Abstract
We present a new paper-based isotachophoresis (ITP) device design for focusing DNA samples having lengths ranging from 23 to at least 1517 bp. DNA is concentrated by more than two orders of magnitude within 4 min. The key component of this device is a 2 mm-long, 2 mm-wide circular paper channel formed by concertina folding a paper strip and aligning the circular paper zones on each layer. Due to the short channel length, a high electric field of ~16 kV/m is easily generated in the paper channel using two 9 V batteries. The multilayer architecture also enables convenient reclamation and analysis of the sample after ITP focusing by simply opening the origami paper and cutting out the desired layers. We profiled the electric field in the origami paper channel during ITP experiments using a nonfocusing fluorescent tracer. The result showed that focusing relies on formation and subsequent movement of a sharp electric field boundary between the leading and trailing electrolyte.
Introduction
Here we report a paper isotachophoresis (ITP)1 platform fabricated using the principles of origami (Japanese paper folding). The device is very inexpensive, easy to assemble and operate, and is able to electrokinetically concentrate DNA. The design of this origami paper analytical device (oPAD)2,3 is illustrated in Scheme 1. Briefly, a piece of wax-patterned paper4 is folded into a concertina configuration, a plastic slip layer5–7 is inserted into one of the folds, and then this assembly is sandwiched between reservoirs for the trailing electrolyte (TE) and leading electrolyte (LE). DNA, present at concentrations on the order of 10−9 M, is initially mixed with the TE solution and then added to the TE reservoir, followed by addition of the LE solution to its reservoir. These solutions flow spontaneously into the paper channel but are prevented from mixing by the slip layer. Next, a voltage bias is applied between electrodes in the reservoirs, and then the slip layer is removed. This results in accumulation of DNA (100-fold concentration amplification) at the interface of the two electrolytes within ~4 min.
Scheme 1.
Four important outcomes emerge from the experiments reported here. First, the power requirements of the oPAD-ITP are supplied by two 9 V batteries, which is a >20-fold lower voltage than previously reported, and therefore true point-of-care (POC) applications are accessible and complications due to Joule heating are minimized.8–10 Second, the origami paper channel is fully enclosed by wax, and therefore solvent (water) evaporation is minimized. Third, the plastic slip layer (Scheme 1) makes it very easy to establish a well-defined initial TE/LE boundary. Fourth, the oPAD-ITP is "digital" in the sense that the enriched product will be on individual paper layers and can be reclaimed by simply cutting off the desired layer(s). This opens up the possibility of coupling the oPAD-ITP with other detection systems to achieve lower limits of detection (LOD).
Our work builds on the original reports of two-11 and three-dimensional12 PADs introduced by the Whitesides group in 2007 and 2008, respectively. Due to their biocompatibility,13 ease of fabrication,4 and low-cost,13, 14 PADs provide a natural entry into the field of POC diagnostic devices and systems.15 A number of detection methods have been developed for PADs, including: electrochemistry,16–18 photography,19, 20 luminescence,21–24 and others.25, 26 Nevertheless, it is still difficult to achieve sufficiently low LODs for some important applications, particularly those involving nucleic acid detection. One way around this problem is sample preconcentration, and although there are many ways to approach this for bulk solutions the number that have been reported for paper platforms is very limited.26
Isotachophoresis (ITP) is an electrophoretic technique that can effectively concentrate ionic samples with minimal sample pretreatment.1, 27, 28 In a typical ITP experiment, the electric field profile across an electrophoretic channel is controlled by using electrolytes having different mobilities: a fast moving LE and a slow moving TE.29 When a voltage is applied across the channel, sample ions, initially present in the TE solution, out-pace the TE and accumulate at the TE/LE boundary resulting in a high local concentration. The ITP technique for nucleic acids28 and proteins30 is well-established for capillaries31 and microfluidic devices.32, 33 For example, Bercovici et al. demonstrated that ITP in capillary can enable >10,000-fold increase in hybridization rates of nucleic acid.34 Garcia-Schwarz and Santiago developed a microfluidic chip-based two-stage ITP assay to detect micro RNA targets from only a few nanogram total RNA.35
In contrast to traditional fluidic platforms, there are only a few examples of ITP being implemented on PADs. For example, Moghadam et al. used a paper-based device for ITP focusing of the dye Alexa Fluor 488. The specific platform in this case was nitrocellulose paper strips, and they were able to demonstrate 900-fold enrichment using an applied voltage in the 250–350 V range.9 Later, the same group integrated lateral flow immunoassays (LFA) into their paper-based ITP device demonstrated an LOD improvement of two orders of magnitude.10 Rosenfeld and Bercovici developed an ITP microfluidic device made of wax-patterned filter paper and transparent tape. Their device enabled 1,000-fold enrichment of a fluorescent tracer using an applied voltage of ~400 V.8
Our group recently reported an oPAD design for electrophoretic separations that is intended to be simpler and operate at much lower voltages than those that have been reported previously.3 In the present paper, we adapted this design for ITP, and specifically for ITP concentration of DNA. We report on four specific experiments. First, we discuss ITP of 23-mer single-stranded DNA labeled with Cyanine5 (ssDNA) using the oPAD-ITP and compare this result to a mathematical model reported by Rosenfeld and Bercovici.8 Second, we show how ITP enrichment is affected by the initial concentration of sample DNA. Third, we report on some fundamental principles of the oPAD-ITP, and in particular the electric field profile during sample focusing. Finally, we demonstrate a specific application of the oPAD-ITP: ITP of a 100 bp dsDNA ladder, which is comprised of 100–1517 bp of double-stranded DNA (dsDNA). The results of these studies suggest that the oPAD-ITP will be able to provide additional functionality for a variety of paper-based detection platforms.
