phosphorylation events, one in the activation loop and the other in the hydrophobic motif (Hergovich et al., 2006). The activation loop is autophosphorylated, while the hydrophobic motif is phosphorylated by an upstream kinase. The essentiality of the hydrophobic site has been well demonstrated as mutation of this site abolishes the kinase activity (Chan et al., 2005; Hergovich et al., 2006; Yin et al., 2013). Although Hpo/Mst is the only known kinase that directly phosphorylates the hydrophobic motif of Wts/Lats, the hydrophobic motif of Wts/Lats was still phosphorylated in Mst1/2 null mouse embryonic fibroblasts (MEFs) or Mst1/2 null hepatocellular carcinoma cells (HCCs), suggesting the existence of alternative Hpo-like kinase(s) targeting this phosphorylation site (Zhou et al., 2009). In agreement with this hypothesis, in the many recent studies that have implicated the actin cytoskeleton as an regulator of Yki/YAP(TAZ) activity, a consistent finding was that F-actin reorganization, whether triggered by cell detachment, certain GPCR ligands or actin polymerization inhibitors, can regulate YAP(TAZ) activity in Mst1/2 null cells (Kim et al., 2013; Yu and Guan, 2013; Yu et al., 2012; Zhao et al., 2012). These studies, however, yielded inconsistent results on the requirement of Wts/Lats – while some studies suggested that cytoskeleton-mediated regulation of YAP (TAZ) is Lats-dependent, others argued for a Lats-independent mechanism (Dupont et al., 2011; Low et al., 2014; Zhao et al., 2012). Notably, since much of these studies were carried out in cultured mammalian cells, the specific requirement of Mst1/2 or Lats1/2 in cytoskeleton-mediated regulation of Hippo signaling in intact tissues remains to be defined.
Here we use Drosophila imaginal discs as an in vivo system to assess the requirement of Hpo and Wts in cytoskeleton-mediated regulation of Yki. We show that Wts, but not Hpo, is genetically indispensable for F-actin-mediated regulation of Yki subcellular localization. Through a systematic screen, we identify the Ste-20 kinase Happyhour (Hppy) and its mammalian counterpart MAP4K(1/2/3/5) as an alternative kinase that phosphorylates the hydrophobic motif of Wts/Lats in a similar manner as Hpo/Mst. Consistent with their redundant function as activating kinases of Wts/Lats, combined loss of Hpo/Mst and Hppy/MAP4K abolishes F-actin-mediated regulation of Yki/YAP subcellular localization, as well as nuclear exclusion of YAP induced by contact inhibition. Our identification of these Hpo/Mst-like kinases provides an expanded view of the Hippo kinase cascade and offers another entry point to understanding the regulation of Hippo signaling in development and physiology.
Results
Wts, but not Hpo, is genetically indispensable for nuclear exclusion of Yki induced by disruption of F-actin cytoskeleton in intact Drosophila tissues
In mammalian cells, F-actin disruption by the actin polymerization inhibitor latrunculin B (LatB) activates Hippo signaling and promotes YAP phosphorylation (Kim et al., 2013; Zhao et al., 2012). LatB treatment also promotes Yki phosphorylation in Drosophila S2R+ cells (Yin et al., 2013), and triggers Yki nuclear exclusion as shown by subcellular fractionation (Figure S1A). Since previous studies examining cytoskeleton-mediated regulation of YAP/TAZ were based on cultured cells, we wished to investigate this question in intact tissues. Drosophila imaginal discs are ideal for this purpose since one can use genetic mosaics, in which the localization of Yki in mutant clones of genetically defined mutations can be compared to neighboring wildtype cells in the same tissue. As in cultured mammalian cells, treatment with the actin polymerization inhibitor latrunculin B (LatB) efficiently disrupted actin polymerization in Drosophila imaginal discs (Figure S1B–C). To test whether F-actin disruption can induce Yki cytoplasmic sequestration in a manner that bypasses Hpo, we generated GFP-positive hpo clones in wing imaginal discs using the MARCM technique. We focused on the wing pouch region because this area is less folded, thus facilitating the analysis of Yki localization by confocal microscopy. Without LatB treatment, Yki was largely excluded from the nucleus in the wildtype cells but showed obvious nuclear staining in the hpo mutant clones (Figure 1A–D and S1E). Consistent with previous observation in Mst1/2 null mammalian cells, LatB treatment induced nuclear exclusion of Yki in hpo mutant cells in the wing discs (Figure 1E–F and S1F–G). We noted that cells located in the peripheral region of the posterior/ventral quadrant of the wing pouch are most sensitive to LatB treatment, as LatB consistently induced nuclear exclusion of Yki in hpo mutant cells residing in this region (Figure S1D–G). For this reason, we focused our analysis of Yki localization on this region of the wing pouch in all the subsequent studies.
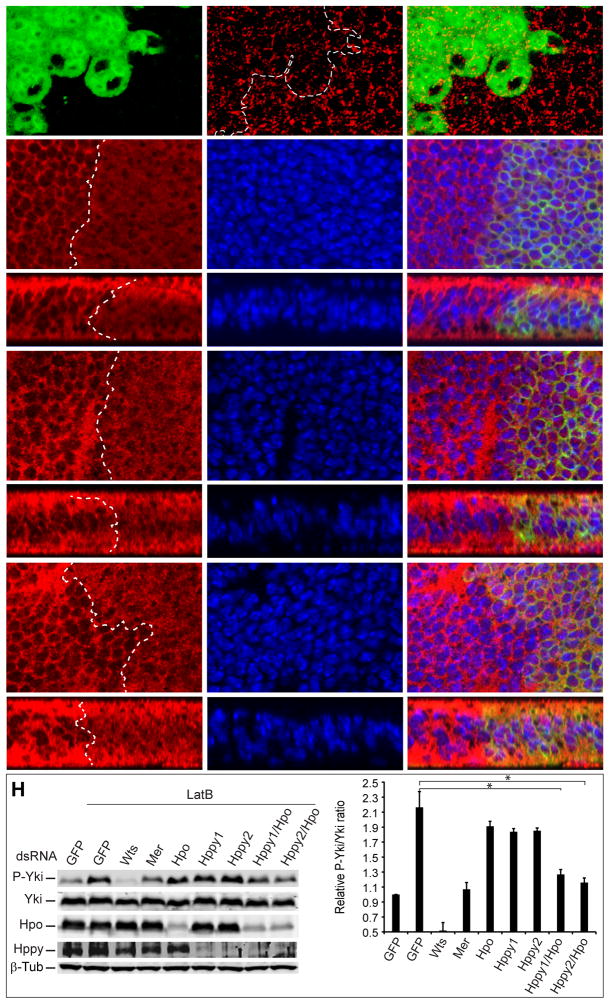
Figure 1. F-actin disruption induces nuclear exclusion of Yki in hpo but not wts mutant clones in Drosophila wing discs.
Third instar larval wing discs containing GFP-positive mutant clones of the indicated genotypes were cultured in Schneider medium supplemented with (+LatB) or without (−LatB) 4μg/mL LatB for 1h and then stained with Yki antibody (Red) and DAPI (Blue). Both horizontal (A, E, G and I) and vertical (B, F, H and J) confocal sections are shown. Enlarged views of the boxed area in A″ and B″ are shown in C–C″ and D–D″, respectively. Without LatB treatment, both hpo and wts mutant clones showed clear nuclear staining of Yki (A, B, G and H). LatB treatment induced nuclear exclusion of Yki in hpo mutant clones (E and F) but not in wts mutant clones (I and J).
See also Figure S1.
In contrast to hpo mutant clones, we found that LatB did not induce nuclear exclusion of Yki in wts clones in the wing imaginal discs (Figure 1G–J), despite that LatB efficiently disrupted F-actin in wts mutant tissues (Figure S1B–C). Taken together, these findings suggest that Wts, but not Hpo, is genetically indispensable for nuclear exclusion of Yki induced by disruption of F-actin cytoskeleton in intact Drosophila tissues.
Identification of Hppy and the KHS subfamily kinases MAP4K1/2/3/5 as activators of Hippo signaling
The genetic requirement of Wts, but not Hpo, in LatB-induced nuclear exclusion of Yki in Drosophila, together with the dispensability of Mst1/2 in LatB-induced nuclear exclusion of YAP in mammalian cells, suggest that an unknown kinase(s) can phosphorylate and activate Wts/Lats in the absence of Hpo.
