Abstract
Double-stranded oligonucleotides with +1 interstrand zipper arrangements of intercalator-functionalized nucleotides are energetically activated for recognition of mixed-sequence double-stranded DNA. Incorporation of nonyl (C9) bulges at specific positions of these probes, results in more highly affine (>5-fold), faster (>4-fold) and more persistent dsDNA recognition relative to conventional Invader probes.
Chemical probes capable of sequence-specific recognition of dsDNA have tremendous potential as tools in diagnostics, structural elucidations, and nanotechnology.1–5 Hybridization-based approaches are particularly interesting due to their predictable binding modes and the resulting ease of design. To realize sequence-specific dsDNA recognition, probes must invade Watson-Crick base pairs or bind via extrahelical contacts such as Hoogsteen base-pairing, with triplex-forming oligonucleotides1,6 and peptide nucleic acids (PNAs)4,7 as prime examples of the latter. However, triplex-based approaches rely on the presence of long polypurine regions, which limits the number of targetable sites. In contrast, conformationally restricted γ-PNAs8 bind to complementary DNA (cDNA) with sufficient affinity to invade Watson-Crick base-pairs of dsDNA targets, albeit only at non-physiologic ionic strengths, resulting in displacement of one target strand and formation of a D-loop.
Double-stranded probes that bind to dsDNA via double-duplex invasion, offer the promise of even more favorable binding thermodynamics and improved specificity, as binding to mismatched dsDNA regions generates two destabilized duplexes.9 However, the probe duplex must dissociate easily for this approach to be effective. One strategy to realize this has been through the use of pseudocomplementary (pc) base pairs such as 2,6-diaminopurine and 2-thiouracil, which form weak base-pairs with each other, while forming stable pairs with thymine and adenine in target strands.10 The energy difference between the double-stranded probe and the resulting probe-target duplexes generates a thermodynamic gradient for dsDNA recognition. While pcDNA only are weakly activated for dsDNA recognition,11 pcPNA have been shown to recognize internal regions of mixed-sequence dsDNA at low ionic strengths.12
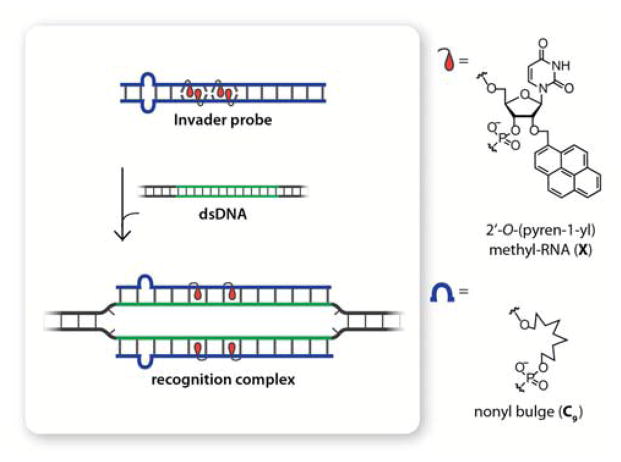
As part of our efforts toward developing new strategies for mixed-sequence dsDNA recognition, we recently introduced so-called Invader probes, which also rely on energy differences between probe duplexes and recognition complexes to drive dsDNA recognition (Figure 1).13 These probes feature 2′-intercalator-functionalized nucleotides that are arranged in +1 interstrand zipper motifs, which force the covalently linked intercalators to compete for the same inter-base-pair region, leading to violation of the nearest-neighbor exclusion principle14 and probe destabilization.13,15–19 In the recognition complex, in which each probe strand is bound to a complementary DNA region, the intercalators no longer compete for the same space, leading to strong duplex stabilization due to efficient π-π-stacking interactions with neighboring base-pairs. In previous studies, we have: i) identified more easily accessible analogs of the N2′-pyrene-functionalized 2′-amino-α-L-LNA (Locked Nucleic Acid) monomers that were used in original Invader designs,15 which include the 2′-O-(pyren-1-yl)methyl-RNA monomer shown in Figure 1, ii) studied the influence that the intercalator, linker, nucleobase, and number and distance between the intercalator-functionalized nucleotides13,15-19 have on dsDNA recognition efficiency, and iii) demonstrated recognition of chromosomal DNA targets at non-denaturing conditions.19
Figure 1.