Experimental Section
Chemicals and materials
What man Grade 1 cellulose paper, HCl, acetic acid, and agarose were purchased from Fisher Scientific (Walthman, MA). Single-stranded DNA (ssDNA, 5’-AGT CAG TGT GGA AAA TCT CTA GC-Cy5-3’) was ordered from Integrated DNA Technologies (Coralville, IA) and purified by HPLC. The 100 bp dsDNA ladder was from New England BioLabs (Ipswich, MA). The following chemicals were from Sigma-Aldrich (St. Louis, MO) and used as received: 2-amino-2-(hydroxymethyl)-1,3-propanediol (tris base), 2-aminoethanesulfonic acid (taurine), Ru(bpy)3Cl2, ethidium bromide (EtBr) solution (10 mg/mL), and EDTA.
Device fabrication
The fabrication of the oPAD-ITP is similar to that of a related electrophoretic device we have reported previously,3 but there are some important differences. What man grade 1 cellulose paper (~180 µm thick) was patterned with wax (CorelDRAW designs shown in Figure S1) using a Xerox ColorQube 8750DN inkjet printer. The key feature of the patterning is the presence of a ~2 mm-diameter wax-free region in the center of each section of the device. The patterned paper was placed in an oven at 120 °C for 45 s to allow the wax to penetrate through the thickness of paper4 and then cooled to 25 ± 2 °C. After folding the paper into an 11-layer origami structure (Scheme 1), a piece of plastic sheet (photo laminating sheets from 3M: 0.5 mm thick, 4.0 cm long, and 1.0 cm in wide) was inserted between the second and third layers. This slip layer is used to form the initial TE/LE boundary and also serves as a switch to initiate the ITP process. The TE and LE reservoirs were fabricated from acrylonitrile butadiene styrene (Figure S1b) using a Flash forge Pro XL 3D printer. The assembled oPAD-ITP was then sandwiched between the two reservoirs. The degree of compression of the concertina fold was controlled using four screws situated at the corners of the reservoirs (Figure S1c).
Device operation
The TE and LE solutions used in these experiments were 2.0 mM tris-taurine (pH 8.7) and 1.0 M tris-HCl (pH 7.3), respectively. After assembling the oPAD-ITP (origami paper, slip layer and reservoirs), the reservoirs were filled with 1.0 mL of the TE or LE buffer as shown in Scheme 1. The circular paper channel was completely wetted within 1 min. A Pt wire was inserted into each reservoir, and then two 9 V batteries were connected in series and used as the power supply (18 V in total).
For the ssDNA concentration experiments, 40.0 nM of the ssDNA was initially present in the TE solution. For the dsDNA ladder experiments, 0.50 µg/mL of the dsDNA ladder was initially mixed in the TE solution. In the electric field profiling experiment, 30.0 µM Ru(bpy)32+ was initially mixed in the LE solution. For these latter experiments, home-made Ag/AgCl electrodes,36 rather than Pt, were used to avoid generation Cl2 arising from the low resistance and high current level (~17 mA). After ITP experiments, the solutions in both reservoirs were removed and the device was disassembled to analyze the content of each paper layer. The oPAD-ITP is a disposable device and used for only a single experiment.
Fluorescence microscopic analysis
A Nikon AZ100 multi-purpose zoom fluorescence microscope with Nikon filters (ssDNA: 590–650 nm excitation and 663–738 nm emission; Ru(bpy)32+: 420–490 nm excitation and 510–700 nm emission) was used to acquire fluorescence images of each fold of the oPAD-ITP device. All images were then processed with ImageJ software to obtain integrated relative fluorescence unit (RFU) intensity for quantification of fluorescent molecules on each layer.
Gel electrophoresis analysis
Gel electrophoresis was used to quantify the dsDNA content on each paper layer after ITP of the dsDNA ladder. Gel electrophoresis was chosen for two reasons. First, most common dsDNA stains, such as SYBR gold or EtBr, exhibit a high background on cellulose paper, and this makes it difficult to visualize and quantify the amount of dsDNA. Second, the dsDNA ladder is comprised of twelve dsDNA components having lengths ranging from 100 to 1517 bp. Gel electrophoresis can separate them and provides quantitative information for each component of the ladder.
The gel electrophoresis analyses were conducted as follows. First, each fold of the paper was cut off, dried, and then inserted into a 1.3% agarose gel containing 10 µg/mL EtBr (Figure S2). Control samples were prepared by drying 1.0 µL of the 500 µg/mL dsDNA ladder stock solution in the paper zone. Gel electrophoresis was run using 1× TAE (containing 40.0 mM tris, 20.0 mM acetic acid, and 1.0 mM EDTA) buffer for 50 min at 100 V (Lambda LLS9120 DC Power Supply). A Typhoon Trio fluorescence scanner (GE Healthcare, Piscataway, NJ) was used to image the gel, followed by ImageJ software analysis.
Results and Discussion
Design and operation of the oPAD-ITP for ssDNA focusing
Complete details regarding the design and operation of the oPAD-ITP are provided in the Experimental Section. Briefly, however, ssDNA focusing experiments were carried out as follows. An 11-layer oPAD-ITP was assembled as shown in Scheme 1, with the insulating slip layer initially placed between the second and third paper folds. Note, however, that the slip layer can be placed between any two folds in the device. Next, two 3D-printed reservoirs were filled with 1.0 mL of 2.0 mM tris-taurine (TE) containing 40.0 nM ssDNA and 1.0 M tris-HCl (LE), respectively. Electrodes were placed into the reservoirs, an 18 V bias was applied, and the slip layer was removed.
