Abstract
The presence of disseminated tumor cells (DTCs) in the bone marrow (BM) of breast cancer patients is prognostic for early relapse. In the present study, we analyzed the gene expression profiles from BM cells of breast cancer patients to identify molecular signatures associated with DTCs and their relevance to metastatic outcome. We analyzed BM from 30 patients with stage II/III breast cancer by gene expression profiling and correlated expression with metastatic disease development. A candidate gene, PITX2, was analyzed for expression and phenotype in breast cancer cell lines. PITX2 was knocked down in the MDAMB231 cell lines for gene expression analysis and cell invasiveness. Expression of various signaling pathway molecules was confirmed by RT–PCR. We found that the expression of Paired-like Homeobox Transcription factor-2 (PITX2) is absent in the BM of normal healthy volunteers and, when detected in the BM of breast cancer patients, is significantly correlated with early metastatic disease development (p = 0.0062). Suppression of PITX2 expression significantly reduced invasiveness in MDAMB231 cells. Three genes—NKD1, LEF1, and DKK4—were significantly downregulated in response to PITX2 suppression. Expression of PITX2 in BM of early-stage breast cancer patients is associated with risk for early disease recurrence. Furthermore, PITX2 likely plays a role in the metastatic process through its effect on the expression of genes associated with the Wnt/beta-Catenin signaling pathway.
Electronic supplementary material
The online version of this article (doi:10.1007/s10549-015-3576-z) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Disseminated tumor cells, Gene expression, Metastasis, PITX2, Wnt signaling, Invasiveness
Introduction
Metastatic spread of breast cancer is the major cause of mortality in patients. Although thousands of cells are shed from primary tumors, only a few cells possess the potential to give rise to metastatic foci [1, 2]. The metastatic process consists of a series of steps, all of which must be successfully completed to form a metastatic focus [3–6]. These include shedding of cells from a primary tumor into the circulation, survival of the cells in the circulation, arrest in the vasculature of a new organ, extravasation into the surrounding tissue, initiation and maintenance of growth, and vascularization of the metastatic tumor [7]. To halt the metastatic process, it is essential to identify those intermediary cells with metastatic potential which are often resistant to chemotherapy, understand their biology through molecular characterization, and target their therapeutic vulnerabilities. The small number of disseminated, pre-metastatic tumor cells found in the circulation has previously hindered studies on the metastatic process. In breast cancer patients, clinical studies suggest that bone marrow (BM) serves as a reservoir for the disseminated tumor cells (DTCs), which are thought to be precursors to metastatic foci [8]. DTCs can survive in the BM for long periods of time, and their presence in BM portends an increased risk of distant disease development [9, 10]. Detection of DTCs in BM after chemotherapy treatment is an indicator of particularly poor prognosis suggesting that those cells which survive cytotoxic chemotherapy have greater metastatic potential [11]. Focusing on the DTC population that is present after chemotherapy treatment could lead to a better understanding of the molecular mechanisms that contribute to metastatic potential of the DTCs.
Earlier studies from our lab identified 67 genes that were overrepresented in DTCs enriched from the BM of stage II/III breast cancer patients after chemotherapy treatment [12]. Among these, expression of five genes, TWIST1, PITX2, S100A3, PDGFRL, and DUSP9, was not detected in BM from healthy volunteers suggesting that expression most likely emanates from DTCs residing in the BM of the breast cancer patients.
Paired-like homeobox transcription factor-2 (PITX2) is a bicoid-related homeobox transcription factor that is involved in pituitary-specific gene regulation and left–right patterning during embryonic development [13–15]. PITX2, as observed with other metastatic disease regulators such as TWIST1, may also be involved in epithelial-mesenchymal transition (12). PITX2 has three alternative transcripts resulting in 3 isoforms 1, 2, and 3 [16, 17]. Promoter P1 drives the transcription of the isoform 3 [18]. Isoforms 1 and 2 are driven by a common promoter, P2 [19]. Expression of isoforms 1 and 2 is regulated by WNT pathway and expression of isoform 3 is regulated by TGF-β family members [18–20]. Methylation of P2 in primary tumors has been reported to be an indicator of high risk for disease recurrence in hormone receptor-positive, node-negative breast cancer patients [19]. In this study, we investigated the role of PITX2 in regulating invasive potential and specific gene expression programs in breast cancer cells which may mediate the pro-invasive action of PITX2.
Materials and methods
Patient population and BM collection
BM collected from 30 patients with clinical stage II/III breast cancer who had been enrolled into a prospective clinical trial March 2003 and March 2006 as previously described between (12). Median follow-up is approximately 10 years. The protocol was approved by the institutional review board of Washington University and all patients signed written informed consent. BM was collected from each iliac crest at the time of diagnosis, after 4 cycles of epirubicin/taxotere, and 1 year after diagnosis (12). BM samples from each iliac crest collected prior to any treatment were used to determine the relationship of gene expression to recurrent disease development. Half of the 30 patients developed recurrent disease within 16 months of diagnosis. Control BM specimens were collected from healthy female volunteers after informed consent.