Schematic representation of dsDNA recognition by Invader probes containing non-nucleosidic bulges and the chemical modifications used for this approach.
Herein, we describe improved dsDNA recognition using a novel Invader probe architecture that contains non-nucleosidic nonyl (C9) bulge inserts (Figure 1). This design was pursued based on the hypothesis that internal C9 bulges would destabilize the probe duplex, promote local denaturation, thus revealing the Watson-Crick face of the probe, and accelerate nucleation with, and invasion of, dsDNA targets.
Bulges have been used to tune the hybridization properties of oligonucleotides.20,21 While they induce minimal perturbation of the global duplex conformation, they do destabilize duplexes by interrupting the π-stack.21 By adjusting the number and position of the C9 bulges, we hypothesized that we could destabilize probe duplexes more than probe-target duplexes, resulting in a more prominent thermodynamic driving force and faster dsDNA recognition.
A library of Invader probes, containing two consecutive +1 interstrand zipper motifs of 2′-O-(pyren-1-yl)methyl-RNA-U monomers at the center and one or two C9 bulges at one or both termini, were synthesized (Table 1). Thermal denaturation temperatures (Tm’s) of these probes and the duplexes with cDNA were compared to conventional Invaders without C9 bulges. As expected from our previous work, reference Invader strands ON1 and ON2 form very stable duplexes with cDNA (ΔTm = 18 °C relative to unmodified ON).19 The insertion of a single C9 bulge into an Invader strand greatly reduces Tm’s (−9 to −12 °C) relative to ON1 or ON2. Insertion of two C9 bulges potentiates these trends (Tm < 15 °C for ON7 or ON8 vs cDNA). The double-stranded Invader probes display significantly lower Tm’s than the corresponding duplexes between individual probe strands and cDNA, verifying our previous observations that +1 interstrand zipper motifs of X monomers are inherently destabilizing (e.g., compare Tm of ON1:ON2 vs ON1:cDNA and ON2:cDNA). Invader probes, in which two C9 bulges either are present on the same strand or on two different strands but the same terminus, are particularly destabilized.
Table 1.
Thermal denaturation temperatures (Tm’s) and thermal advantages (TA’s) for modified DNA duplexes.a
|
Tm [ΔTm] (°C)
|
|||||
|---|---|---|---|---|---|
| Probe | Sequence | 5′-Inv: 3′-Inv | 5′-Inv: cDNA | 3′-Inv: cDNA | TA(°C) |
| 1:2 |
|
45.0 [+7.5] | 55.5 [+18.0] | 55.5 [+18.0] | 28.5 |
| 3:2 |
|
31.5 [−6.0] | 44.0 [+6.5] | 55.5 [+18.0] | 30.5 |
| 5:2 |
|
33.0 [−4.5] | 44.5 [+7.0] | 55.5 [+18.0] | 29.5 |
| 1:4 |
|
35.0 [−2.5] | 55.5 [+18.0] | 46.5 [+9.0] | 29.5 |
| 1:6 |
|
28.5 [−9.0] | 55.5 [+18.0] | 43.5 [+6.0] | 33.0 |
| 3:4 |
|
<15.0 | 44.0 [+6.5] | 46.5 [+9.0] | >38.0 |
| 5:6 |
|
<15.0 | 44.5 [+7.0] | 43.5 [+6.0] | >35.5 |
| 7:2 |
|
<15.0 | <15.0 | 55.5 [+18.0] | - |
| 1:8 |
|
<15.0 | 55.5 [+18.0] | <15.0 | - |
| 3:6 |
|
28.5 [−9.0] | 44.0 [+6.5] | 43.5 [+6.0] | 21.5 |
| 5:4 |
|
32.5 [−5.0] | 44.5 [+7.0] | 46.5 [+9.0] | 21.0 |
ΔTm is calculated relative to the corresponding unmodified dsDNA (Tm = 37.5 °C; Thermal denaturation curves were recorded in medium salt phosphate buffer ([Na+] = 110 mM, [Cl−] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4), [EDTA] = 0.2 mM) and each [ON] = 0.5 μM; see main text for definition of TA.