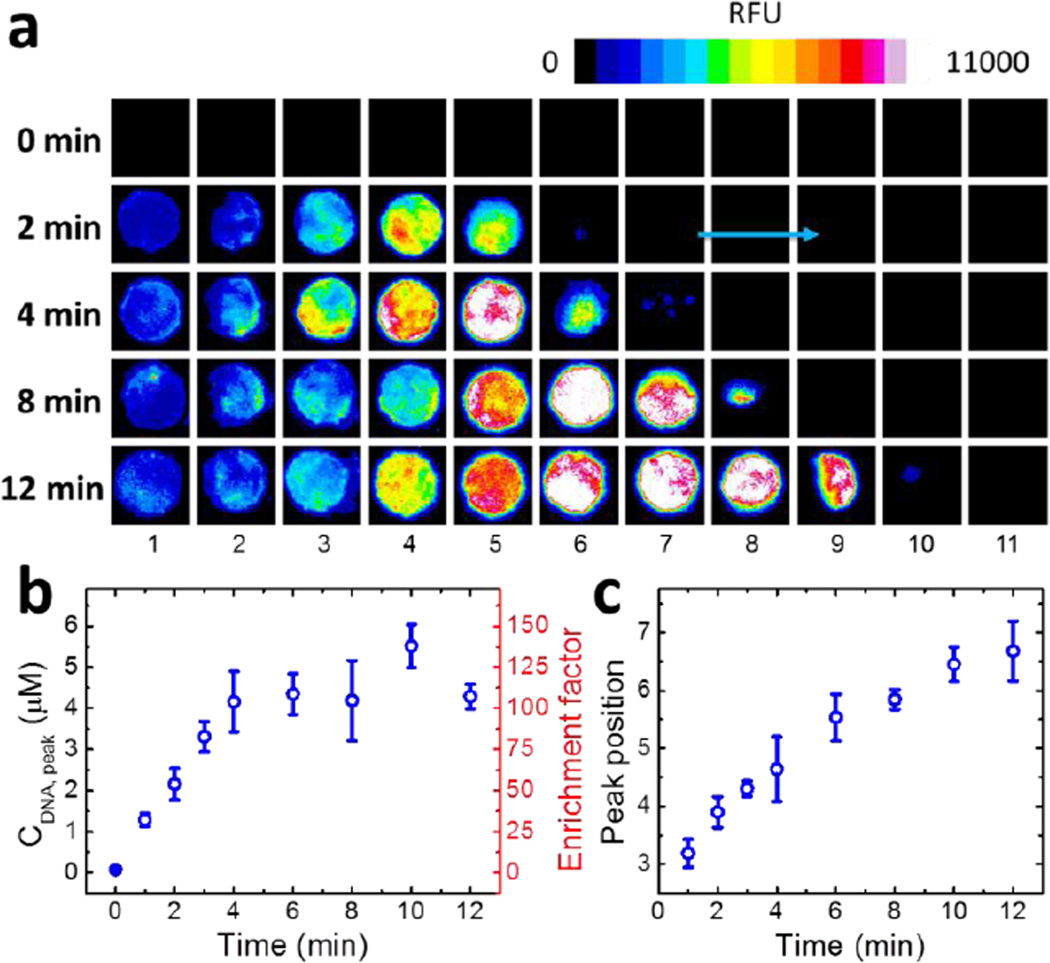
Figure 1a shows fluorescence micrographs of each paper layer as a function of time during ITP focusing of ssDNA. The fluorescence intensities increase with time, indicating successful accumulation of ssDNA from the buffer. The majority of the concentrated ssDNA is distributed between 2 to 4 paper layers, which correspond to a width of ~0.4 – 0.7 mm (the average thickness of a single layer of paper is ~180 µm).4 The precise concentration of ssDNA in each layer is determined by integrating the fluorescence intensity and then comparing it to a standard calibration curve (Figure S3). Typical concentration histograms of ssDNA as a function of position and time are shown in Figure S4. Figure 1b shows that the peak (maximum) concentration of ssDNA (CDNA, peak) grows linearly at a rate of ~1 µM/min until it reaches a plateau of ~4 µM at ~4 min. This corresponds to a ~100-fold enrichment of ssDNA from the initial 40.0 nM concentration. This enrichment factor is smaller than previously reported values (~several hundred to 1000) using other paper-based ITP devices.8–10 The main reason is that the value of CDNA, peak here is averaged over a thickness of 180 µm, and so it does not represent a true peak concentration. In other words, the oPAD-ITP is a digital device, with each paper fold representing one bin.
Figure 1.
ITP focusing of ssDNA using an 11-layer oPAD-ITP. (a) Time-resolved fluorescence micrographs of ssDNA during focusing at 18 V. The TE and LE were 2.0 mM tris-taurine (pH 8.7) and 1.0 M tris-HCl (pH 7.3), respectively, and the initial TE/LE boundary was between Layers 2 and 3. The initial ssDNA concentration in the TE solution was 40.0 nM. (b) Plot of the ssDNA peak concentration as a function of time. The peak concentrations were calculated from the images in (a). (c) Plot of peak position (in terms of layer number) as a function of time. The peak positions were obtained by Gaussian fitting of the ssDNA distributions shown in Figure S4. The error bars in (b) and (c) represent the standard deviation for three independent replicates.
In microfluidic ITP experiments, sample ions are focused at the TE/LE boundary, where there is a sharp change in the magnitude of the electric field. As the experiment progresses, this boundary migrates toward the LE reservoir. Figure 1a shows that this behavior is qualitatively replicated in the oPAD-ITP. That is, the location of maximum ssDNA concentration migrates from left to right, mirroring the location of the TE/LE boundary. This result is quantified in Figure 1c. Here, the peak positions were determined by Gaussian fitting of the ssDNA distributions (Figure S4), and then plotted as a function of time. This relationship is nearly linear, and the slope of this plot (~0.3 layers/min) represents the velocity of the TE/LE boundary. The mobility of the TE/LE boundary (µITP) can be estimated using eq 1.