RNA isolation and qRT–PCR
Whole BM samples were subjected to hypotonic RBC lysis, 5 × 106 nucleated cells were pelleted and snap frozen for subsequent RNA isolation. Total RNA was isolated using Trizol reagent (Invitrogen) according to manufacturer’s recommendations. RNA from cell lines was isolated using the same protocol. 1 μg of RNA was used for synthesis of first strand cDNA using the Retroscript (Ambion) kit with random hexamers. Resulting cDNA was diluted to an equivalent of 10 ng/μL of input RNA.
qRT–PCR for the indicated genes was performed as described previously [12]. Primer/probe sets were purchased from Applied Biosystems. Each reaction consisted of 2 μL of cDNA, TaqMan Master Mix (Applied Biosystems), and primer/probe set in a total volume of 20 μL. The expression of the three PITX2 transcript variants was determined by qRT–PCR using SybrGreen Universal master mix (ABI, California). Primer sets for isoform 1 and isoform 2 were designed as described in Nimmrich et al. [19] (isoform 1 forward GCGTGTGTGCAATTAGAGAAAG and reverse CCGAAGCCATTC TTGCATAG; isoform 2 forward GCCGTTGAATGTCTCTTCTC and reverse CCTTTGCCGCTTCTTCTTAG). Isoform 3 was detected using the primers—forward GCT TCACTTGGAGCACCGAGCA, and reverse GAGGTGTCGGAGATGGTGTG. For each transcript/sample, triplicate reactions were analyzed in an ABI 7500 FAST Sequence Detection System. The Ct values of each gene were normalized to GAPDH Ct values for the same sample (dCt) and compared with that of BM of four normal healthy volunteers to calculate the ddCt and fold overexpression.
Cell culture and stable transductions
Breast cancer cell lines MDAMB231, MCF7, MCF10A, T47D, ZR75, and SKBR3 were purchased from ATCC. MDAMB231 cells were grown in DMEM containing 10 % fetal bovine serum (FBS). MCF7 and MCF 10A cells were grown in a mixture of 50 % MEGM (Lonza) and DMEM with 10 % FBS, ZR75 was grown in RPMI containing 10 % FBS. SKBR3 and T47D were grown in McCoy’s media with 10 % FBS. All cells were grown at 37 °C with 5 % CO2.
RNA expression level of each of the 3 PITX2 isoforms was quantified by qRT–PCR using specific primers designed for each isoform in human breast cancer cell lines listed above. PITX2 expression was stably silenced in MDAMB231 cell line, which had high endogenous levels of all three PITX2 transcript isoforms, using Mission lentiviral transduction particles (Sigma, St. Louis) and selecting the stably transduced cells with puromycin. The construct targeted all three isoforms (TRCN000002479). Individual clones were selected and tested for PITX2 expression and several PITX2 knockdown clonal lines were established.
Matrigel invasion assay
MDAMB231 cells with near total knockdown of the PITX2 gene were used for matrigel invasion assays (BD Biosciences) and performed according to the manufacturer’s recommendations. Briefly, cells were suspended in growth factor-free media. 3 × 104 cells were added to each well of an invasion chamber which contained growth factor-free media. Complete growth medium was added to the lower chamber as chemoattractant. The number of invasive cells was counted at 24 h and 48 h after seeding. MDAMB231 cells transfected with empty vector, a non-targeting sequence, or shRNA against the unrelated gene, beta-2 microglobulin, served as negative controls. The number of invasive cells was counted from five random fields from each well and total cells counted for each well were calculated for subsequent statistical analysis. All experiments were repeated three times.
Wnt pathway expression analysis
MDAMB231 cells with PITX2 gene knockdown and controls were used for gene expression analysis. Three different clonal lines of stably knocked down PITX2 gene were grown in two wells of a six-well plate. A similar set of cells that were stably transduced with a non-targeting sequence were grown in parallel. Two wells of the same clonal line of PITX2 knockdown or non-targeting sequence were pooled and used for RNA extraction as described earlier. Samples were analyzed for gene expression using Taqman 96 well Array plates (Applied Biosystems) according to manufacturer’s protocol. The relative expression of each gene with respect to GAPDH levels of corresponding samples was determined and the average of four samples in each group was used for subsequent ddCt and fold difference calculation.
Analysis of data
For PCR analysis, fivefold increased expression over normal BM was considered positive. Patient BM was considered positive if BM from at least one side (right or left) over-expressed the gene. Gene expression from patient samples and survival were analyzed using Kaplan–Meier model and Cox proportional hazards models. All estimates are derived from single-variable models, i.e., fold change in only 1 gene was included in each model. For the invasion experiments, percentage invasion was calculated as the percentage of number of cells passed through the Matrigel to the total number of cells present before harvest. Significant difference between samples was analyzed using paired t test.
Results
TWIST1 and PITX2 are expressed in BM of patients who develop recurrent disease
We had earlier identified a 67-gene signature associated with the presence of DTCs in the BM of breast cancer patients using gene array analysis (Supplemental Table 1) [12]. Within the 67-gene signature, 5 genes, TWIST1, PITX2, S100A3, PDGFRL, and DUSP9, were highly expressed in breast cancer cell lines and had no detectable expression in normal human BM by qRT–PCR (Supplemental Table 2). Expression of these 5 genes as well as Keratin 19 (KRT19) and 5 other genes from the 67-gene signature (HSP27, IGF, PIR, SlAC2, SNAIL1) were examined in 50 BM specimens collected from 30 patients with clinical stage II/III breast cancer prior to any treatment. 20 patients had BM collected from both the right and left iliac crests which were analyzed individually and 10 patients had the right and left BM pooled prior to analysis (Table 1). Thirteen (43 %) of these patients developed recurrent disease within 16 months of diagnosis. All sites of visceral metastases were visceral except for one patient who developed contralateral axillary metastases. Approximately half of the patients (47 %), in the metastatic group and non-metastatic group, had triple negative cancers (Table 2). Using a threshold of fivefold higher than pooled BM controls, TWIST1 was elevated in 3 BM specimens from 2 patients, both of whom developed distant recurrent breast cancer and PITX2 was elevated in 7 BM specimens from 5 patients, 4 of whom developed distant recurrent breast cancer within 16 months of diagnosis. TWIST1 and PITX2 expression in patient BM were found to correlate significantly with the development of early recurrent disease (p = 0.005 and 0.0062, respectively) (Table 3).
Table 1.