The thermodynamic dsDNA recognition potential of a specific Invader probe can be estimated by the term thermal advantage, given as TA = Tm (5′-Inv:cDNA) + Tm (3′-Inv:cDNA) − Tm (Invader probe) − Tm (dsDNA target), with large positive values signifying a strongly activated probe. Invader probe ON1:ON2, which is based on a traditional probe architecture without bulges, has a prominent TA value of 28.5 °C due to the high Tm’s of probe:cDNA duplexes and low Tm of the probe duplex.
Invader probes with a single C9 bulge (e.g., ON3:ON2) display similar or slightly higher TAs since the bulge destabilizes probe:cDNA and Invader probe duplexes to similar degrees. Probes ON3:ON4 and ON5:ON6, which have two C9 bulges at one of the termini, display significantly increased dsDNA recognition potential (TAs > 35.5 °C), because the probe duplexes are very strongly destabilized, while the probe-target duplexes only are mildly destabilized; presumably, this is because two adjacent C9 bulges (as in probe duplexes) have a more detrimental effect on base-pairing cooperativity than two separate C9 bulges (as in probe-target duplexes). In line with this, Invader probes with two C9 bulges on separate strands and termini (ON3:ON6 and ON5:ON4) display lower dsDNA recognition potential because the probe duplexes are not as destabilized. TA values for Invader probes with two C9 bulges on one strand (ON7:ON2 and ON1:ON8) could not be determined due to the low stability of probe-target duplexes.
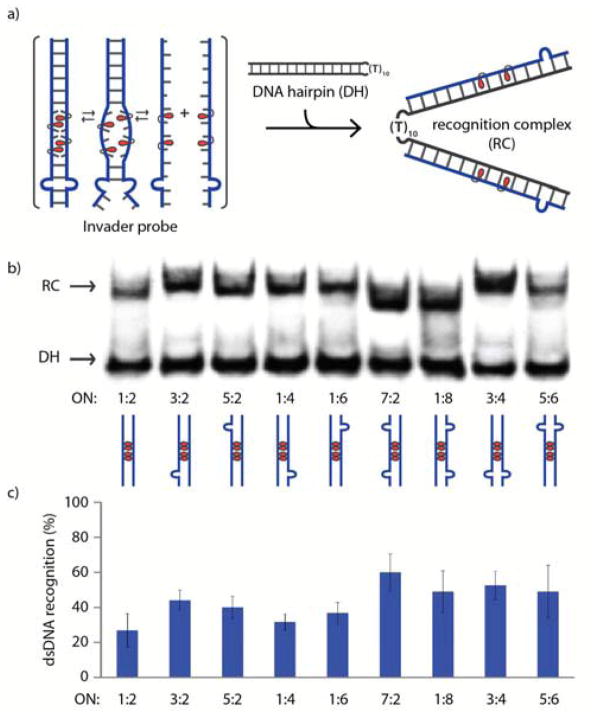
TA values provide an estimate for the thermodynamic dsDNA recognition potential of specific Invader probes.‡ However, other factors, including the experimental temperatures used, likely influence recognition efficiency and kinetics. To elucidate this, an electrophoretic mobility shift assay (EMSA) was performed. Pre-annealed Invader probes were incubated with DNA hairpin DH1, in which the double-stranded target region is linked via a decameric thymidine loop (Figure 2a). Recognition of this model target results in the formation of a recognition complex, which is observed as a slower moving band on non-denaturing polyacrylamide gel electrophoresis (Figure 2b). A 200-fold molar excess of Invader probes was incubated with DH1 at 8 °C for 17 h. At these conditions, the conventional Invader probe ON1:ON2 only results in ~22% recognition, whereas single bulge Invaders result in more efficient recognition (30–42%) (Figure 2c and Table S2†). Invader probes with two C9 bulges at one terminus (ON3:ON4 and ON5:ON6) or two C9 bulges on the same strand (ON1:ON8 and ON7:ON2) recognize the dsDNA target even more efficiently (41–55%). The recognition complexes formed with ON1:ON8 and ON7:ON2 have slightly greater electrophoretic mobilities than those formed with other Invader probes. This is almost certainly because binary, rather than ternary, recognition complexes are formed, as ON7 and ON8 have very low cDNA affinity (Tm < 15 °C for ON7/ON8:cDNA, Table 1 – see also Figure S2†). Invader probes with two C9 bulges on separate strands and termini (ON3:ON6 and ON5:ON4) do not result in detectable dsDNA recognition, suggesting that the process is energetically unfavorable (Figure S3†). For similar reasons, Invader probes with three or four bulge insertions also do not result in detectable dsDNA recognition (Figure S3 and Table S3†).