| (1) |
Here, a is the slope of the linear fit in Figure 1c, d is the thickness of one paper layer (~180 µm), and E is the electric field strength. E can be estimated by dividing the applied voltage (18 V) by the total thickness of the 11-layer origami paper. This assumes that the majority of the potential drop occurs in the paper channel rather than the reservoirs, which given the higher resistance of the paper is reasonable. The calculated value of µITP is 1.08 × 10−10 m2/s·V, one order of magnitude smaller than its counterpart in conventional microfluidic channels or other paper ITP devices (~10−9 m2/s·V).1, 8 A possible explanation is that all ions are forced to travel through the three-dimensional cellulose matrix in the oPAD-ITP. This might increase the true migrational distance (tortuous path), and in addition could result in specific interactions between the ions and the cellulose fibers. This view is consistent with our previously reported finding that the mobilities of ions through paper are about one order of magnitude smaller than in free solution.3 In other paper-based ITP devices, ions migrate laterally across the paper (rather than normal to it, which is the case for the oPAD-ITP), which can lead to different migrational pathways.8, 9
Collection and extraction efficiency
In ITP, the collection efficiency (C%) is defined as the percentage of the original sample that is accumulated by an ITP device during a defined period of time. We calculated C% for the oPAD-ITP using eq 2.28
| (2) |
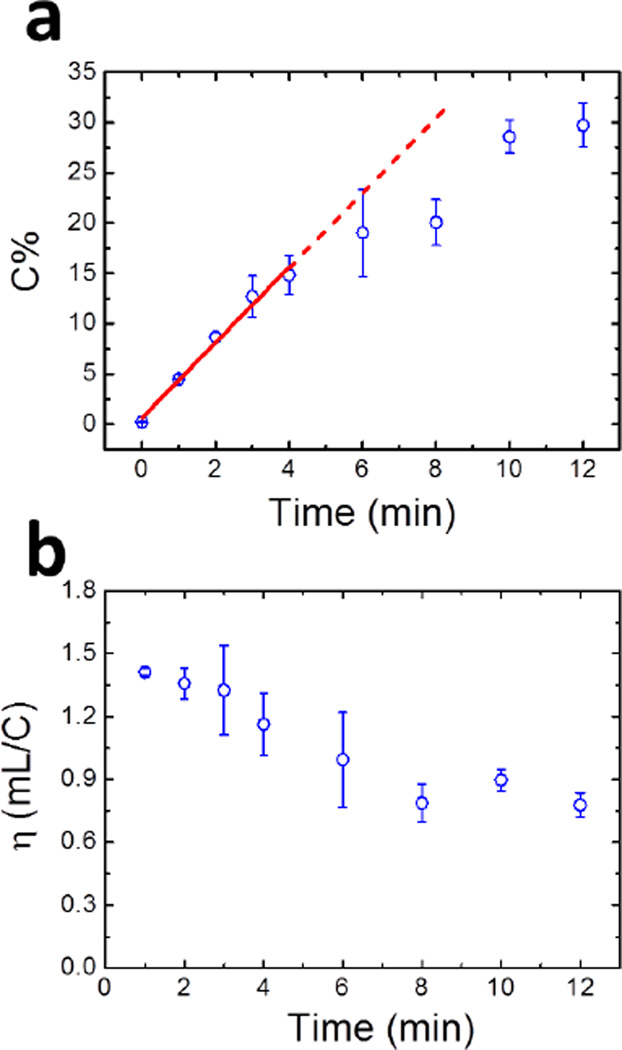
Here, Cj is the concentration of ssDNA on the jth layer and Vj is the liquid capacity of one paper layer, ~0.5 µL. C0 and VTE are the original sample concentration and the volume of the TE solution, respectively. The calculated value of C% is plotted as a function of time in Figure 2a. Between 0 and 4 min, C% increases linearly with time at a rate of ~4%/min. At longer times, however, C% increases at a lower rate. The maximum value of C% is ~30%, which is obtained after 12 min. The main reason for the slower accumulation rate at long times is probably related to the lowering of the ssDNA concentration in the reservoir as the experiment progresses. Another possible reason is that the broadened distribution of the focused ssDNA after 4 min (Figures 1a and S4) affects the ion concentration profile near the TE/LE boundary, thus disrupting the local electric field.
Figure 2.
Focusing data for ssDNA using an 11-layer oPAD-ITP. (a) Plot of collection efficiency (C%) as a function of time. C% was calculated by comparing the amount of ssDNA injected into the device with the sum of the amount of ssDNA present on each paper layer following ITP. (b) Plot of extraction efficiency (η) as a function of time. The definition of η is given in eq 3. The raw data used to generate the plots in (a) and (b) are shown in Figures 1 and S4. The error bars represent the standard deviation for three independent replicates.
In ITP, the extraction efficiency is the ability of the device to concentrate a defined sample volume per unit electrical charge consumed. Rosenfeld and Bercovici used a descriptor, η, to represent the extraction efficiency during focusing of small fluorescent molecules by their paper-based ITP device.8 The value of η is calculated using eq 3. A high value of η means less energy is required to concentrate a sample.
| (3) |
Here, NDNA(t) is the total moles of ssDNA focused by the oPAD-ITP after t min. In our experiments, the current, i(t), remained almost constant at ~0.53 mA during the focusing process (Figure S5), and NDNA(t) is linearly correlated with time for the first 4 min of the experiment (Figure 2a). Therefore, η is 0.9–1.5 mL/C (Figure 2b), which is 3–5 times higher than the value reported by Bercovici (0.3 mL/C).8 The higher η value observed in our experiments is caused primarily by the lower TE concentration (2.0 mM taurine/2.0 mM tris), required for operation of the oPAD-ITP, compared to that used by Rosenfeld and Bercovici (10 mM tricine/20 mM bistris). Specifically, when lower concentrations of TE are used, the sample ions carry a higher percentage of the total current, and this leads to better extraction efficiencies.
Effect of initial sample concentration on enrichment
According to classical peak-mode ITP theory, when the sample concentration is negligible compared with the concentration of either electrolyte (TE and LE), the maximum peak sample concentration (Csample, peak) depends solely on the TE and LE composition and is independent of the initial sample concentration (C0).1 Therefore, the enrichment factor (EF), defined as the value of Csample, peak divided by C0, will be inversely proportional to the value of C0. Accordingly, we examined the oPAD-ITP performance using different initial ssDNA concentrations (CDNA,0), but otherwise the same experimental procedure described in the previous section.