Relative expression levels of TWIST1, PITX2, and KRT19 in BM specimens from clinical stage II/III breast cancer patient bone marrows collected prior to therapy
| Patient # | Site | Histology | ER | PR | Her-2 | Recurrence site | *Twist-1 | *PITX2 | *KRT19 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | R | IDC | neg | neg | pos | NT | 1.55 | 0.42 | |
| L | IDC | neg | neg | pos | 0.85 | 1.24 | 1.35 | ||
| 2 | L | IDC | neg | neg | neg | Lung | 0.62 | 0.30 | 0.71 |
| R | IDC | neg | neg | neg | Lung | 0.95 | 0.51 | 0.33 | |
| 3 | R | IDC | pos | neg | pos | 0.92 | 0.53 | 7.47 | |
| L | IDC | pos | neg | pos | 0.52 | 0.73 | 5.59 | ||
| 4 | L | IDC | neg | pos | neg | Lung | 3.08 | 0.94 | 2.65 |
| R | IDC | neg | pos | neg | Lung | 0.23 | 3.35 | 0.34 | |
| 5 | L | IDC | neg | neg | neg | Liver | 9.05 | 42.19 | 0.57 |
| R | IDC | neg | neg | neg | Liver | 27.86 | 91.71 | 1.75 | |
| 6 | X | IDC | neg | neg | neg | 0.93 | 1.52 | 0.23 | |
| X | IDC | neg | neg | neg | 0.47 | 1.65 | 3.66 | ||
| 7 | R | IDC | neg | neg | neg | Liver | ND | 1.25 | 0.53 |
| L | IDC | neg | neg | neg | Liver | 0.41 | 0.89 | 3.98 | |
| 8 | R | IDC | pos | pos | neg | ND | 3.47 | 0.76 | |
| 9 | R | IDC | pos | neg | neg | Liver, bone | 1.16 | 13.99 | 7.85 |
| L | IDC | pos | neg | neg | Liver, bone | 1.91 | 23.75 | 0.23 | |
| 10 | R | IDC | neg | neg | neg | 0.67 | 0.71 | 3.05 | |
| L | IDC | neg | neg | neg | 1.33 | 5.82 | 0.56 | ||
| 11 | R | IDC | neg | neg | neg | ND | ND | 0.40 | |
| L | IDC | neg | neg | neg | 0.34 | 0.98 | 1.11 | ||
| 12 | L | IDC | neg | neg | pos | 0.47 | 1.93 | 0.93 | |
| R | IDC | neg | neg | pos | 0.51 | 0.51 | 0.19 | ||
| 13 | L | IDC | pos | pos | neg | ND | ND | ND | |
| R | IDC | pos | pos | neg | 0.79 | 2.23 | 0.60 | ||
| 14 | L | IDC | neg | neg | pos | Brain | 0.68 | 2.28 | 0.68 |
| R | IDC | neg | neg | pos | Brain | 0.61 | 16.71 | 0.47 | |
| 15 | L | IDC | neg | neg | neg | 0.83 | 1.42 | 5.82 | |
| R | IDC | neg | neg | neg | 0.56 | 1.40 | 0.12 | ||
| 16 | L | ILC | neg | neg | neg | Liver, pancreas | 0.42 | 6.24 | 3.28 |
| 17 | X | IDC | pos | pos | neg | 0.96 | 0.41 | 1.09 | |
| 18 | R | IDC | pos | neg | neg | Liver, lung, bone | 0.37 | 0.24 | 0.17 |
| L | IDC | pos | neg | neg | Liver, lung, bone | 1.49 | 0.30 | 3.25 | |
| 19 | X | IMC | neg | neg | neg | 0.32 | 0.42 | 0.87 | |
| 20 | L | ILC | neg | neg | neg | Liver, bone | 0.78 | 0.68 | 0.10 |
| R | ILC | neg | neg | neg | Liver, bone | 1.20 | 0.91 | 0.44 | |
| 21 | R | IDC | neg | neg | pos | Contralateral axilla | ND | 0.75 | 0.55 |
| L | IDC | neg | neg | pos | Contralateral axilla | 0.32 | 0.36 | 0.54 | |
| 22 | L | IDC | neg | neg | neg | 0.91 | 0.69 | 0.17 | |
| R | IDC | neg | neg | neg | 0.62 | 1.77 | 0.35 | ||
| 23 | L | IMC | pos | pos | neg | Brain, liver | 1.16 | 1.70 | 0.64 |
| R | IMC | pos | pos | neg | Brain, liver | ND | 2.08 | 0.12 | |
| 24 | X | ILC | pos | neg | neg | 1.10 | 1.16 | 0.96 | |
| 25 | R | ILC | neg | pos | pos | Liver | 1.30 | 0.92 | 1.12 |
| 26 | R | IDC | neg | pos | neg | ND | 2.55 | 0.52 | |
| 27 | X | IDC | pos | pos | neg | 0.21 | 3.13 | 0.33 | |
| 28 | L | IMC | pos | pos | neg | 0.19 | 0.53 | 3.39 | |
| R | IMC | pos | pos | neg | 0.47 | 3.60 | 2.01 | ||
| 29 | R | IDC | neg | neg | pos | Liver, lung | 1.63 | 0.56 | 6.27 |
| 30 | X | IDC | neg | neg | neg | 0.34 | 0.90 | 3.29 |
Site- site of BM collection, right or left iliac crest, Histology- IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; IMC, invasive mammary carcinoma. PCR values are expressed as fold change in gene expression relative to normal human BM
Table 2.
Tumor biomarkers of the patient specimens analyzed
| No mets | Mets | |
|---|---|---|
| Number (%) | Number (%) | |
| Total number | 17 | 13 |
| ER+ | 6 (35) | 3 (23) |
| Her2+ | 2 (12) | 4 (30) |
| ER+/Her+ | 1 (5) | 0 (0) |
| TN | 8 (47) | 6 (46) |
Table 3.