Figure 2.
(a) Schematic representation of the EMSA used to evaluate dsDNA recognition of Invader probes. (b) Representative electrophoretograms for recognition of model dsDNA target DH1 (34.4 μM) by different Invader probes (6.88 μM) at 8 °C. (c) Histogram showing the average of three experiments; error bars represent standard deviation. DIG-labeled DH1 (5′-GGTATATATAGGC-T10-GCCTATATATACC-3′) was incubated with pre-annealed Invader probe in HEPES buffer (50 mM HEPES, 100 mM NaCl, 5 mM MgCl2, pH 7.2, 10% sucrose, 1.44 mM spermine tetrahyrdochloride) for 17 h.
While conventional Invader strands ON1 and ON2 result in some recognition of DH1 when used as single-stranded probes, none of the C9-containing single-stranded probes result in significant recognition of DH1 (Figure S4†). Interestingly, ON7:ON2 results in more pronounced dsDNA recognition than single-stranded ON2, indicating that the presence of ON7 is advantageous despite its low cDNA affinity (Figure S2†).
Dose-response assays were performed at 8 °C or ambient temperature (22 °C) for representative Invader probes (Figure 3). At ambient temperature, single bulge Invader ON3:ON2 and Invader ON3:ON4, which has two bulges at the same terminus, display similar dose-response profiles and sub-micromolar C50 values (i.e. the probe concentration resulting in 50% recognition of DH1; ~0.3 μM, Table 2). Conventional Invader probe ON1:ON2 has a significantly higher C50 value (~1.6 μM), whereas Invader ON7:ON2, with two bulges on the same strand, has an intermediate C50 value of ~1.0 μM. Incubation at 8 °C results in slightly different dose-response trends (compare Figures 3a and 3b). Thus, double bulge Invaders ON3:ON4 and ON7:ON2 display lower C30 values, than single bulge Invader ON3:ON2 or conventional Invader ON1:ON2. These observations suggest that probes with large thermodynamic driving forces result in more efficient dsDNA recognition at higher experimental temperatures, whereas probes with low Tm’s result in efficient dsDNA recognition at low experimental temperatures where breathing of base-pairs is minimal. Probes with low Tm’s are likely partially or even fully dissociated at low experimental temperatures, thereby enabling the Watson-Crick face of the probe strands to be available for nucleation with DNA targets.
Figure 3.
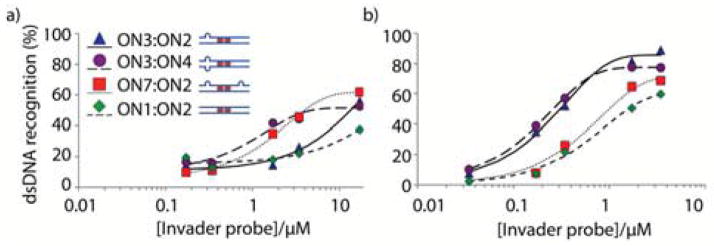
Dose-response curves for recognition of dsDNA by Invader probes ON3:ON2, ON3:ON4, ON7:ON2, and ON1:ON2 at (a) 8 °C or (b) 22 °C. Experimental conditions otherwise as described in Figure 2.