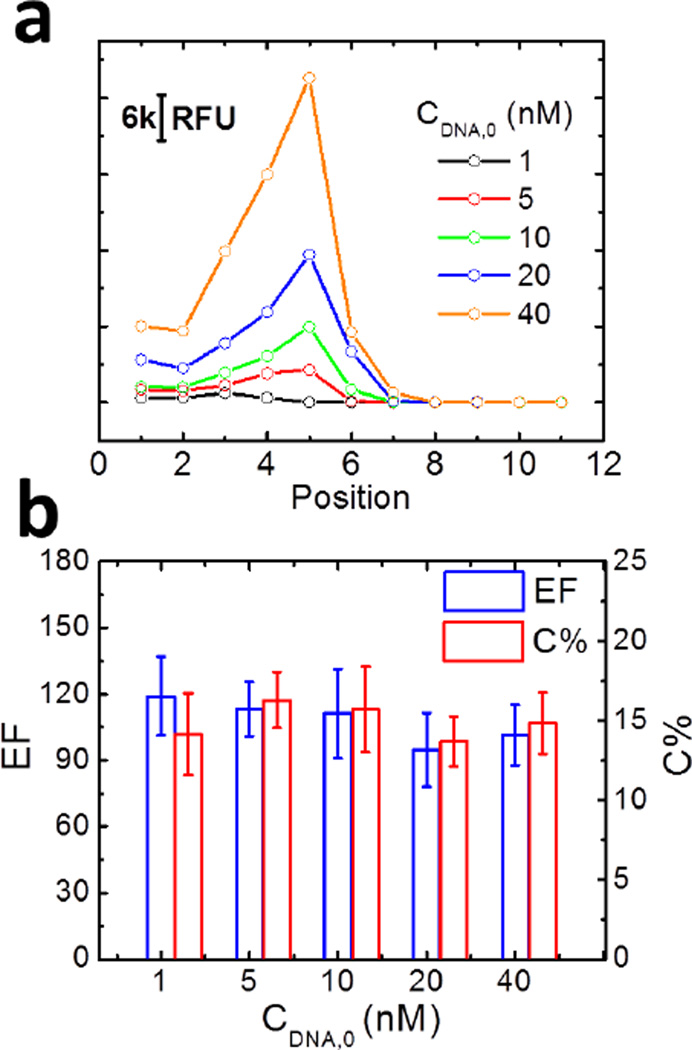
Figure 3a shows the distribution of accumulated ssDNA as a function of its position within the oPAD-ITP and CDNA,0 after 4 min of ITP focusing. The concentration profiles shown in this figure are similar in shape, indicating that the electric field in the devices is not a strong function of CDNA,0. EFs and C%s as a function of CDNA,0 are presented in Figure 3b. Surprisingly, both quantities have roughly constant values of ~100 and 15%, respectively, as CDNA,0 varies from 1.0 nM to 40.0 nM. This finding is in contrast to the expectation that both quantities should be inversely related to CDNA,0. There are several possible explanations for this observation. First, and most likely, the accumulation process during the first 4 min was limited by the migration of ssDNA within the cellulose matrix and did not reach the theoretical maximum accumulation. Therefore, ssDNA accumulates in the channel at a constant rate regardless of the initial concentration CDNA, 0, leading to constant EF and C% values. Second, as mentioned before, the value of EF is calculated from CDNA, peak, which is averaged over the thickness of a 180 µm paper fold. This introduces some uncertainty into the determination of EFs. Third, electroosmotic flow (EOF) may play a role in ITP focusing by generating a counter flow in the paper channel. This would slow ssDNA migration and broaden the peak. However, a control experiment shown in Figure S6 does not support this idea, because the EF is unchanged in presence and absence of EOF.
Figure 3.
Focusing data as a function of the initial concentration of ssDNA for an 11-layer oPAD-ITP. (a) Distribution of ssDNA as a function of position (paper layer number) and initial ssDNA concentration after 4.0 min of ITP at 18 V. Each data point was calculated by integrating the RFU of the fluorescence image of each paper layer. (b) Enrichment factor (EF) and collection efficiency (C%) as a function of the initial ssDNA concentration. The ssDNA concentration was calculated by comparing integrated RFU value with the calibration curve shown in Figure S3. The error bars represent the standard deviation for three independent replicates.
Electric field in the oPAD-ITP
In microfluidic devices, focusing of analyte at the TE/LE boundary results from a sharp transition of the electric field between the TE and LE.37 We assume that the same is true for the oPAD-ITP, but to be certain we measured the electric field profile during ITP focusing. For this study, we adopted an approach reported by Santiago and coworkers.37 Specifically, we added Ru(bpy)32+, a nonfocusing fluorescent tracer (NFT),37 to the LE solution, and then determined its distribution across the paper folds in the oPAD-ITP after focusing. In principle, the NFT will migrate through the channel during ITP and leave behind a concentration distribution that is inversely proportional to the local electric field strength.
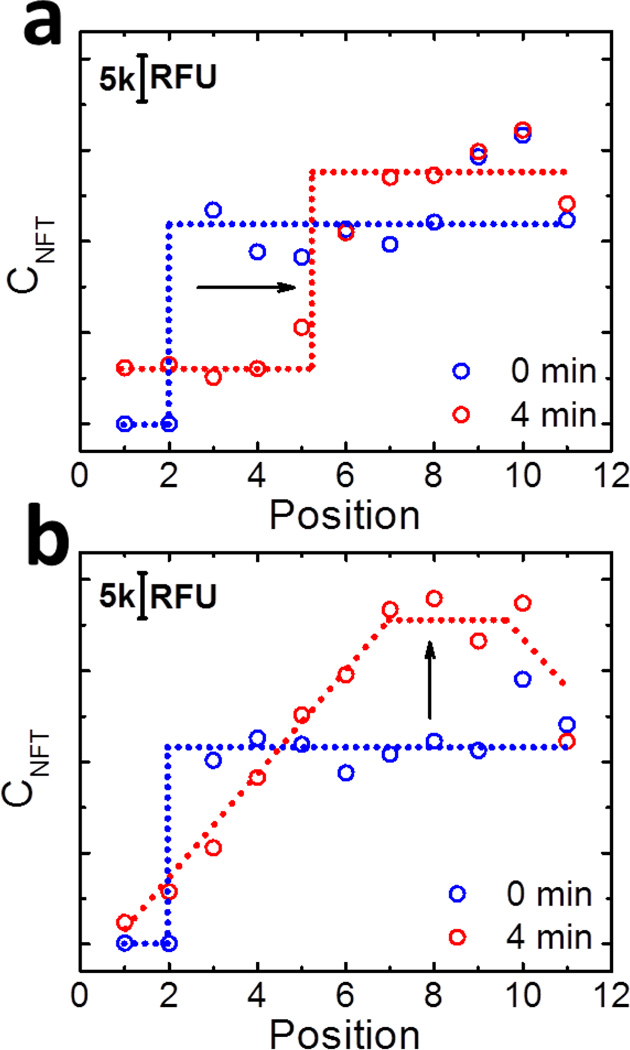
Figure 4a shows the distribution of the NFT concentration (CNFT) in an oPAD-ITP at t = 0 and 4 min. These data were obtained using the same experimental conditions used for the ssDNA focusing experiments (e.g., Figure 1). At t = 0 min, the NFT is only present in the LE zone (Layers 3 to 11), and there is a concentration step between Layer 2 and 3. This step represents the TE/LE boundary where the slip layer was initially located. At t = 4 min, the concentration step is still present, but it has moved three positions to the right. This is the same location where ssDNA accumulated after 4 min of ITP (Figure 1a), thereby confirming that ssDNA accumulation occurs at the TE/LE boundary. Based on the value of CNFT in the channel, the electric field strength in the LE and TE zones can be approximated as 3 and 14 kV/m, respectively.