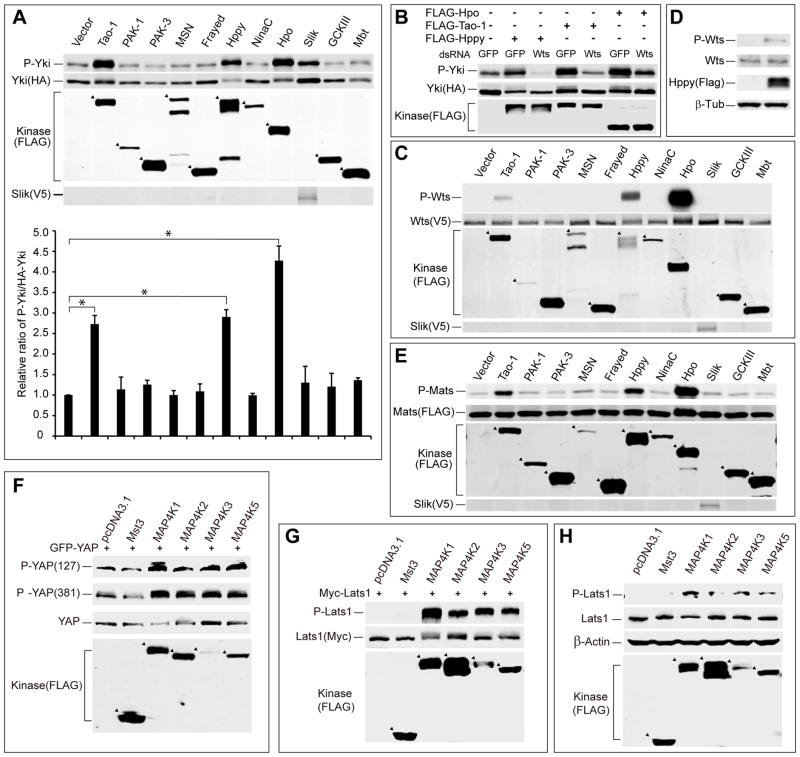
In order to identify this putative Wts/Lats kinase, we turned to Drosophila because it has a much smaller genome and thus less genetic redundancy than mammals. We reasoned that the hypothetical Hpo-like kinase is most likely a member of the Ste20 kinase family, which include 12 members in Drosophila. We therefore conducted an overexpression screen in Drosophila S2R+ cells by expressing each Ste20 family member (except for the pseudokinase Stlk), followed by the examination of Yki S168 phosphorylation or Wts hydrophobic motif (T1077) phosphorylation as specific readout for the Hippo pathway (Dong et al., 2007; Yu et al., 2010). Three kinases, including Hpo, Tao-1, and Happyhour (Hppy), were found to induce Yki S168 phosphorylation (Figure 2A). RNAi of Wts greatly attenuated the increased Yki phosphorylation induced by Hpo, Tao-1 or Hppy, consistent with Wts being the kinase for Yki S168 phosphorylation (Figure 2B). Interestingly, these three kinases were the only Ste-20 family members that induced Wts hydrophobic motif (T1077) phosphorylation in our assay (Figure 2C). Hppy also promoted phosphorylation of endogenous Wts (Figure 2D). Since Mats phosphorylation by Hpo can increase its affinity towards Wts as well as Wts kinase activity (Wei et al., 2007), we also systematically assessed all Ste20-like kinases on Mats phosphorylation. Consistent with the results of Yki and Wts phosphorylation, Hpo, Tao-1, and Hppy were the only Ste-20 kinases that induced Mats phosphorylation in our assay (Figure 2E). These results suggest that Hpo, Tao-1, and Hppy all act upstream of the Wts-Mats complex to promote Yki phosphorylation.
Figure 2. Identification of Hppy/MAP4K(1/2/3/5) as activators of Hippo signaling.
(A) Eleven Drosophila Ste-20 kinases were individually expressed in S2R+ cells with HA-Yki. Cell lysates were probed for P-Yki S168, HA-Yki, FLAG-Ste20 kinases, or V5-Slik (upper panel). The relative ration of P-Yki versus total Yki was quantified (lower panel; mean±SD, n=3). Tao-1, Hppy, and Hpo increased the relative P-Yki/Yki ratio. Slik increased both Yki and P-Yki levels, but relative P-Yki/Yki ratio was not increased. * denotes a p-value<0.05.
(B) wts RNAi abolished Yki phosphorylation induced by Hppy, Tao-1 or Hpo in S2R+ cells. Cell lysates expressing the indicated constructs were probed with the indicated antibodies.
(C) Eleven Drosophila Ste-20 kinases were individually expressed in S2R+ cells with V5-Wts. Cell lysates were probed for P-Wts T1077, V5-Wts, FLAG-Ste20 like kinases, and V5-Slik. Tao-1, Hppy, and Hpo induced Wts phosphorylation.
(D) Hppy promoted phosphorylation of endogenous Wts in S2R+ cells. Cell lysates expressing FLAG-Hppy were probed for P-Wts T1077.
(E) Eleven Drosophila Ste-20 kinases were individually expressed in S2R+ cells with FLAG-Mats. Cell lysates were probed for P-Mob T12, FLAG-Mats, FLAG-Ste20 kinases, and V5-Slik. Tao-1, Hppy, and Hpo induced Mats phosphorylation.
(F) 293T cells expressing the indicated constructs were probed for the phosphorylation of co-transfected GFP-YAP at YAPS127 and YAPS381 sites.
(G) 293T cells expressing the indicated constructs were probed for the phosphorylation of co-transfected Myc-Lats1.
(H) 293T cells expressing the indicated constructs were probed for the phosphorylation of endogenous Lats1.
See also Figure S2.
Hpo is a canonical Hippo pathway component that directly phosphorylates Wts and Mats (Wei et al., 2007; Wu et al., 2003; Yin et al., 2013; Yu et al., 2010). Tao-1 has been implicated as a kinase that functions upstream of Hpo (Boggiano et al., 2011; Poon et al., 2011). In contrast, neither Hppy nor its human counterpart has been linked to the Hippo pathway. The kinase activity of Hppy is required for stimulating Hippo signaling, as a kinase dead mutant of Hppy failed to promote Wts phosphorylation (Figure S2A). The Human counterpart of Hppy is the KHS subfamily, which includes four kinases, namely MAP4K1, MAP4K2, MAP4K3, and MAP4K5 (Manning et al., 2002). Of note, MAP4K4 belongs to a different kinase subfamily called the MSN subfamily (Manning et al., 2002). In keeping with the results in Drosophila, MAP4K1/2/3/5 markedly induced YAP S127 and S381 phosphorylation when expressed in 293T cells (Figure 2F). Of note, although MAP4K3 was expressed at much lower levels, it induced YAP phosphorylation to a similar extent as the other KHS kinases (Figure 2F). In contrast, Mst3, a Ste20-like kinase more closely related to Mst1/2 than the KHS kinases based on protein sequence alignment (Manning et al., 2002), had no effect on YAP phosphorylation (Figure 2F). Consistent with their effects on YAP phosphorylation, MAP4K1/2/3/5, but not Mst3, also strongly induced Lats1 phosphorylation in 293T cells (Figure 2G), and a kinase dead mutant of MAP4K1 abolished this activity (Figure S2B). MAP4K1/2/3/5 also induced the phosphorylation of endogenous Lats1 (Figure 2H). Taken together, these findings implicate Hppy and its mammalian counterpart as potential activators of Hippo signaling in both Drosophila and mammalian cells.