Table 2.
Summary of parameters for dsDNA recognition by representative Invader probes.
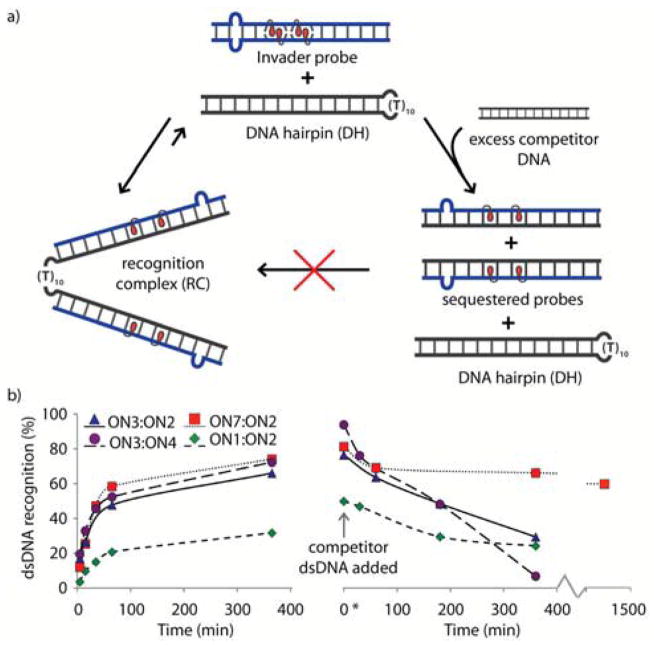
The kinetics of Invader-mediated dsDNA recognition were determined in experiments in which a 100-fold molar excess of probe was incubated with DH1 at 22 °C (Figure 4). All of the bulge-containing Invaders display much faster recognition kinetics than conventional Invader probe ON1:ON2 (pseudo-first order rate constants shown in Table 2). Invader probes ON3:ON2, ON3:ON4 and ON7:ON2 display 2.3, 2.7 and 4.1-fold faster kinetics, respectively. Presumably, the bulges promote partial or even full denaturation of the Invader probes, thus revealing their Watson-Crick face for faster target binding.
Figure 4.
a) Assays used to determine kinetic parameters for dsDNA recognition using representative Invader probes. b) Left: Kinetics of recognition complex formation at 22 °C using 100-fold molar excess of Invader probes. Right: Competitive dissociation kinetics of recognition complexes between DNA hairpins and Invader probes (for representative gel electrophoretograms, see Figure S6†). 100-fold molar excess of Invader probes (3.44 μM) was incubated with DH1 for 24 h, followed by addition of a 1000-fold molar excess of linear competitor dsDNA target (34.4 μM – sequence: 5′-GGTATATATAGGC:3′-CCATATATATCCG). T = 22 °C.
The persistence of dsDNA-binding was evaluated in a competition assay,19 in which pre-formed complexes (24 h incubation at 22 °C) were challenged with a 1000-fold excess of linear dsDNA target (Figure 4). Dissociating Invader strands bind to this competitor target,13 resulting in formation of a faster moving band in non-denaturing gel electrophoresis consistent with re-formation of DH1. Approximately 25% of the recognition complexes between DH1 and ON1:ON2 or ON3:ON2 remain intact 6 h post-challenge. The recognition complex between DH1 and ON3:ON4, undergoes rapid dissociation (>90% within 6 h), likely due to the low cDNA affinity of ON3 and ON4. Surprisingly, the recognition complex between DH1 and ON7:ON2 is remarkably stable (~60% of complex intact after 24 h). This construct is unique, as only one probe strand (i.e., ON2) is firmly bound to the target in the recognition complex (Figure S2†). Given the slower dissociation of DH1:(ON7):ON2 relative to DH1:ON1:ON2, it is clear that the unbound ON7 plays a role in slowing down dissociation, possibly due to transient binding to the binary complex and/or weak affinity toward the target competitor strand.