Estimate of the effect of fold change in gene expression on time to recurrence in the 30 patients
| Gene tested | p value | Hazard ratio | Lower limit of 95 % confidence interval for hazard ratio | Upper limit of 95 % confidence interval for hazard ratio |
|---|---|---|---|---|
| PITX2 | 0.0062 | 1.09 | 1.02 | 1.16 |
| TWIST1 | 0.005 | 1.71 | 1.18 | 2.48 |
| HSPB27 | 0.0139 | 6.66 | 1.47 | 30.18 |
| DUSP9 | 0.6 | 0.76 | 0.27 | 2.13 |
| ID1 | 0.99 | 0.99 | 0.85 | 1.17 |
| IGF | 0.61 | 1.11 | 0.73 | 1.69 |
| KRT19 | 0.79 | 1.04 | 0.78 | 1.38 |
| PDGFRL | 0.27 | 1.13 | 0.91 | 1.4 |
| PIR | 0.23 | 0.71 | 0.4 | 1.24 |
| S100A3 | 0.74 | 0.9 | 0.5 | 1.63 |
| SLAC2 | 0.44 | 0.85 | 0.58 | 1.27 |
| SNAIL1 | 0.2 | 0.44 | 0.13 | 1.53 |
All estimates are derived from single-variable models; in other words, fold change in only 1 gene has been included in each model. PITX2, TWIST1, and HSPB27 are statistically significantly related to time to recurrence
PITX2 is expressed in breast cancer cell lines with invasive phenotype
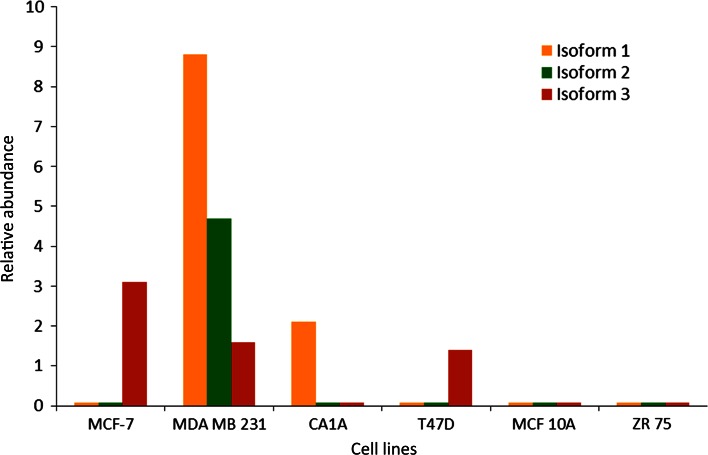
Since PITX2 was not expressed in the normal BM and its expression in BM from stage II/III breast cancer patients significantly correlated with early recurrent disease development, we sought to determine the expression levels and physiological function in established breast cancer cell lines. PITX2 has 3 isoforms (Fig. 1). The expression of each isoform in six breast cancer cell lines with varying metastatic potential was determined by qRT–PCR. The highly metastatic breast cancer cell line MDAMB231 expressed all three isoforms of PITX2 at different relative levels. Isoform 1 was fourfold higher in MDAMB 231 than in the next highest PITX2 expressing cell line, CA1A. In this cell line, which is also can form metastases [21] [12], only isoform 1 was detected. The expression of all isoforms of PITX2 in MCF10A and ZR75, both of which are non-invasive, was very low to undetectable. MCF-7 and T47D, both of which are non-metastatic, expressed isoform 3. The two invasive cells lines tested expressed high levels of isoform 1, which has been reported to be important in the Wnt/beta-catenin pathway while the non-invasive cell lines expressed either no/low levels of PITX2 or isoform 3, which is in the TGF-beta pathway. The relative level of expression of the 3 isoforms of PITX2 in cell lines is shown in Fig. 2.
Fig. 1.
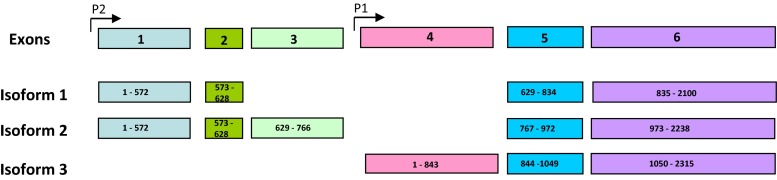
Diagram showing the alternate splice variants of PITX2. The general exon arrangement and promotor location is shown in the top and exon structure of each isoforms are shown subsequently
Fig. 2.