Hppy/MAP4K directly phosphorylate the hydrophobic motif of Wts/Lats independently of Hpo/Mst
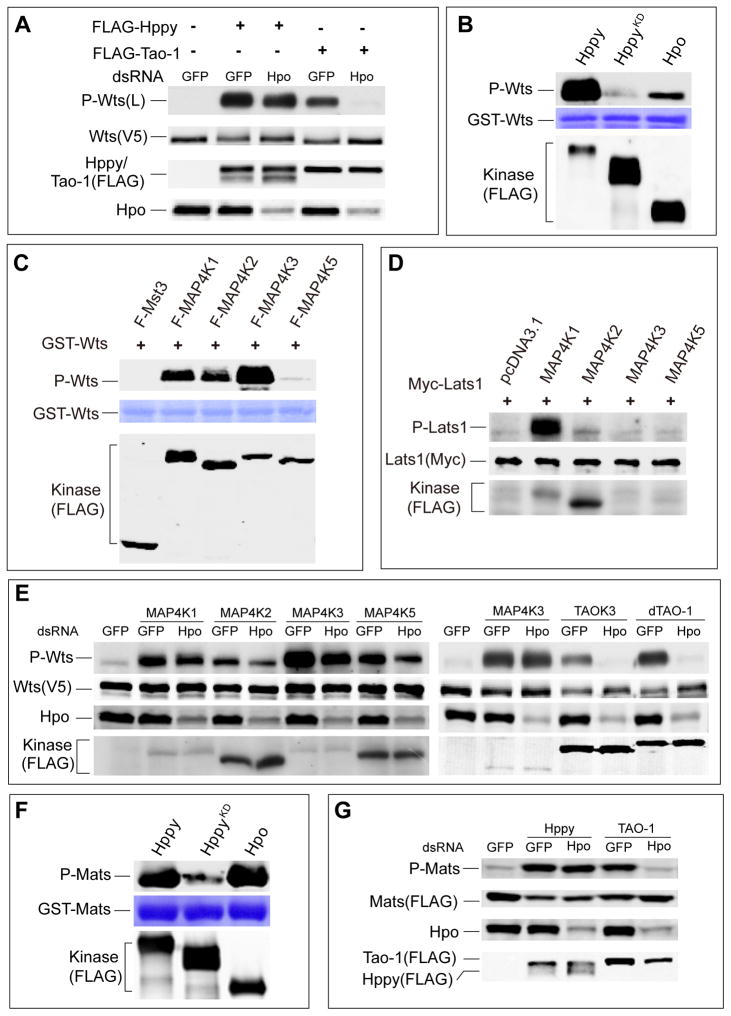
Having demonstrated that Hppy/MAP4K1/2/3/5 can stimulate hydrophobic motif phosphorylation of Wts/Lats, we sought to investigate the underlying mechanism. We first explored this in Drosophila. Since Hpo is the only known kinase that directly phosphorylates the hydrophobic motif of Wts, we assessed whether Hppy, like Tao-1 (Boggiano et al., 2011; Poon et al., 2011), acts through Hpo to activate Wts in S2R+ cells. Consistent with Tao-1 acting through Hpo (Boggiano et al., 2011; Poon et al., 2011), Tao-1-induced Wts phosphorylation was abolished by hpo RNAi (Figure 3A). In contrast, Hppy-induced Wts phosphorylation was largely insensitive to hpo RNAi (Figure 3A). These results suggest that Hppy likely functions in parallel with, not upstream of, Hpo, to activate Wts.
Figure 3. Hppy/MAP4K(1/2/3/5) directly phosphorylates the hydrophobic motif of Wts/Lats.
(A) S2R+ cells treated with the indicated dsRNAs were transfected with the indicated plasmids and probed for P-Wts, V5-Wts, FLAG-Tao-1, FLAG-Hppy, or endogenous Hpo. Note that hpo RNAi abolished Tao-1 induced but had little effect on Hppy induced Wts phosphorylation.
(B) Hppy and Hpo phosphorylate Wts in vitro. Hppy, HppyKD, and Hpo expressed in S2R+ cells were purified by immunoprecipitation and incubated with GST-Wts, and the phosphorylation products were detected with P-Wts T1077 antibody.
(C) In vitro kinase assay of MAP4K1/2/3/5. MAP4K1/2/3/5 expressed in 293T cells was purified by immunoprecipitation and incubated with GST-Wts, and the reaction products were probed for P-Wts.
(D) MAP4K1 and MAP4K2 induce Lats1 phosphorylation in Mst1/2 null HCCs. MAP4K1, MAP4K2, MAP4K3, and MAP4K5 were individually expressed with Myc-Lats1 in Mst1/2 null HCCs, and followed by the analysis of Lats1 phosphorylation.
(E) MAP4K1/2/3/5 and TAOK3 induces Wts phosphorylation in S2R+ cells. Note that hpo RNAi only slightly reduced Wts phosphorylation induced by MAP4K1/2/3/5 but abolished TAOK3 induced Wts phosphorylation.
(F) Hppy and Hpo phosphorylate GST-Mats in vitro. Hppy, HppyKD, and Hpo expressed in S2R+ cells were purified by immunoprecipitation and incubated with GST-Mats, and the phosphorylation products were detected with P-Mob T12 antibody.
(G) S2R+ cells treated with the indicated dsRNAs were transfected with the indicated plasmids and probed for P-Mob, FLAG-Mats, FLAG-Tao-1, or FLAG-Hppy. Note that hpo RNAi abolished Tao-1 induced but had little effects on Hppy induced Mats phosphorylation.
See also Figure S3.
Since previous studies have implicated Hppy/MAP4K as activators of the JNK and TOR pathways (Bryk et al., 2010; Findlay et al., 2007; Lam et al., 2010; Resnik-Docampo and de Celis, 2011), we examined whether these effector pathways are required for Hppy to promote Hippo signaling. Inhibition of JNK or TOR pathway did not affect Hppy-induced Wts or Mats phosphorylation (Figure S3A–D), suggesting that Hppy may directly phosphorylate and activate Wts. To test whether Hppy is indeed a Wts kinase, we carried out in vitro kinase assay using bacterially purified GST-Wts fusion protein as substrate and FLAG-tagged Hppy purified from S2R+ cells by immuneprecipitation as kinase. Hppy strongly phosphorylated Wts in vitro as detected by an antibody that is specific for phosphorylated T1077 of Wts (Yu et al., 2010) (Figure 3B). FLAG-tagged Hpo also phosphorylated Wts in the same assay (Figure 3B). In contrast, a kinase dead mutant of Hppy did not phosphorylate Wts (Figure 3B). Similar in vitro kinase activity was demonstrated using FLAG-tagged MAP4K1, MA4K2, MAP4K3 or MAP4K5 immunoprecipitated from 293T cells using GST-Wts as substrate (Figure 3C; note that the hydrophobic motif is identical in Drosophila and human Wts/Lats). The ability of Hpo/Mst and Hppy/MAP4K to directly phosphorylate the hydrophobic motif of Wts/Lats further supports the notion that they act in parallel with each other to activate Wts/Lats.
To further corroborate that the mammalian counterpart of Hppy can function independently of Mst1/2 to stimulate Lats phosphorylation, we expressed MAP4K1, MA4K2, MAP4K3, and MAP4K5 individually in the Mst1/2 null HCCs and examined their effects on Lats1 hydrophobic motif phosphorylation. MAP4K1 strongly induced Lats1 phosphorylation in Mst1/2 null HCCs, suggesting that MAP4K1 can activate Lats1 independently of Mst1/2 (Figure 3D). MAP4K2 also induced Lats1 phosphorylation, albeit to a lesser extent (Figure 3D). Our analyses of MAP4K3 and MAP4K5 were inconclusive since we could not express these two kinases to detectable levels (Figure 3D). To circumvent this technical difficulty, we expressed MAP4K1/2/3/5 individually in Drosophila S2R+ cells. Each of these kinases induced Wts phosphorylation, and such phosphorylation was largely insensitive to hpo RNAi (Figure 3E). In contrast, hpo RNAi completely abolished Wts phosphorylation induced by Tao-1 or its human homologue TAOK3 (Figure 3E). Taken together, these results suggest that Hppy/MAP4K functions in parallel with Hpo/Mst to directly phosphorylate the hydrophobic motif of Wts/Lats.
Hppy directly phosphorylates Mats independently of Hpo
Besides its well characterized role as Wts kinase, Hpo also directly phosphorylates Mats and this phosphorylation has been shown to further stimulate Wts activity (Wei et al., 2007). Given the strong Mats phosphorylation induced by Hppy in S2R+ cells, we examined whether Hppy also functions as a Mats kinase. An in vitro kinase assay was conducted using FLAG-tagged Hppy or Hpo immunoprecipitated from S2R+ cells as kinase and bacterially purified GST-Mats fusion protein as substrate. Both Hppy and Hpo, but not a kinase dead mutant of Hppy, strongly phosphorylated Mats in vitro as detected by an antibody that specifically recognized T12 phosphorylation of Mats (Praskova et al., 2008) (Figure 3F). Thus, Hppy and Hpo display similar kinase activity towards Mats.