Figure 4.
Electric field profiles in an oPAD-ITP obtained using Ru(bpy)32+ as a NFT. Each data point represents the integrated RFU value of the fluorescence image of each paper layer. The dash lines represent the guidelines for the NFT distributions, which corresponding to electric field strength in the channel. (a) Distributions of the NFT before application of the voltage and at t = 4 min. Initially, the LE solution contained 30.0 µM Ru(bpy)32+. (b) Distribution of the NFT before application of the voltage and at t = 4 min. For this experiment both reservoirs were filled with LE solution, but the NFT was only present in the anodic reservoir adjacent to Layer 11. To avoid oxidation of Cl− at the anode, Ag/AgCl wire electrodes replaced the Pt wire electrodes used in (a). For all experiments the applied voltage was 18 V.
There is a small possibility that the step-shaped distribution of NFT shown in Figure 4a (t = 4 min) could arise from slow migration of NFT in the paper matrix. Accordingly, we carried out a control experiment to rule out this possibility. Specifically, instead of using two different electrolytes (TE and LE), the same electrolyte (1.0 M tris-HCl) was loaded into both reservoirs, though the NFT is only placed in the LE reservoir. This condition should result in a uniform electric field across the entire channel, and therefore we expected the NFT concentration step to disappear after applying the voltage. Figure 4b shows the result of this experiment. At t =0 min, a step-shaped concentration profile is observed, just as in Figure 4a. After 4 min, however, the NFT concentration step is replaced by a trapezoidal distribution: a linear increase from Layer 1 to 7, a plateau from Layer 7 to 10, and finally a decrease at Layer 11. This unusual distribution is mainly caused by the mobility differences of the NFT in the paper medium and in free solution,3 which determines the value of CNFT near the paper/reservoir boundary (boundary effect). As shown in Figure 4b, accumulation of NFT is observed near the right paper/reservoir boundary, due to larger influx of NFT from that reservoir into the paper. In the contrast, depletion of NFT near the left paper/reservoir boundary results from larger out-flow of NFT from paper into that reservoir.
Based on the results in this section, we conclude that ssDNA focusing is caused primarily by the electric field transition at the TE/LE boundary, and to a much lesser extent by boundary effect.
ITP focusing of a dsDNA ladder
Even though short DNA strands (usually several tens of bases) are widely used as model targets for developing DNA sensing technologies, real-world DNA, for example in viruses or bacteria, is usually composed of thousands of base pairs.38, 39 Accordingly, it is essential for an ITP device to be capable of focusing DNA strands longer 100 bp. In this section, we demonstrate the use of the oPAD-ITPs for focusing a 100 bp dsDNA ladder containing 100–1517 bp dsDNA. The same experimental setup and buffer conditions used in the previous section were used for these experiments. That is, the dsDNA ladder was loaded into the TE buffer, and the voltage was switched on for 10 min.
After ITP, each fold of paper was removed from the channel, and gel electrophoresis was used to elute its dsDNA content (Figure S2). The dsDNA on the gel was stained by EtBr and then imaged using a fluorescence scanner. A raw fluorescence image of a typical gel is shown in Figure 5a. Here, each lane represents one paper layer. The right-most lane was used as a standard (the dsDNA ladder solution was dropcast onto a single paper fold, but it was not exposed to ITP). Clearly, the concentrations for the different dsDNA lengths achieve their maximum values at Layers 5 and 6. Recall that the peak position for the shorter ssDNA was at Layer 7 (Figure S4). The slightly reduced mobility of the longer dsDNA ladder is probably due to its stronger affinity for the cellulose matrix.40
Figure 5.
ITP focusing of a 100 bp dsDNA ladder. Initially, 1.0 µL of a 500 µg/mL solution of the 100 bp dsDNA ladder was dissolved in 1.0 mL of the TE solution (final dsDNA concentration: 0.5 µg/mL). Following 10 min of ITP of this solution at 18 V, each fold of the 11-layer oPAD-ITP was cut from the device, and gel electrophoresis was used to elute the dsDNA in that layer (Figure S2). (a) The left panel is a fluorescence micrograph of the gel after electrophoresis and staining with 10 µg/mL EtBr. The numbers under the lanes of the gel correspond to the Layer numbers of the oPAD-ITP. The right panel is a control experiment showing the result of gel electrophoresis of a paper fold onto which 1.0 µL of a 500 µg/mL dsDNA ladder solution was dispensed (no ITP). The gel electrophoresis conditions for all paper folds were: 1.3% agarose gel, 100 V, and 50 min. (b) The black lines are fluorescence line profiles of the stained gels in each lane of the left panel in (a). The integral of the profiles is represented by the blue bar. (c) The black line is the fluorescence line scan of the gel in the right panel in (a). The blue hollow and red solid bars represent the total dsDNA placed in the reservoir prior to ITP and the total collected dsDNA on the oPAD-ITP, respectively. The calculated C% values for each dsDNA length are shown in the right-most column.