Relative expression pattern of PITX2 isoforms in breast cancer cell lines by qRT–PCR. Expression of each isoform relative to four normal human BM samples
Knockdown of PITX2 reduces invasiveness in MDAMB231 cells
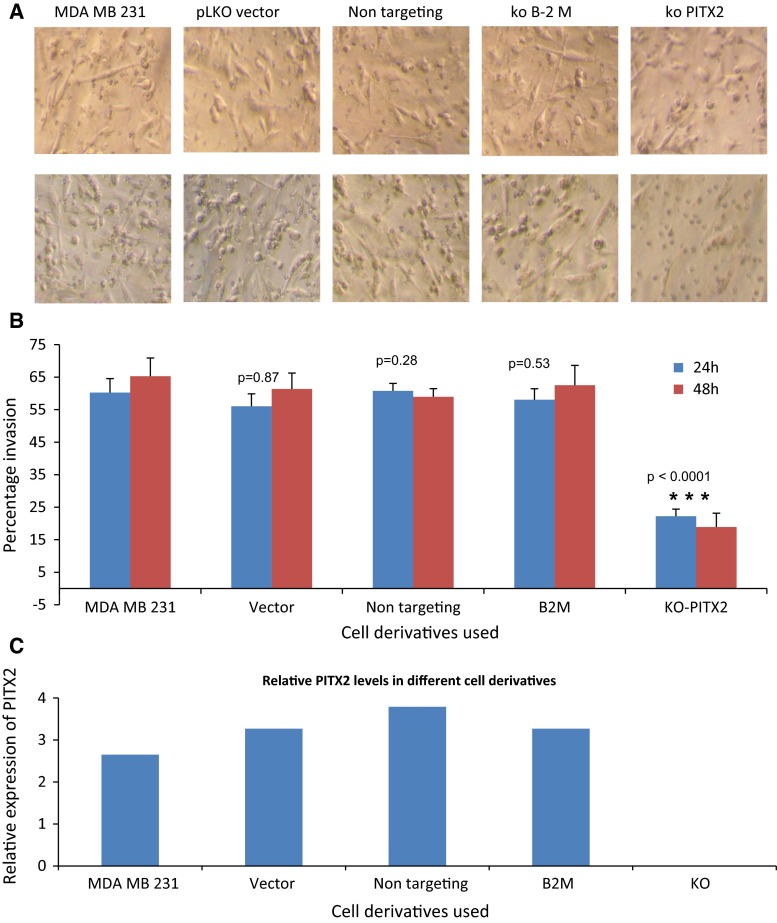
To determine whether PITX2 plays a role in cell invasiveness, we performed gene knockdown experiments using the MDAMB231 cell line. MDAMB231 is highly invasive and has relatively high expression of all three of PITX2 isoforms. Using a lentivirus shRNA system, PITX2 expression was reduced to near undetectable level as determined by qRT–PCR. Clonal cell lines with the highest level of knockdown were selected for invasion assays. MDAMB231 cells stably transduced with empty vector, a non-targeting sequence, or shRNA targeting an unrelated gene, beta-2 microglobulin, were included as controls. The relative expression of PITX2 in each cell type used is shown in Fig. 3c. The knockdown cells (KO) had undetectable expression of PITX2, while the control cell lines expressed similar levels of PITX2. The percentage of cells which migrated into the lower chamber of the invasion cassette is shown in Fig. 3b. Cells with reduced PITX2 expression showed a significant reduction in invasion compared to all control cells, both at 24 and 48 h after plating (Fig. 3a, b). There was a 63.8 % reduction in invasion in PITX2 deficient cells at 24 h and 72.6 % reduction at 48 h compared to the parental cells (p < 0.0001). These data suggest that loss of PITX2 expression attenuates the invasive phenotype of MDAMB231 cells.
Fig. 3.
Matrigel invasion assays showing reduced invasion in MDAMB231 cells with PITX2 knockdown. a Top panel shows the cells in the matrigel invasion chamber prior to the clearing of the top chamber. Cells present both at the top chamber and invaded cells in the bottom chamber are visible. Bottom panel shows cells after clearing the top chamber; thus, only those cells invaded to bottom chamber are visible. Different derivatives of cells used in the experiment are indicated. Statistical significance by paired t test in comparison with non-transduced MDAMB231 cells are shown. (MDAMB231- non-transduced parental cells, pLKO vector—transduced with empty vector, Non-targeting—transduced with a non-targeting sequence, B2M—transduced with shRNA of an unrelated gene beta-2 microglobulin, ko- PITX2—transduced with shRNA of PITX2.) b Percentage of cells present in bottom chamber at 24 and 48 h. c Expression levels of PITX2 in the cells used for experiment as determined by qRT–PCR
PITX2 mediates the expression of genes associated with aggressive tumors
PITX2 isoform 1 is a component of the Wnt/beta-Catenin pathway and is a downstream target of LEF1 [13]. The Wnt/beta-Catenin pathway is known to contribute to tumor invasion and metastasis [22, 23]. We hypothesized that loss of the invasive phenotype associated with downregulation of PITX2 in MDAMB231 cells was mediated through the Wnt pathway. To test the effect of PITX2 knockdown on the Wnt pathway signaling program, we analyzed the gene expression pattern of genes in the Wnt pathway as well as other pathways, by qRT–PCR using modified Wnt pathway arrays. Four sets of independent PITX2 knockdown cells and four sets of mock transfected cells were used for analysis. The expression of 96 genes was analyzed which represented the Wnt/beta-catenin pathway, EMT, and TGF-beta pathways (Supplemental Table 3). The statistical significance in the expression of each gene in the two experimental groups was determined. Of the 96 genes examined, only the expression of three genes, NKD1, LEF1, and DKK4, was significantly reduced as a result of PITX2 knockdown (Table 4). All three of these genes have been reported to contribute to an aggressive tumor phenotype and metastases development and are in the Wnt/beta-Catenin pathway. Our results suggest that PITX2 plays a role in mediating invasiveness in cancer cells through the Wnt/beta-Catenin pathway and not the TGF-B or through EMT.
Table 4.
Relative expression of genes associated with Wnt pathway
| Gene | Fold difference between control cells and KO PITX2 cells | p value |
|---|---|---|
| NKD1 | 29.5 | 0.009 |
| LEF1 | 2.05 | 0.04 |
| DKK4 | 4.4 | 0.04 |
The expression levels of various genes involved in Wnt pathway in PITX2 depleted and control MBAMB231 cells were determined by qRT-PCR. Four independent sets of PITX2 depleted clones and mock transduced cells were used in the analysis. The values were normalized to GAPDH and the fold difference calculated by ddCt method. The statistical significance in PITX2 expression was calculated by t Test
Discussion
Identification of the molecular mechanisms behind the metastatic process is essential for elimination of minimal residual disease and preventing cancer recurrence. The presence of DTCs in the BM of breast cancer patients before and after chemotherapy has been reported as an independent predictor of prognosis [9, 11]. However, the rarity and biochemical heterogeneity of DTCs have hindered studies focused on defining their molecular features and therapeutic vulnerabilities. Previously, we analyzed the BM of breast cancer patients and identified a molecular signature associated with early disease relapse. In this manuscript, we demonstrate that one of the genes in this signature, PITX2, identifies patients with who are at risk for developing early recurrent disease and which likely plays a role in the invasiveness of a breast cancer cells through the Wnt/beta-Catenin pathway.