If Hpo and Hppy act in parallel to directly phosphorylate Mats, we would expect Hppy-induced Mats phosphorylation to be largely independent of Hpo. Indeed, hpo RNAi had little effect on Hppy-induced Mats phosphorylation (Figure 3G). In contrast, Mats phosphorylation induced by Tao-1, a kinase that activates Hpo (Boggiano et al., 2011; Poon et al., 2011), was completely abolished by hpo RNAi (Figure 3G). Taken together, these results suggest that Hppy/MAP4K and Hpo/Mst activate Hippo signaling through a similar mechanism, by directly phosphorylating Wts and Mats.
Hppy overexpression restricts Drosophila tissue growth by activating the Hippo pathway
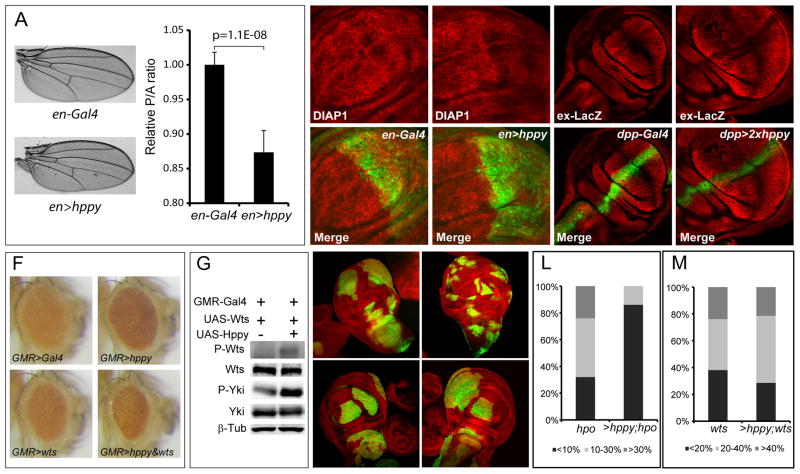
After demonstrating that Hppy can act in a similar manner as Hpo to promote Hippo signaling in vitro and in cultured cells, we examined whether Hppy can promote Hippo signaling in Drosophila imaginal discs through a similar mechanism. hppy was initially identified as homozygous mutant adult flies that display increased resistance to ethanol-induced sedation (Corl et al., 2009). It was later reported that Hppy overexpression can reduce wing size by promoting apoptosis (Lam et al., 2010; Resnik-Docampo and de Celis, 2011). Consistent with the previous studies, overexpression of Hppy resulted in smaller wing size when driven by the poster compartment-specific en-Gal4 driver (Figure 4A). Interestingly, Diap1, a well-established Hippo pathway target gene, was decreased by Hppy overexpression in the wing discs (Figure 4B–C). Expanded (Ex), another Hippo pathway target gene, was also decreased by Hppy overexpression (Figure 4D–E). Hppy overexpression by the eye-specific GMR-Gal4 driver (GMR>hppy) resulted in no obvious phenotype in the eye (Figure 4F). However, when co-expressed with Wts by the GMR-Gal4 driver, Hppy greatly synergized with Wts to decrease eye size (Figure 4F). Analysis of protein extracts from adult heads revealed increased phosphorylation of Wts and endogenous Yki in flies overexpressing Hppy plus Wts compared to flies overexpressing Wts only (Figure 4G). Thus, as in cultured cells, Hppy can activate the Hippo kinase cascade in Drosophila tissues.
Figure 4. Hppy overexpression activates Hippo signaling in vivo and restricts growth of hpo but not wts mutant clones.
(A) Hppy overexpression in the posterior compartment by en-Gal4 decreases wing size (left) and the relative ratio of posterior to anterior compartment (right; mean±SD, n=10).
(B–C) Wing discs expressing GFP only (B) or GFP plus Hppy (C) by the en-Gal4 driver were stained for Diap1. Note the down-regulation of Diap1 expression in the poster compartment in the wing disc with Hppy overexpression (C).
(D–E) Wing discs expressing GFP only (D) or GFP plus hppy (E) by the dpp-Gal4 driver were stained for the expression of ex-lacZ. Note the down-regulation of ex-lacZ expression upon hppy overexpression (arrows). The genotypes are: dpp-Gal4; UAS-GFP
(D) and dpp-Gal4; UAS-GFP; UAS-hppy/UAS-hppy (E).
(F) Hppy synergizes with Wts to reduce Drosophila eye size. Fly eyes from control (GMR-Gal4), Hppy-overexpression (GMR>hppy), Wts-overexpression (GMR>wts) or Hppy and Wts co-overexpression (GMR>hppy&wts) were imaged under the same magnification. Note that the fly eye overexpressing both Hppy and Wts is smaller than eyes overexpressing either Hppy or Wts.
(G) Hppy promotes Wts and Yki phosphorylation in Drosophila eye tissues. Extracts made from fly heads overexpressing the indicated transgenes by the GMR-Gal4 driver were analyzed by western blotting.
(H–K) Third instar larval wing discs containing GFP-positive clones of the indicated genotypes. hpo clones (H), hpo clones with Hppy overexpression (I), wts clones (J), wts clones with Hppy overexpression (K). Note that Hppy overexpression decreased the size of hpo clones.
(L–M) Clone size distribution of the indicated genotypes. 20 discs were analyzed for each genotype. Clone size was quantified as the percentage of clone area relative to total wing area. Note that Hppy overexpression significantly decreased the size of hpo but not wts mutant clones.
If Hppy directly phosphorylates Wts in Drosophila tissues independently of Hpo, we would expect the growth suppression induced by Hppy overexpression to be dependent on Wts but not Hpo. To test this prediction, we used the MARCM technique to generate hpo or wts mutant clones with or without concomitant Hppy overexpression and quantified clone size. The majority of hpo mutant clones occupied over 10% of the wing area (Figure 4H and 4L). Hppy overexpression restricted the growth of hpo mutant clones as the majority of clones had size less than 10% of the wing disc (Figure 4I and 4L). In contrast, Hppy had no effect on the size of wts mutant clones, since the size distribution of wts mutant clones was similar with or without Hppy overexpression (Figure 4J, 4K and 4M). These results are consistent with our in vitro characterization implicating Hppy and Hpo as direct, independent activators of Wts.
Hppy and Hpo play redundant roles in Hippo signaling induced by F-actin disruption
The similar biochemical activity of Hppy and Hpo suggests that these Ste-20 kinases may function together to regulate Hippo signaling, at least in some cellular contexts. Given our observation that the F-actin inhibitor LatB induced nuclear exclusion of Yki in the wing discs in a Wts-dependent, but Hpo-independent manner, we considered the possibility that Hppy may play a redundant role with Hpo in this process. To test this hypothesis, we generated hppy mutant flies using CRISPR/Cas9 system mediated genome editing (Gratz et al., 2014; Port et al., 2014). Two gRNAs were designed and used to delete genomic DNA encompassing exons 4–7 of the hppy gene. Multiple independent lines were obtained and the deletions were verified by DNA sequencing and western blotting (Figure S4A). Two sequence-confirmed mutant lines, hppyΔ1 and hppyΔ2 were used for subsequently analysis. Since exon 4 encodes essential part of the kinase domain, we consider these mutants as null alleles. As reported before (Resnik-Docampo and de Celis, 2011), hppy mutant flies are viable and fertile without noticeable morphological malformations. Despite that, hppy mutant clones in pupal retina consistently showed a modest increase of the Hippo target gene Ex (Figure 5A), consistent with enhanced Yki activity upon loss of hppy.
Figure 5. Hppy functions redundantly with Hpo to regulate Yki in response to F-actin disruption.
(A) Pupal eye discs containing GFP-negative hppy clones were stained with Ex antibody (Red). Clone boundary is marked by dotted line in (A′). Note the increased Ex expression in hppy mutant clone compared to the neighboring wildtype cells.
(B–G) Third instar larval wing discs containing GFP-positive mutant clones of the indicated genotypes were cultured in Schneider medium supplemented with 4μg/mL LatB for 1h and then stained with Yki antibody (Red) and DAPI (Blue). Both horizontal (B, D, and F) and vertical (C, E, and G) confocal sections are shown. hppy;hpo mutant clone showed nuclear accumulation of Yki (B–C), and LatB treatment did not trigger Yki nuclear exclusion in hppy;hpo mutant clones (D–G). D–E and F–G represent two different hppy alleles recombined with hpo.