In conclusion, probes with appropriately positioned non-nucleosidic bulges display faster, more efficient, and longer-lasting recognition of mixed-sequence dsDNA targets than conventional Invader probes. The robustness and simplicity of design render these optimized probes amenable to a variety of applications in molecular diagnostics and DNA nanotechnology.
Supplementary Material
Acknowledgments
Support for this study was provided by Award GM088697 from the National Institute of General Medical Sciences (NIH), and Awards IF13-001 and IF14-012 from the Higher Education Research Council, Idaho State Board of Education
Footnotes
Electronic Supplementary Information (ESI) available: Experimental protocols; MS data for modified ONs; representative thermal denaturation curves; additional gel electrophoretograms, kinetics plots, and Tm and dsDNA-recognition data. See DOI: 10.1039/x0xx00000x
Thermodynamic data could not be obtained via the van’t Hoff method as denaturation curves lacked clear base lines.
Notes and references
- 1.Besch R, Giovannangeli C, Degitz K. Curr Drug Targets. 2004;5:691. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann D, Famulok M. Nucleic Acids Res. 2013;41:4729. doi: 10.1093/nar/gkt121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiba Y, Sumaoka J, Komiyama M. Chem Soc Rev. 2011;40:5657. doi: 10.1039/c1cs15039a. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen PE. Chem Biodiv. 2010;7:786. doi: 10.1002/cbdv.201000005. [DOI] [PubMed] [Google Scholar]; Blackledge MS, Melander C. Bioorg Med Chem. 2013;21:6101. doi: 10.1016/j.bmc.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gai T, Gersbach CA, Barbas CF., III Trends Biotechnol. 2013;31:397. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1497. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 8.Bahal R, Sahu B, Rapireddy S, Lee CM, Ly DH. Chem Bio Chem. 2012;13:56. doi: 10.1002/cbic.201100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen SX, Zhange DY, Seelig G. Nature Chem. 2013;5:782. doi: 10.1038/nchem.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutyavin IV, Rhinehart RL, Lukhtanov EA, Gorn VV, Meyer RB, Jr, Gamper HB., Jr Biochemistry. 1996;35:11170. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 11.Smolina IV, Demidov VV. Chem Biol. 2003;10:591. doi: 10.1016/s1074-5521(03)00150-9. [DOI] [PubMed] [Google Scholar]
- 12.Lohse J, Dahl O, Nielsen PE. Proc Natl Acad Sci USA. 1999;96:11804. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sau SP, Kumar TS, Hrdlicka PJ. Org Biomol Chem. 2010;8:2028. doi: 10.1039/b923465a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crothers DM. Biopolymers. 1968;6:575. doi: 10.1002/bip.1968.360060411. [DOI] [PubMed] [Google Scholar]
- 15.Sau SP, Madsen AS, Podbevsek P, Andersen NK, Kumar TS, Andersen S, Rathje RL, Anderson BA, Guenther DC, Karmakar S, Kumar P, Plavec J, Wengel J, Hrdlicka PJ. J Org Chem. 2013;78:9560. doi: 10.1021/jo4015936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karmakar S, Guenther DC, Hrdlicka PJ, Org J. Chem. 2013;78:12040. doi: 10.1021/jo402085v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karmakar S, Madsen AS, Guenther DC, Gibbons BC, Hrdlicka PJ. Org Biomol Chem. 2014;12:7758. doi: 10.1039/c4ob01183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson BA, Onley JJ, Hrdlicka PJ, Org J. Chem. 2015;80:5395. doi: 10.1021/acs.joc.5b00742. [DOI] [PubMed] [Google Scholar]
- 19.Guenther DC, Anderson GH, Karmakar S, Anderson BA, Didion BA, Guo W, Verstegen JP, Hrdlicka PJ. Chem Sci. 2015;6:5006. doi: 10.1039/c5sc01238d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braunlin W, Volker J, Plum GE, Breslauer KJ. Biopolymers. 2013;99:408. doi: 10.1002/bip.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyshnyi DV, Lomzov AA, Pyshnaya IA, Ivanova EM. J Biomol Struct Dyn. 2006;23:567. doi: 10.1080/07391102.2006.10507082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.