Figures 5b presents line profiles (black lines) of the RFU corresponding to the dsDNA bands in Figure 5a. The integrated RFU values of each dsDNA band, representing the dsDNA amount, are shown as filled blue bars aligned with the line profiles. Figure 5c is the same analysis for the ladder standard (right side of Figure 5a): the black lines present the RFU line profiles, the hollow blue bars show the integrated RFU value of each dsDNA band, and filled red bars equal the sum of the dsDNA amount on individual paper folds (equivalent to the filled blue bars in Figure 5b). The EF for each dsDNA length was calculated as the area of filled blue bars in Figure 5b divided by the area of hollow blue bars in Figure 5c. Figure S7 is a plot of the EF as a function of the layer number for different dsDNA lengths. The maximum EFs are found at Layer 5 and they vary from 60 to 120, which is consistent with the ssDNA results. The C% values of each dsDNA length are obtained as the area of filled red bars divided by the area of hollow blue bars in Figure 5c. The right column of Figure 5c shows that C% for all dsDNA lengths ranges from ~15–20%. These results support our conclusion that focusing of dsDNA having different lengths (up to 1517 bp) in the oPAD-ITP yields a consistent C% of ~20% and EF of ~100.
Summary and Conclusions
In this article we presented a novel origami paper-based device, the oPAD-ITP, suitable for carrying out low-voltage ITP focusing of DNA. Our approach resolves several issues inherent to previously reported paper-based ITP designs, including high operating voltage, solvent evaporation, and difficult sample reclamation. Using the oPAD-ITP, we demonstrated >100-fold enrichment of ssDNA and dsDNA having lengths of up to 1517 bps. The time required for enrichment is ~10 min, the paper device can accommodate solution volumes of up to 1.0 mL, and it is battery operated (18 V). The collection efficiency ranges from ~15–20%. The electric field profiling experiments, using Ru(bpy)32+ as a tracer, clearly show that the focusing mechanism in the oPAD-ITP is the same as in bulk liquid solutions: accumulation of sample at the boundary between the TE and LE.
Currently, we are working to couple the oPAD-ITP with other paper-based detection system to achieve lower LODs.17 Also, we are interested in tailoring the structure of the paper channels to achieve better sample enrichment. The results of these experiments will be reported in due course.
Supplementary Material
Acknowledgements
We gratefully acknowledge the National Science Foundation (Grant No. 1402242) and the National Institutes of Health (Grant No. 1R21HL128199-01) for support of this project. We also thank the Robert A. Welch Foundation (Grant F-0032) for sustained support of our research program.
Footnotes
Electronic Supplementary Information
Electronic supplementary information (ESI) available: Design and photographs of the oPAD-ITP, photograph of the gel electrophoresis arrangement, ssDNA calibration curve, distributions of ssDNA in the oPAD-ITP as a function of time, current vs time curves for a typical ITP experiment, electroosmotic flow control experiment, EF analysis of ITP of a dsDNA ladder. See DOI: 10.1039/x0xx00000x
References
- 1.Jung B, Bharadwaj R, Santiago JG. On-chip millionfold sample stacking using transient isotachophoresis. Anal. Chem. 2006;78:2319–2327. doi: 10.1021/ac051659w. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Crooks RM. Three-Dimensional Paper Microfluidic Devices Assembled Using the Principles of Origami. J. Am. Chem. Soc. 2011;133:17564–17566. doi: 10.1021/ja2071779. [DOI] [PubMed] [Google Scholar]
- 3.Luo L, Li X, Crooks RM. Low-Voltage Origami-Paper-Based Electrophoretic Device for Rapid Protein Separation. Anal. Chem. 2014;86:12390–12397. doi: 10.1021/ac503976c. [DOI] [PubMed] [Google Scholar]
- 4.Carrilho E, Martinez AW, Whitesides GM. Understanding Wax Printing: A Simple Micropatterning Process for Paper-Based Microfluidics. Anal. Chem. 2009;81:7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- 5.Du WB, Li L, Nichols KP, Ismagilov RF. SlipChip. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Li X, Crooks RM. Paper-Based SlipPAD for High-Throughput Chemical Sensing. Anal. Chem. 2013;85:4263–4267. doi: 10.1021/ac4008623. [DOI] [PubMed] [Google Scholar]
- 7.Shen F, Du WB, Kreutz JE, Fok A, Ismagilov RF. Digital PCR on a SlipChip. Lab Chip. 2010;10:2666–2672. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld T, Bercovici M. 1000-fold sample focusing on paper-based microfluidic devices. Lab Chip. 2014;14:4465–4474. doi: 10.1039/c4lc00734d. [DOI] [PubMed] [Google Scholar]
- 9.Moghadam BY, Connelly KT, Posner JD. Isotachophoretic Preconcenetration on Paper-Based Microfluidic Devices. Anal. Chem. 2014;86:5829–5837. doi: 10.1021/ac500780w. [DOI] [PubMed] [Google Scholar]
- 10.Moghadam BY, Connelly KT, Posner JD. Two Orders of Magnitude Improvement in Detection Limit of Lateral Flow Assays Using Isotachophoresis. Anal. Chem. 2015;87:1009–1017. doi: 10.1021/ac504552r. [DOI] [PubMed] [Google Scholar]
- 11.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem., Int. Ed. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez AW, Phillips ST, Whitesides GM. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 14.Carrilho E, Phillips ST, Vella SJ, Martinez AW, Whitesides GM. Paper microzone plates. Anal. Chem. 2009;81:5990–5998. doi: 10.1021/ac900847g. [DOI] [PubMed] [Google Scholar]