(H) LatB-induced Yki phosphorylation in S2R+ cells was reduced by depletion of both hppy and hpo. S2R+ cells depleted with the indicated genes were treated with 1μg/mL LatB for 1 hr and then analyzed by western blotting (left). Relative P-Yki/Yki ratio was quantified in the graph (right; mean±SD, n=3)). Two non-overlapping dsRNAs against hppy were used (Hppy1 and Hppy2), alone or in combination with Hpo RNAi. * denotes a p-value<0.05.
See also Figure S4.
To examine whether Hppy and Hpo play redundant roles in LatB-induced Hippo signaling, we combined hppyΔ1 or hppyΔ2 with a null allele of hpo to generate two independent hppy;hpo double mutants. Loss of hppy does not affect the subcellular localization of Yki, regardless of LatB treatment (Figure S4B–C). Similar to hpo mutant clones, hppy;hpo mutant clones showed apparent nuclear accumulation of Yki (Figure 5B–C). In contrast to hpo mutant clones, however, hppy;hpo double mutant clones were largely irresponsive to LatB-induced nuclear exclusion of Yki, as the majority of hppyΔ1;hpo and hppyΔ2;hpo mutant clones (17 out of 20 clones from 20 discs) still showed robust nuclear Yki accumulation after LatB treatment (Figure 5D–G). We noticed that in occasional hppy;hpo clones (3 out of 20 clones from 20 discs), nuclear exclusion of Yki was still observed in the periphery region as defined in Figure S1D (Figure S4D–G). These results suggest that Hppy and Hpo play redundant roles in Hippo signaling induced by the disruption of F-actin.
To further corroborate our findings from imaginal discs, we examined the requirement of Hppy and Hpo in LatB-induced Hippo signaling in cultured cells. We have previously shown that LatB induced Yki phosphorylation in Drosophila S2R+ cells (Yin et al., 2013) (Figure 5H). In keeping with the essential role of Wts in LatB-induced nuclear exclusion of Yki in vivo, LatB-induced Yki phosphorylation was reduced to lower than the basal level by Wts RNAi, suggesting that Wts is the major Yki kinase in S2R+ cells (Figure 5H). As reported before (Yin et al., 2013), RNAi of the upstream Hippo pathway regulator Merlin also attenuated Yki phosphorylation elicited by LatB treatment (Figure 5H). In contrast, Hpo or Hppy RNAi had little effect on LatB-induced Yki phosphorylation (Figure 5H). However, co-depletion of Hppy and Hpo resulted in a marked decrease of LatB-induced Yki phosphorylation (Figure 5H). These data further support our model Hppy and Hpo play redundant roles in Hippo signaling induced by F-actin disruption.
MAP4K1/2/3/5 is required for Hpo/Mst-independent phosphorylation and nuclear exclusion of YAP induced by F-actin disruption
Having demonstrated the requirement of Hppy and Hpo in F-actin-mediated regulation of Hippo signaling, we wished to examine whether a similar mechanism operates in mammalian cells. It was previously reported that F-actin disruption by LatB treatment still induced YAP phosphorylation in Mst1/2 null cells (Kim et al., 2013; Zhao et al., 2012). We confirmed this finding using two different Mst1/2 null cell lines, Mst1/2 null MEF cells and HCCs (Figure S5A). As in Drosophila, we hypothesized that the mammalian counterpart of Hppy may contribute to Mst1/2-independent activation of Hippo signaling in the Mst1/2 null cells.
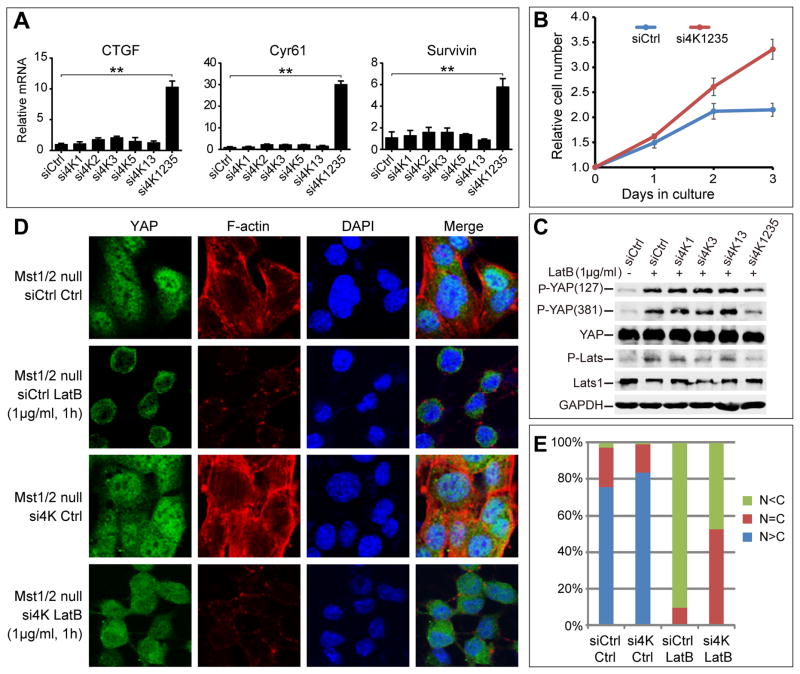
To test this hypothesis, we used RNAi to knockdown the KHS kinases MAP4K1/2/3/5 in Mst1/2 null HCCs. Knocking down each KHS kinase alone had no effect on the expression of three widely used YAP target genes ctgf, cyr61, and survivin (Figure 6A), nor did co-depletion of MAP4K1 and MAP4K3 (data not shown). We confirmed that all four KHS kinases were expressed in HCCs and efficiently knocked down by RNAi (Figure S5B). Strikingly, simultaneous knockdown of all four kinases resulted in a dramatic increase in the expression of ctgf, cyr61, and surviving (Figure 6A), as well as increased proliferation of the Mst1/2 null cells (Figure 6B), suggesting that these KHS kinases are required as a whole to suppress YAP activity in the Mst1/2 null cells in steady state.
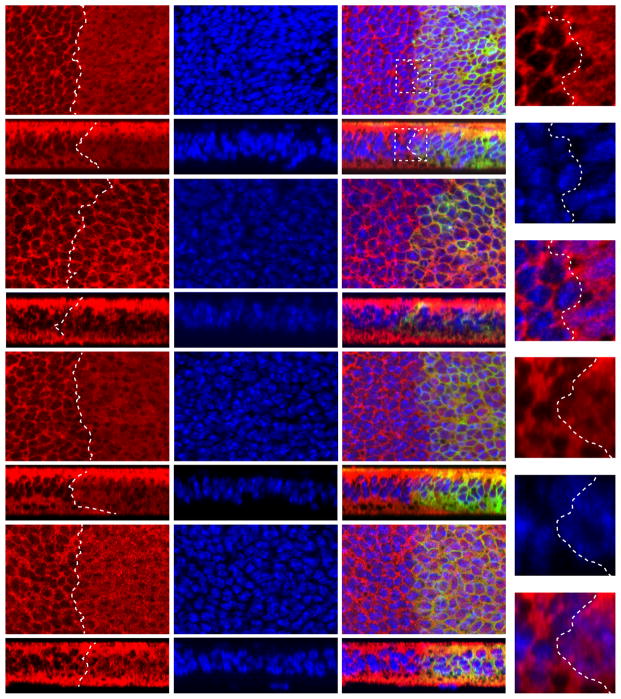
Figure 6. MAP4K1/2/3/5 is required for Hpo/Mst-independent phosphorylation and nuclear exclusion of YAP induced by F-actin disruption.
(A) Concomitant depletion of all four MAP4Ks leads to elevation in the expression levels of three Hpo pathway target genes ctgf, cyr61, and survivin in the Mst1/2 null HCCs. Total RNAs were prepared from HCCs transfected with the indicated siRNAs and the relative mRNA levels of indicated genes were measured by qPCR (mean±SD, n=3). Note the significant increase of gene expression when all four kinases were depleted. ** denotes a p-value<0.01.
(B) Concomitant depletion of all four MAP4Ks promotes the proliferation of Mst1/2 null HCCs.