We employed gene expression analysis of BMs collected from breast cancer patients to determine the specific molecular features of DTCs. Those genes which were expressed in the BM of patients but had no expression in the BM from healthy volunteers were considered to be derived from cancer cells lodged in the BM. Since DTCs are known to be heterogeneous, it is likely that only a subpopulation of the detectable DTCs have the potential to progress to a metastatic focus. Therefore, we correlated the expression of DTC-associated genes to metastatic outcome and time to recurrence. Of the genes examined, expressions of TWIST1 and PITX2 in the BM of breast cancer patients were found to be significantly associated with metastatic disease development in this analysis. The number of patients with detectable TWIST1 or PITX2 in their BM was 14 % (7 of 30 patients) which identified 46 % (6 of 13 patients) of the patients who subsequently developed metastatic disease.
The role of TWIST1 is well documented in the EMT process and metastasis of cancers [24] and we have previously reported on its significance in the BM of breast cancer patients [12]. The role of PITX2 in tumorigenesis in general and its association with metastatic process in particular are not as well understood. PITX2 is a pituitary homeobox transcription factor responsible for embryonic development of organs and morphogenesis. One of the transcript variants, isoform 3 is responsible left right symmetry during development. There are several recent reports on the association of PITX2 expression in various neoplasms. Upregulation of PITX2 has been reported in pituitary adenomas, colorectal cancers, and Wilms tumors [25–27]. A recent study demonstrated that PITX2 expression activates tumor progression and invasion in ovarian cancer, possibly independent of Wnt/beta-Catenin pathway [28]. In contrast, promoter methylation and subsequent silencing has been associated with certain prostate [29] and breast [19, 30] cancers specifically estrogen receptor-positive cancers. These data suggest that the mechanism of PITX2 in tumor progression may vary depending on the tumor type.
Our results indicate that PITX2 could play a direct role in the metastatic phenotype of breast cancer. Presumptive expression by DTCs is significantly correlated with early disease recurrence and experimental evidence that gene suppression reduces the invasive phenotype in MDAMB231 cells reinforces this notion. Interestingly, we were unable to develop stable long-term knockdowns of PITX2 in MDAMB231 cells and thus were unable to perform animal experiments. This may indicate an essential role of PITX2 in the stability of the cancer cells.
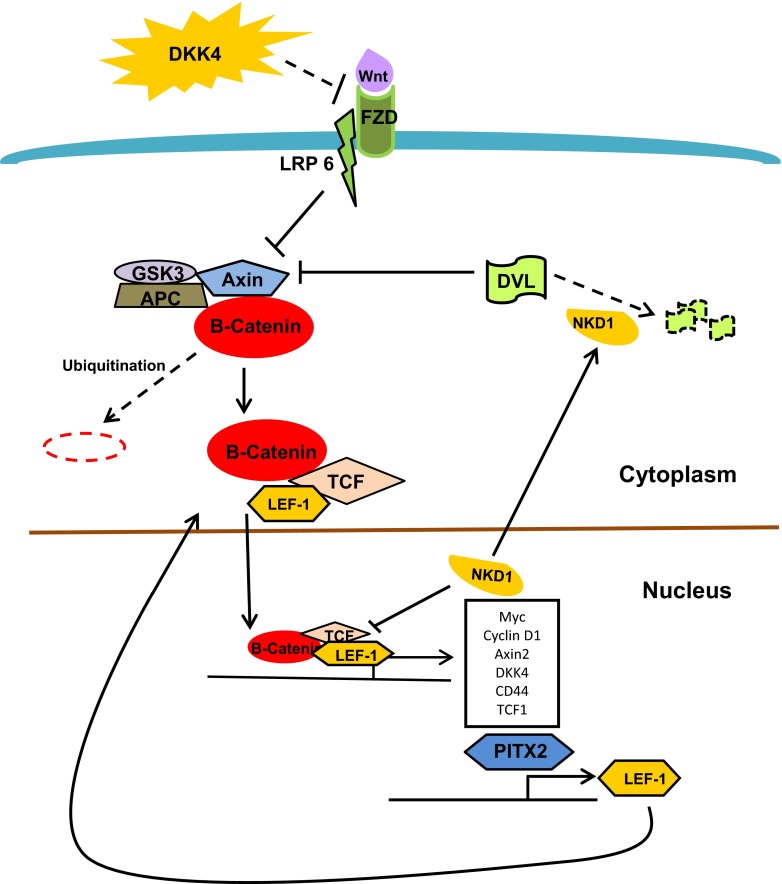
In our system, PITX2 appears to mediate its effects through the Wnt/beta-Catenin pathway rather than other pathways associated with PITX2 such as the TGF-beta pathway. Three genes, NKD1, LEF1, and DKK4 significantly downregulated by PITX2 knockdown, are known to contribute to tumorigenesis and metastasis through activation of Wnt pathway (Fig. 4). Activation of the Wnt/beta-Catenin results in high levels of NKD1 [31]. It has been reported that loss of function mutation in NKD1 can alter Wnt signaling and contribute to enhanced tumorigenesis [32]. However, the full pathway between Wnt/beta-Catenin and NKD1 activation has not yet been elucidated. High levels of DKK4 have been reported to increase migration and invasion properties in colon cancer cells and its downregulation may have anti-cancer action [33]. DKK4 is downstream target of TCF/beta Catenin, and its high expression indicates activated state of Wnt signaling pathway [34]. It is known that PITX2 transactivates the LEF1 promoter and also enhances the endogenous expression of the full length beta-Catenin-dependent isoform. LEF1 is a downstream component of Wnt signaling and is known to contribute to invasion in breast cancer cells [35]. The Wnt signaling pathway mediated through the TCF/LEF1 transcriptional activator complex is activated in lung adenocarcinoma metastases [22]. Thus, PITX2 likely contributes to cell division, proliferation, and migration through mediating differential LEF1 isoform expression and subsequent interactions with LEF1 and beta-Catenin [36].