- 15.Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Point of care diagnostics: status and future. Anal. Chem. 2011;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 16.Scida K, Li BL, Ellington AD, Crooks RM. DNA Detection Using Origami Paper Analytical Devices. Anal. Chem. 2013;85:9713–9720. doi: 10.1021/ac402118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scida K, Cunningham JC, Renault C, Richards I, Crooks RM. Simple, Sensitive, and Quantitative Electrochemical Detection Method for Paper Analytical Devices. Anal. Chem. 2014;86:6501–6507. doi: 10.1021/ac501004a. [DOI] [PubMed] [Google Scholar]
- 18.Dungchai W, Chailapakul O, Henry CS. Electrochemical detection for paper-based microfluidics. Anal. Chem. 2009;81:5821–5826. doi: 10.1021/ac9007573. [DOI] [PubMed] [Google Scholar]
- 19.Ellerbee AK, Phillips ST, Siegel AC, Mirica KA, Martinez AW, Striehl P, Jain N, Prentiss M, Whitesides GM. Quantifying colorimetric assays in paper-based microfluidic devices by measuring the transmission of light through paper. Anal. Chem. 2009;81:8447–8452. doi: 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W, Ali MM, Aguirre SD, Brook MA, Li Y. Paper-based bioassays using gold nanoparticle colorimetric probes. Anal. Chem. 2008;80:8431–8437. doi: 10.1021/ac801008q. [DOI] [PubMed] [Google Scholar]
- 21.Ge L, Yan J, Song X, Yan M, Ge S, Yu J. Three-dimensional paper-based electrochemiluminescence immunodevice for multiplexed measurement of biomarkers and point-of-care testing. Biomaterials. 2012;33:1024–1031. doi: 10.1016/j.biomaterials.2011.10.065. [DOI] [PubMed] [Google Scholar]
- 22.Ge L, Wang S, Ge S, Yu J, Yan M, Li N, Huang J. Electrophoretic separation in a microfluidic paper-based analytical device with an on-column wireless electrogenerated chemiluminescence detector. Chem. Commun. 2014;50:5699–5702. doi: 10.1039/c3cc49770d. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Li J, Chen C, Lou B, Zhang L, Wang E. A self-powered microfluidic origami electrochemiluminescence biosensing platform. Chem. Commun. 2013;49:3866–3868. doi: 10.1039/c3cc40905h. [DOI] [PubMed] [Google Scholar]
- 24.Delaney JL, Hogan CF, Tian J, Shen W. Electrogenerated chemiluminescence detection in paper-based microfluidic sensors. Anal. Chem. 2011;83:1300–1306. doi: 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- 25.Parolo C, Merkoçi A. Paper-based nanobiosensors for diagnostics. Chem. Soc. Rev. 2013;42:450–457. doi: 10.1039/c2cs35255a. [DOI] [PubMed] [Google Scholar]
- 26.Yetisen AK, Akram MS, Lowe CR. Paper-based microfluidic point-of-care diagnostic devices. Lab Chip. 2013;13:2210–2251. doi: 10.1039/c3lc50169h. [DOI] [PubMed] [Google Scholar]
- 27.Walker PA, Morris MD, Burns MA, Johnson BN. Isotachophoretic separations on a microchip. Normal Raman spectroscopy detection. Anal. Chem. 1998;70:3766–3769. doi: 10.1021/ac980195h. [DOI] [PubMed] [Google Scholar]
- 28.Persat A, Marshall LA, Santiago JG. Purification of nucleic acids from whole blood using isotachophoresis. Anal. Chem. 2009;81:9507–9511. doi: 10.1021/ac901965v. [DOI] [PubMed] [Google Scholar]
- 29.Everaerts FM, Beckers JL, Verheggen TP. Isotachophoresis: theory, instrumentation and applications. Elsevier; 2011. [Google Scholar]
- 30.Qu YT, Marshall LA, Santiago JG. Simultaneous Purification and Fractionation of Nucleic Acids and Proteins from Complex Samples Using Bidirectional Isotachophoresis. Anal. Chem. 2014;86:7264–7268. doi: 10.1021/ac501299a. [DOI] [PubMed] [Google Scholar]
- 31.Malá Z, Gebauer P, Boček P. Recent progress in analytical capillary isotachophoresis. Electrophoresis. 2013;34:19–28. doi: 10.1002/elps.201200323. [DOI] [PubMed] [Google Scholar]
- 32.Smejkal P, Bottenus D, Breadmore MC, Guijt RM, Ivory CF, Foret F, Macka M. Microfluidic isotachophoresis: A review. Electrophoresis. 2013;34:1493–1509. doi: 10.1002/elps.201300021. [DOI] [PubMed] [Google Scholar]
- 33.Bahga SS, Santiago JG. Coupling isotachophoresis and capillary electrophoresis: a review and comparison of methods. Analyst. 2013;138:735–754. doi: 10.1039/c2an36150g. [DOI] [PubMed] [Google Scholar]
- 34.Bercovici M, Han CM, Liao JC, Santiago JG. Rapid hybridization of nucleic acids using isotachophoresis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:11127–11132. doi: 10.1073/pnas.1205004109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Schwarz G, Santiago JG. Rapid High-Specificity micro RNA Detection Using a Two-stage Isotachophoresis Assay. Angew. Chem., Int. Ed. 2013;125:11748–11751. doi: 10.1002/anie.201305875. [DOI] [PubMed] [Google Scholar]
- 36.Lathrop DK, Ervin EN, Barrall GA, Keehan MG, Kawano R, Krupka MA, White HS, Hibbs AH. Monitoring the escape of DNA from a nanopore using an alternating current signal. J. Am. Chem. Soc. 2010;132:1878–1885. doi: 10.1021/ja906951g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chambers RD, Santiago JG. Imaging and Quantification of Isotachophoresis Zones Using Nonfocusing Fluorescent Tracers. Anal. Chem. 2009;81:3022–3028. doi: 10.1021/ac802698a. [DOI] [PubMed] [Google Scholar]
- 38.Lodish HF, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. Molecular cell biology. Citeseer; 2000. [Google Scholar]
- 39.Davis L. Basic methods in molecular biology. Elsevier; 2012. [Google Scholar]
- 40.Araújo AC, Song Y, Lundeberg J, Ståhl PL, Brumer H., III Activated paper surfaces for the rapid hybridization of DNA through capillary transport. Anal. Chem. 2012;84:3311–3317. doi: 10.1021/ac300025v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.