(C) Depletion of MAP4K1/2/3/5 by RNAi reduces LatB-induced YAP and Lats phosphorylation in the Mst1/2 null HCCs. HCCs cultured on fibronectin-coated surface at low cell density were treated with 1μg/mL LatB for 1 hr and then analyzed by western blotting. Note that LatB induced YAP and Lats phosphorylation (compare lanes 1 and 2). Both inductions were diminished when four kinases were depleted (compare lanes 2 and 6).
(D–E) Depletion of MAP4K1/2/3/5 by RNAi attenuates LatB-induced nuclear exclusion of YAP in the Mst1/2 null HCCs. HCCs transfected with control siRNAs or siRNAs against MAP4K1/2/3/5 were cultured on chamber slides coated with fibronectin at low cell density. Cell were treated with 1μg/mL LatB for 1 hr and then fixed and stained with YAP antibody, phalloidin for F-actin, and DAPI for cell nuclei (C). 100 cells were quantified for YAP localization and the results were shown in (D).
See also Figure S5.
The Mst1/2 null cells with MAP4K1/2/3/5 knockdown allowed us to test whether the KHS kinases contribute to LatB-induced Hippo signaling. Indeed, co-depletion of all four KHS kinases, but not the depletion of individual kinases, resulted in a marked decrease of YAP (at both S127 and S381) and Lats phosphorylation induced by LatB treatment in Mst1/2 null HCCs (Figure 6C). Consistent with these findings, co-depletion of all four KHS kinases also suppressed LatB-induced nuclear exclusion of YAP (Figure 6D–E). Thus, in both Drosophila and mammalian cells, Hppy/MAP4K contributes to Hpo/Mst-independent activation of Hippo signaling induced by F-actin disruption.
MAP4K1/2/3/5 is required for Hpo/Mst-independent cytoplasmic sequestration of YAP induced by high cell density
After establishing a functional role for Hpo/Mst and Hppy/MAP4K in LatB-induced Hippo signaling, we wished to examine the requirement of these kinases in a more physiological setting. In Drosophila, Hippo signaling restricts the growth of imaginal discs. Unlike its requirement in LatB-induced Yki nuclear exclusion, loss of hppy did not increase the size of hpo mutant clones (Figure S6). The interpretation of this finding is complicated by the fact that Hppy is a pleiotropic kinase involved in multiple pathways, some of which are growth-promoting, such as TOR pathway (Bryk et al., 2010; Resnik-Docampo and de Celis, 2011) and some of which are growth-suppressing, such as the Hippo pathway. Thus, the size of double mutant clones does not only reflect defects in Hippo signaling. We therefore turned to mammalian cells for a more physiological context. A hallmark of Hippo signaling in cultured mammalian cells is density-dependent subcellular localization of YAP, whereby YAP is nuclear in sparsely cultured cells and translocates to the cytoplasm in confluence cultures (Aragona et al., 2013; Varelas et al., 2010; Zhao et al., 2007). It was suggested that this density-dependent localization of YAP is due to low mechanical stress and low F-actin levels in confluent cultures, as increasing cellular F-actin levels could shuttle YAP to the nucleus at high cell density (Aragona et al., 2013). Since high cell density still induced cytoplasmic sequestration/nuclear exclusion of YAP in Mst1/2 null cells, it was suggested that Hippo signaling induced by high cell density may also involve an Mst1/2-independent mechanism (Zhou et al., 2009).
To examine whether the KHS kinases contribute to Hippo signaling induced by high cell density, we took advantage of the Mst1/2 null cells with or without MAP4K1/2/3/5 knockdown described above by culturing them at low or high density. Consistent with previous suggestion of an Mst1/2-indepent mechanism, Mst1/2 null HCCs showed nuclear YAP localization at low cell density and cytoplasmic translocation at high cell density (Figure 7). This nuclear-to-cytoplasmic translocation was greatly diminished in Mst1/2 null cells with MAP4K1/2/3/5 knockdown (Figure 7). Taken together, these results suggest that Hppy/MAP4K contributes to Hpo/Mst-independent activation of Hippo signaling induced by both F-actin disruption and high cell density.
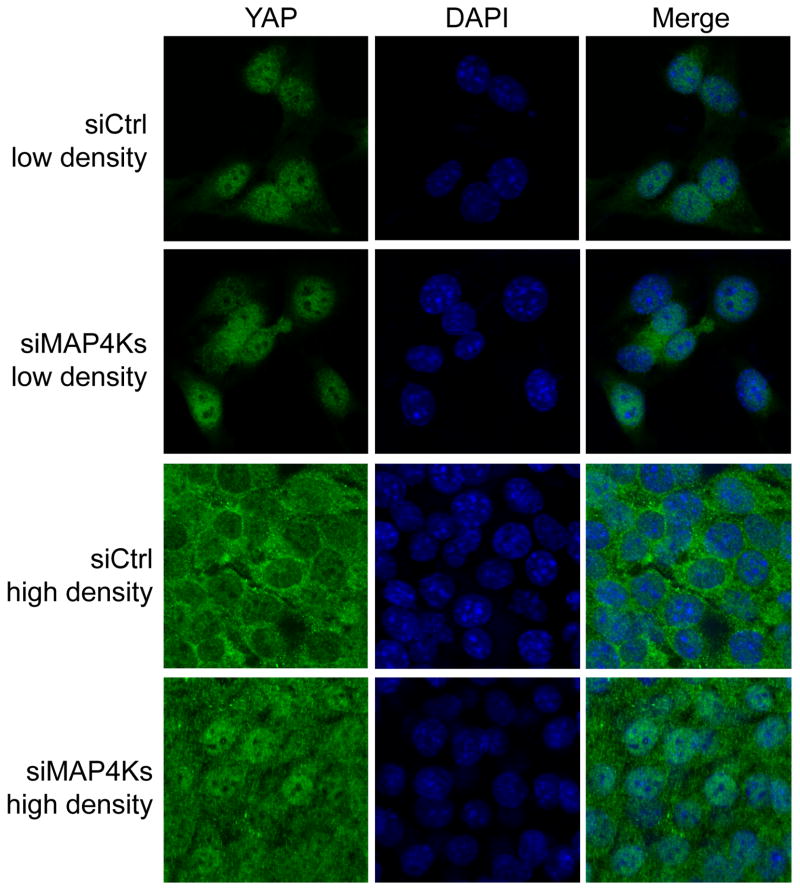
Figure 7. MAP4K1/2/3/5 is required for Hpo/Mst-independent nuclear exclusion of YAP induced by high cell density.
Mst1/2 null HCCs cultured at low (upper two rows) or high (lower two rows) cell density were transfected with control or MAP4K1/2/3/5 siRNAs for 3 days, fixed, and stained with YAP antibody and DAPI. Both types of cells showed nuclear YAP signal at low density. At high density, cells with control RNAi showed cytoplasmic localization of YAP while cells with MAP4K1/2/3/5 depletion showed more nuclear localization of YAP.
See also Figure S6.
Discussion
Our understanding of the core kinase cascade of the Hippo pathway has been aided by multiple lines of investigation. First, genetic screens for tumor suppressors using mosaic flies have identified Hpo, Sav, Wts and Mats as main constituents of the core kinase cassette (Harvey et al., 2003; Justice et al., 1995; Kango-Singh et al., 2002; Lai et al., 2005; Pantalacci et al., 2003; Tapon et al., 2002; Udan et al., 2003; Wu et al., 2003; Xu et al., 1995). Second, biochemical studies of the activation mechanism of NDR family kinases, which include Lats1/2 and NDR1/2, demonstrate the importance of regulatory phosphorylation sites on the activation loop and the hydrophobic motif (Hergovich et al., 2006). The realization that the hydrophobic motif of Wts is phosphorylated by Hpo provides a fitting molecular explanation for the linear genetic pathway uncovered by in vivo studies (Chan et al., 2005; Hergovich et al., 2006; Wu et al., 2003; Yu et al., 2010). The simplicity of this linear pathway begun to be challenged based on the observation that Mst1/2 null cells still showed high level of Lats phosphorylation on the hydrophobic motif (Zhou et al., 2009). Indeed, in many subsequent reports, various signals have been reported to still regulate YAP/TAZ activity in Mst1/2 null cells (Kim et al., 2013; Yu et al., 2012; Zhao et al., 2012; Zhou et al., 2009). While semantically these observations were implied to support the existence of “Mst1/2-independent” mechanisms, the molecular underpinning of this phenomenon has been elusive. It was also unclear whether this represents a mammalian-specific phenomenon as there has been no evidence to date that a similar mechanism operates in Drosophila.