Fig. 4.
Canonical Wnt pathway illustrating the roles of PITX2 dependant genes. Solid lines indicate activated state and discontinuous lines indicated inhibited state of the pathway. Genes highlighted in yellow shapes showed significant reduction upon PITX2 downregulation. When activated by the Wnt ligands, the Frizzled/LRP1/6 receptor complex inhibits the GSK3/Axin/APC complex and stabilizes beta-Catenin. Subsequently beta-Catenin facilitates the nuclear translocation of TCF/LEF complex resulting in the transactivation of target genes including PITX2, NKD1, DKK4, and several other genes contributing to invasion and metastasis. NKD1 antagonizes the Wnt pathway as transcriptional repressor and also by mediating the degradation of DVL1
As illustrated in Fig. 4, all three genes, NKD1, LEF1, and DKK4, found to be downregulated with PITX2 knockdown are consistent with activation of the Wnt signaling pathway by PITX2. Although DKK4 and NKD1 are Wnt pathway antagonists [37, 38], their elevated levels are indicative of activated Wnt/Beta-catenin signaling as both of them are transcriptionally activated by the TCF/LEF complex. Since LEF1 is a downstream target of PITX2 transactivation, reduced levels of NKD1 and DKK4 are expected as consequence of PITX2 downregulation. Even though the expression of two Wnt pathway antagonists are diminished as a result of PITX2 knockdown, the Wnt pathway remains abrogated primarily due to the reduced transcription of LEF1 which is the major transcriptional co-activator effecting the pathway [39].
Our results suggest that PITX2 expression in the BM of breast cancer patients is indicative of the presence of DTCs and is a predictor of high risk for metastatic disease development. PITX2 contributes to the invasiveness of breast cancer cells, and this appears to be mediated through the Wnt/beta-Catenin pathway. It is possible that therapeutically targeting PITX2 or its downstream activators could dismantle the invasive process and would be a step towards eliminating minimal residual disease and micrometastases in high risk women.
Electronic supplementary material
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Weiss L. Metastatic inefficiency. Adv Cancer Res. 1990;54:159–211. doi: 10.1016/S0065-230X(08)60811-8. [DOI] [PubMed] [Google Scholar]
- 2.Wong CW, Lee A, Shientag L, Yu J, Dong Y, Kao G, Al-Mehdi AB, Bernhard EJ, Muschel RJ. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61(1):333–338. [PubMed] [Google Scholar]
- 3.Chambers AF, Naumov GN, Varghese HJ, Nadkarni KV, MacDonald IC, Groom AC. Critical steps in hematogenous metastasis: an overview. Surg Oncol Clin N Am. 2001;10(2):243–255. [PubMed] [Google Scholar]
- 4.Fidler IJ. Critical determinants of cancer metastasis: rationale for therapy. Cancer Chemother Pharmacol. 1999;43(Suppl):S3–S10. doi: 10.1007/s002800051091. [DOI] [PubMed] [Google Scholar]
- 5.Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3(2):65–71. [PubMed] [Google Scholar]
- 6.Woodhouse EC, Chuaqui RF, Liotta LA. General mechanisms of metastasis. Cancer. 1997;80(8 Suppl):1529–1537. doi: 10.1002/(SICI)1097-0142(19971015)80:8+<1529::AID-CNCR2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 7.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 8.Pantel K, Woelfle U. Detection and molecular characterisation of disseminated tumour cells: implications for anti-cancer therapy. Biochim Biophys Acta. 2005;1756(1):53–64. doi: 10.1016/j.bbcan.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, Pierga JY, Marth C, Oruzio D, Wiedswang G, Solomayer EF, Kundt G, Strobl B, Fehm T, Wong GY, Bliss J, Vincent-Salomon A, Pantel K. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 10.Janni W, Hepp F, Rjosk D, Kentenich C, Strobl B, Schindlbeck C, Hantschmann P, Sommer H, Pantel K, Braun S. The fate and prognostic value of occult metastatic cells in the bone marrow of patients with breast carcinoma between primary treatment and recurrence. Cancer. 2001;92(1):46–53. doi: 10.1002/1097-0142(20010701)92:1<46::AID-CNCR1290>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Braun S, Kentenich C, Janni W, Hepp F, de Waal J, Willgeroth F, Sommer H, Pantel K. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol. 2000;18(1):80–86. doi: 10.1200/JCO.2000.18.1.80. [DOI] [PubMed] [Google Scholar]
- 12.Watson MA, Ylagan LR, Trinkaus KM, Gillanders WE, Naughton MJ, Weilbaecher KN, Fleming TP, Aft RL. Isolation and molecular profiling of bone marrow micrometastases identifies TWIST1 as a marker of early tumor relapse in breast cancer patients. Clin Cancer Res. 2007;13(17):5001–5009. doi: 10.1158/1078-0432.CCR-07-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/beta-Catenin→ Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111(5):673–685. doi: 10.1016/S0092-8674(02)01084-X. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, Ohuchi H, Suehiro A, Motegi Y, Nakahara Y, Kondo S, Yokoyama M. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126(24):5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 15.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94(3):307–317. doi: 10.1016/S0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 16.Arakawa H, Nakamura T, Zhadanov AB, Fidanza V, Yano T, Bullrich F, Shimizu M, Blechman J, Mazo A, Canaani E, Croce CM. Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc Natl Acad Sci U S A. 1998;95(8):4573–4578. doi: 10.1073/pnas.95.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14(4):392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 18.Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol Cell. 2001;7(1):137–149. doi: 10.1016/S1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- 19.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, Koenig T, Hartmann O, Kluth A, Dietrich D, Magdolen V, Portengen H, Look MP, Klijn JG, Lesche R, Schmitt M, Maier S, Foekens JA, Martens JW. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res Treat. 2008;111(3):429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 20.Schweickert A, Campione M, Steinbeisser H, Blum M. Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mech Dev. 2000;90(1):41–51. doi: 10.1016/S0925-4773(99)00227-0. [DOI] [PubMed] [Google Scholar]