Our current study addresses these issues in several significant ways. We provide definitive evidence supporting an alternative Hpo-independent mechanism of Hippo pathway activation in Drosophila by demonstrating the genetic requirement of Wts, but not Hpo, in LatB-induced nuclear exclusion/cytoplasmic sequestration. Through a systematic screen, we identify the Ste-20 family kinase Hppy/MAP4K as a plausible molecular explanation for Mst1/2-independent regulation of Hippo signaling. Not only does Hppy/MAP4K directly phosphorylate the hydrophobic motif of Wts/Lats in vitro and in cell cultures, loss of Hppy/MAP4K also abolishes LatB-induced Yki/YAP cytoplasmic translocation in Hpo/Mst null cells in both Drosophila tissues and mammalian cell cultures. Our findings support the view that Hpo/Mst and Hppy/MAP4K act as redundant kinases targeting the hydrophobic motif of Wts/Lats. We note that our analysis of Hpo/Mst and Hppy/MAP4K in F-actin-mediated Hippo signaling was largely based on LatB treatment. Thus, it remains to be determined how these kinases cooperate with each other in a more physiological setting of cytoskeleton modulation. Nevertheless, the fact that MAP4K mediates Mst-independent regulation of YAP target gene expression and contact inhibition of YAP nuclear localization suggests that these kinases co-regulate Wts/Lats in multiple contexts beyond LatB-induced F-actin disruption. Since the hydrophobic motif of NDR1/2 can be phosphorylated by both Mst1 and Mst3 (Hergovich et al., 2009; Stegert et al., 2005), we suggest that phosphorylation of hydrophobic motif by multiple Ste-20 kinases may be a common feature of the NDR family kinases. We note that two other kinases, CK2 and MSN/MAP4K4, were recently reported to promote Wts/Lats activity towards Yki. However, neither kinase was shown to directly phosphorylate the hydrophobic motif of Wts/Lats (Hu et al., 2014; Li et al., 2014a). Furthermore, although MSN was shown to promote Yki phosphorylation when Wts was co-expressed in S2 cells, MSN alone did not affect Yki phosphorylation (Li et al., 2014a), as we observed (Figure 2A). Thus, the mechanisms by which these kinases promote Hippo signaling remain to be determined.
Recent studies have implicated cellular mechanical forces as a regulator of Yki/YAP(TAZ) activity (Aragona et al., 2013; Dupont et al., 2011; Rauskolb et al., 2014; Wada et al., 2011). Reorganization of F-actin cytoskeleton has been suggested as the common mediator of mechanical forces arising from cell-cell and cell-matrix interactions (Gaspar and Tapon, 2014; Halder et al., 2012). However, the underlying mechanism by which F-actin controls Yki/YAP(TAZ) activity remains poorly understood and/or controversial. While some studies suggested that cytoskeleton-mediated regulation of YAP(TAZ) is independent of the Hippo kinase cascade (Aragona et al., 2013; Dupont et al., 2011), others suggested that it requires the Hippo kinase cascade (Wada et al., 2011; Yu et al., 2012; Zhao et al., 2012). Our observation that LatB-induced Yki cytoplasmic localization is Wts-dependent is more consistent with a Hippo signaling-dependent mechanism. An important modification brought by our current study is that the canonical Hippo kinase cascade should be expanded to include Hppy/MAP4K at the level of Hpo. This expanded Hippo kinase cascade may also include NDR1/2 at the level of Lats1/2, given the recent report of NDR1/2 as Lats1/2-like kinases capable of phosphorylating YAP (Zhang et al., 2015).
Finally, we suggest that the Hippo-signaling dependent and independent regulation of YAP/TAZ by F-actin may be potentially reconciled with each other. A major discrepancy between the two models came from the analysis of mutant YAP/TAZ that lacks all the Lats phosphorylation sites (YAP5SA or TAZ4SA) (Zhao et al., 2007). We note, however, different readout was used to assay the regulation of these YAP/TAZ mutants in the different studies (Dupont et al., 2011; Zhao et al., 2012). A luciferase reporter assay was used to show that the transcriptional activity of YAP5SA or TAZ4SA still responded to F-actin reorganization (Dupont et al., 2011), whereas subcellular localization was used to show that YAP5SA no longer responded to cytoplasmic localization of YAP induced by F-actin disruption (Zhao et al., 2012). These results may reflect the functionality of different subcellular pools of F-actin -- inasmuch as YAP/TAZ localization is regulated by F-actin through the Hippo pathway, F-actin may also play a separate role in regulating the transcriptional activity of YAP/TAZ in the nucleus, especially given the increasing appreciation of a more direct role of nuclear F-actin in transcriptional regulation (Miralles and Visa, 2006; Miyamoto and Gurdon, 2013; Olson and Nordheim, 2010).
Experimental Procedures
Plasmids and antibodies
HA-Yki, V5-Wts, FLAG-Tao-1, FLAG-Pak1, FLAG-Pak3, GFP-YAP, and Myc-Lats1expression constructs have been described previously (Boggiano et al., 2011; Dong et al., 2007; Duan et al., 2012; Huang et al., 2005). FLAG-Hpo was generated from the Myc-Hpo construct described before (Wu et al., 2003). FLAG-Hppy (RH10407), FLAG-Msn (LD34191), FLAG-Frayed (RE53265), FLAG-NinaC (GH10824), FLAG-GckIII (RE38276), FLAG-Mbt (LD47563), and FLAG-Mats (LD47553) expression plasmids were generated based on cDNA clones obtained from DGRC. V5-Slik was made from the cDNA clone LP09626 with the original mutations in the cDNA clone corrected by site-directed mutagenesis. FLAG-Mst3, FLAG-MAP4K1, FLAG-MA4K2, FLAG-MAP4K3, and FLAG-MAP4K5 were obtained from Addgene. FLAG-HppyKD and FLAG-MAP4K1KD were generated by changing K55 to E and K46 to E using site-directed mutagenesis, respectively.
Primary antibody used in this study include: FLAG and HA (Sigma-Aldrich); Myc (Calbiochem); V5 (Invitrogen); YAP (Novus; for immunostaining); Yap (Sigma-Aldrich; for western blotting); Lats1, P-YAP S127, and P-YAP S381 (Cell signaling); Hpo and Wts (Wu et al., 2003); Yki and P-Yki (Dong et al., 2007); P-Wts T1077 (Yu et al., 2010); P-Mob T12 (Praskova et al., 2008); β-Tubulin and Lamin A/C (Developmental Biology Hybridoma Bank). Rabbit anti-Mats antibody was generated by immunizing a host animal with the peptide KNIPEGTHQYDLMKH. Rabbit anti-Hppy antibody was raised against the peptide KIGSGTYGDVYKAKRIQS.
Drosophila and mammalian cell culture
Drosophila S2R+ cells were cultured in Schneider’s medium supplemented with 10% FBS and antibiotics at 25°C. 293T, MEF, and HCC cells were maintained at 37°C in DMEM medium supplemented with 10% FBS and antibiotics (Invitrogen). Transfection, immunopreciptation, and western blotting were carried out as described previously (Yu et al., 2010). RNAi in S2R+ cells was carried out using the bathing protocol obtained from DRSC (http://www.flyrnai.org/DRSC-PRR.html). For chemical treatment, HCCs were incubated with Latrunculin B (1 μg/ml, obtained from Sigma-Aldrich) according to Zhao et al (Zhao et al., 2012). Ethanol (0.1%) was used as vehicle control.
Drosophila genetics
Rok RNAi (34324) and Rho RNAi (29002) fly lines were obtained from the Bloomington Drosophila Stock Center. To generate a mutation in hppy, two gRNAs were used to delete the exons 4–7 of hppy by CRISPR/Cas9 according to a described protocol (Port et al., 2014). To facilitate the identification of targeted deletions, a strategy described by Gratz et al was adopted and the two gRNAs was designed to flank a P-element insertion P[GawB]hppyNP3139 whose removal would generate a white eye phenotype (Gratz et al., 2014). The two CRISPR target sites (GGCTCCTGAGGTGGCAGCCG and GCGACCCACCATCATATCGG) were selected by CRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). Mutants were first selected based on eye color and then further verified by DNA sequencing. Representative genotypes used in this study:
-
GFP+ hpo− clones:
UAS-GFP hs-Flp; tub-Gal80 FRT42D/FRT42 hpo42–47; tub-Gal4.
-
GFP+ hpo− clones overexpressing Hppy:
UAS-GFP hs-Flp/UAS-hppy; tub-Gal80 FRT42D/FRT42 hpo42–47; tub-Gal4.
-
GFP+ wts− clones:
UAS-GFP hs-Flp; tub-Gal4; tub-Gal80 FRT82B/FRT82B wtsx1.
-
GFP+ wts− clones overexpressing Hppy:
UAS-GFP hs-Flp/UAS-hppy; tub-Gal4; tub-Gal80 FRT42D/FRT42D wtsx1.
-
GFP+ hppy− hpo− clones:
UAS-GFP hs-Flp; tub-Gal80 FRT42D/FRT42 hppyΔ1 hpo42–47; tub-Gal4.
UAS-GFP hs-Flp; tub-Gal80 FRT42D/FRT42 hppyΔ2 hpo42–47; tub-Gal4.
LatB treatment of Drosophila imaginal discs
Drosophila wing imaginal discs were dissected in Schneider medium and then cultured in Schneider medium supplemented with 4ug/mL LatB for 1h on a rocking platform. Ethanol (0.4%) was used as vehicle control. The discs were then fixed in 4% PFA and incubated with anti-Yki (1:500) for O/N at 4°C.
Nuclear and cytoplasmic fractionation
Fractionation of proteins into nuclear and cytoplasmic fractions was carried out using the REAP protocol as described by Suzuki et al (Suzuki et al., 2010).
In vitro kinase assay
GST-Wts was described previously (Yin et al., 2013). To generated GST-Mats, the ORF of mats was PCR-amplified from FLAG-Mats construct and inserted into pGEX-6p-1 and expressed in BL21-CodonPlus (DE3)-RIPL cells and the products were purified. In vitro kinase assay was carried out as previously described and phosphorylated products were detected using P-Wts T1077 or P-Mob T12 antibody (Yin et al., 2013).
siRNA transfection and cell proliferation assay
Cells were cultured on Lab-Tek II Chamber Slide (Thermo Scientific) to 80–85% confluent, and then siRNA transfection was performed using Lipofectamine RNAiMAX. Cells were processed 3 days post-transfection. SMARTpool siRNA oligonucleotides toward mouse MAP4K1, MAP4K2, MAP4K3, MAP4K5 and Non-Targeting siRNA Pool #2 (control siRNA) were purchased from GE Dharmacon. Relative cell proliferation rate was calculated based on cell numbers counted in a Hemacytometer.
Reverse Transcription and Real-time PCR
RNA samples (1 μg) were reverse-transcribed to complementary DNA (cDNA) using iScript reverse transcriptase (Bio-Rad). cDNA was then diluted and used for quantification by real-time PCR, which was performed using iQ™ SYBR® Green supermix (Bio-Rad) and the CFX96™ real-time system (Bio-Rad). All values were normalized to that of GAPDH, and the normalized values of siCtrl were set as 1. Error bars represent the standard deviations of three independent experiments.
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.3% Triton X-100. After blocking in 5% goat serum for 1 hour, slides were incubated with anti-YAP antibody (Novus, 1:300 dilution) diluted in 5% goat serum for overnight at 4°C. After washing with PBS, slides were incubated with Alexa Fluor 488-conjugated secondary antibodies (1:500 dilution) for 1 hour, washed, and then Alexa Fluor 568-conjugated phalloidin for 40 min. The slides were incubated with DAPI (1:1000 dilution) for 5 min, then washed and mounted.
Supplementary Material
Highlights.
Hppy/MAP4K directly phosphorylates the hydrophobic motif of Wts/Lats
hppy functions genetically in parallel with hpo and upstream of wts
Hppy/MAP4K functions redundantly with Hpo/Mst in F-actin-mediated Hippo signaling
MAP4K mediates Mst-independent contact inhibition of YAP nuclear localization
Acknowledgments
We thank Dr. Richard Fehon for pUAST-FLAG-Tao-1 construct and Anti-Ex antibody, Dr. Elizabeth Chen for pUAST-FLAG-PAK1 and pUAST-FLAG-PAK3 constructs, Dr. Joseph Avruch for Mst1/2 null HCCs and P-Mob T12 antibody, and Dr. Shian Wu for advice on CRISPR. We also thank Bloomington Drosophila Stock Center, Kyoto Drosophila Genetic Resource Center, and Drosophila Genomics Resource Center for fly strains and reagents. This study was supported in part by grants from the National Institutes of Health (EY015708). D.P. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
Y.Z. conceived the project; Y.Z. and W.W. designed experiments; Y.Z., W.W., B.L., H.D. and E.U. performed experiments; D.P. supervised the whole study; Y.Z. and D.P. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Current opinion in cell biology. 2013;25:247–253. doi: 10.1016/j.ceb.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Developmental cell. 2012;22:695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Developmental cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk B, Hahn K, Cohen SM, Teleman AA. MAP4K3 regulates body size and metabolism in Drosophila. Developmental biology. 2010;344:150–157. doi: 10.1016/j.ydbio.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nature genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Jin P, Luo F, Zhang G, Anderson N, Chen EH. Group I PAKs function downstream of Rac to promote podosome invasion during myoblast fusion in vivo. The Journal of cell biology. 2012;199:169–185. doi: 10.1083/jcb.201204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Enderle L, McNeill H. Hippo gains weight: added insights and complexity to pathway control. Science signaling. 2013;6:re7. doi: 10.1126/scisignal.2004208. [DOI] [PubMed] [Google Scholar]
- Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. The Biochemical journal. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar P, Tapon N. Sensing the local environment: actin architecture and Hippo signalling. Current opinion in cell biology. 2014;31:74–83. doi: 10.1016/j.ceb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature reviews Molecular cell biology. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nature reviews Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Current biology: CB. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nature reviews Molecular cell biology. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hu L, Huang H, Li J, Yin MX, Lu Y, Wu W, Zeng R, Jiang J, Zhao Y, Zhang L. Drosophila casein kinase 2 (CK2) promotes warts protein to suppress Yorkie protein activity for growth control. The Journal of biological chemistry. 2014;289:33598–33607. doi: 10.1074/jbc.M114.580456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes & development. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim M, Lee S, Kuninaka S, Saya H, Lee H, Lee S, Lim DS. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. The EMBO journal. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, Pan D. The Hippo effector Yorkie controls normal tissue growth by antagonizing scallopedmediated default repression. Developmental cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lam D, Shah S, de Castro IP, Loh SH, Martins LM. Drosophila happyhour modulates JNK-dependent apoptosis. Cell death & disease. 2010;1:e66. doi: 10.1038/cddis.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, et al. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Developmental cell. 2014a;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cooper J, Zhou L, Yang C, Erdjument-Bromage H, Zagzag D, Snuderl M, Ladanyi M, Hanemann CO, Zhou P, et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer cell. 2014b;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS letters. 2014;588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Miralles F, Visa N. Actin in transcription and transcription regulation. Current opinion in cell biology. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Gurdon JB. Transcriptional regulation and nuclear reprogramming: roles of nuclear actin and actin-binding proteins. Cellular and molecular life sciences: CMLS. 2013;70:3289–3302. doi: 10.1007/s00018-012-1235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nature reviews Molecular cell biology. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes & development. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nature cell biology. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Poon CL, Lin JI, Zhang X, Harvey KF. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Developmental cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2967–2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Current biology: CB. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M, de Celis JF. MAP4K3 is a component of the TORC1 signalling complex that modulates cell growth and viability in Drosophila melanogaster. PloS one. 2011;6:e14528. doi: 10.1371/journal.pone.0014528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegert MR, Hergovich A, Tamaskovic R, Bichsel SJ, Hemmings BA. Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Molecular and cellular biology. 2005;25:11019–11029. doi: 10.1128/MCB.25.24.11019-11029.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC research notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature cell biology. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Developmental cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. The EMBO journal. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013a;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes & development. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Developmental cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tang F, Terracciano L, Hynx D, Kohler R, Bichet S, Hess D, Cron P, Hemmings BA, Hergovich A, et al. NDR functions as a physiological YAP1 kinase in the intestinal epithelium. Current biology: CB. 2015;25:296–305. doi: 10.1016/j.cub.2014.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Current opinion in cell biology. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.