- 21.Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE. Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Res. 2009;69(3):735–740. doi: 10.1158/0008-5472.CAN-08-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138(1):51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Previdi S, Maroni P, Matteucci E, Broggini M, Bendinelli P, Desiderio MA. Interaction between human-breast cancer metastasis and bone microenvironment through activated hepatocyte growth factor/Met and beta-catenin/Wnt pathways. Eur J Cancer. 2010;46(9):1679–1691. doi: 10.1016/j.ejca.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 24.Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S, Maestro R, Voeltzel T, Selmi A, Valsesia-Wittmann S, Caron de Fromentel C, Puisieux A. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65(22):10214–10222. doi: 10.1158/0008-5472.CAN-05-0884. [DOI] [PubMed] [Google Scholar]
- 26.Meeh PF, Farrell CL, Croshaw R, Crimm H, Miller SK, Oroian D, Kowli S, Zhu J, Carver W, Wu W, Pena E, Buckhaults PJ. A gene expression classifier of node-positive colorectal cancer. Neoplasia. 2009;11(10):1074–1083. doi: 10.1593/neo.09808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zirn B, Samans B, Wittmann S, Pietsch T, Leuschner I, Graf N, Gessler M. Target genes of the WNT/beta-catenin pathway in Wilms tumors. Genes Chromosom Cancer. 2006;45(6):565–574. doi: 10.1002/gcc.20319. [DOI] [PubMed] [Google Scholar]
- 28.Fung FK, Chan DW, Liu VW, Leung TH, Cheung AN, Ngan HY. Increased expression of PITX2 transcription factor contributes to ovarian cancer progression. PLoS One. 2012;7(5):e37076. doi: 10.1371/journal.pone.0037076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, Haefliger C, Lesche R, Hartmann A, Corman J, Wheeler T. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181(4):1678–1685. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- 30.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen C, Mueller V, Nahrig J, Schittulli F, Kates R, Lesche R, Schwope I, Kluth A, Marx A, Martens JW, Foekens JA, Schmitt M, Harbeck N. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients–Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43(11):1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 31.Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, Garcia PD, Randazzo FM, Escobedo J, Fantl WJ, Williams LT. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98(26):14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, Cagatay T, Zhou G, Chan CC, Blythe S, Suyama K, Zheng L, Pan K, Qian C, Hamelin R, Thibodeau SN, Klein PS, Wharton KA, Liu W. Mutations in the human naked cuticle homolog NKD1 found in colorectal cancer alter Wnt/Dvl/beta-catenin signaling. PLoS One. 2009;4(11):e7982. doi: 10.1371/journal.pone.0007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pendas-Franco N, Garcia JM, Pena C, Valle N, Palmer HG, Heinaniemi M, Carlberg C, Jimenez B, Bonilla F, Munoz A, Gonzalez-Sancho JM. DICKKOPF-4 is induced by TCF/beta-catenin and upregulated in human colon cancer, promotes tumour cell invasion and angiogenesis and is repressed by 1alpha,25-dihydroxyvitamin D3. Oncogene. 2008;27(32):4467–4477. doi: 10.1038/onc.2008.88. [DOI] [PubMed] [Google Scholar]
- 34.Matsui A, Yamaguchi T, Maekawa S, Miyazaki C, Takano S, Uetake T, Inoue T, Otaka M, Otsuka H, Sato T, Yamashita A, Takahashi Y, Enomoto N. DICKKOPF-4 and -2 genes are upregulated in human colorectal cancer. Cancer Sci. 2009;100(10):1923–1930. doi: 10.1111/j.1349-7006.2009.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen A, Rosner A, Milovanovic T, Hope C, Planutis K, Saha B, Chaiwun B, Lin F, Imam SA, Marsh JL, Holcombe RF. Wnt pathway component LEF1 mediates tumor cell invasion and is expressed in human and murine breast cancers lacking ErbB2 (her-2/neu) overexpression. Int J Oncol. 2005;27(4):949–956. [PubMed] [Google Scholar]
- 36.Amen M, Liu X, Vadlamudi U, Elizondo G, Diamond E, Engelhardt JF, Amendt BA. PITX2 and beta-catenin interactions regulate Lef-1 isoform expression. Mol Cell Biol. 2007;27(21):7560–7573. doi: 10.1128/MCB.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CA, Christiano AM. The Wnt inhibitor, Dickkopf 4, is induced by canonical Wnt signaling during ectodermal appendage morphogenesis. Dev Biol. 2007;305(2):498–507. doi: 10.1016/j.ydbio.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 38.Angonin D, Van Raay TJ. Nkd1 functions as a passive antagonist of Wnt signaling. PLoS One. 2013;8(8):e74666. doi: 10.1371/journal.pone.0074666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295(1):219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.