Abstract
Thymus is involved in induction of self-tolerance in T lymphocytes, particularly due to Aire activity. In peripheral tissues, Treg cells and immunomodulatory molecules, like the major histocompatibility complex (MHC) class Ib molecules, are essential for maintenance of autotolerance during immune responses. Viral infections can trigger autoimmunity and modify thymic function, and YFV17D immunization has been associated with the onset of autoimmunity, being contraindicated in patients with thymic disorders. Aiming to study the influence of YFV17D immunization on the transcriptional profiles of immunomodulatory genes in thymus, we evaluated the gene expression of AIRE, FOXP3, H2-Q7 (Qa-2/HLA-G), H2-T23 (Qa-1/HLA-E), H2-Q10, and H2-K1 following immunization with 10,000 LD50 of YFV17D in C57BL/6 and BALB/c mice. The YFV17D virus replicated in thymus and induced the expression of H2-Q7 (Qa-2/HLA-G) and H2-T23 (Qa-1/HLA-E) transcripts and repressed the expression of AIRE and FOXP3. Transcriptional expression varied according to tissue and mouse strain analyzed. Expression of H2-T23 (Qa-1/HLA-E) and FOXP3 was induced in thymus and liver of C57BL/6 mice, which exhibited defective control of viral load, suggesting a higher susceptibility to YFV17D infection. Since the immunization with YFV17D modulated thymus gene expression in genetically predisposed individuals, the vaccine may be related to the onset of autoimmunity disorders.
1. Introduction
Autoimmune disorders have a complex and multifactorial etiopathogenesis, arising from an imbalance between the mechanisms involved in maintenance of immune tolerance and those related to the generation of an effective immune response [1]. The thymus is the primary lymphoid organ related to establishment of central tolerance and related to functional and phenotypical maturation of thymocytes [2]. The main events in central tolerance of thymocytes are the positive and negative selections [2, 3], characterized, respectively, by the maintenance of cortical double-positive thymocytes that recognize with low affinity/avidity self-antigens presented by self-histocompatibility molecules and by the apoptotic clonal deletion of medullary thymocytes that recognize self-antigens with high affinity/avidity [2–5].
The major regulator of negative selection is the AIRE (autoimmune regulator) gene [2, 6], which imposes the expression of clusters of hundreds of tissue-related (TRAs) genes from peripheral tissues inside the thymic medulla, by the phenomenon known as Promiscuous Gene Expression (PGE) [6–8]. Therefore, PGE represents a remarkable feature to ensure the exposure of the majority of tissue antigens from the body in thymic medulla to prevent autoimmunity [2, 6]. During negative selection, subpopulations of thymocytes can differentiate in natural regulatory T (nTregs) cells by the expression of FOXP3 during clonal deviation [9, 10]. Both Aire and FOXP3 are transcription factors essential to prevent autoimmunity and to maintain immunological homeostasis [2, 11, 12]. Single nucleotide variations in these genes are related to severe systemic autoimmune syndromes, such as APECED (autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy) [13] and IPEX (immunodysregulation polyendocrinopathy enteropathy X-linked syndrome) [12, 14].
Variations in expression of AIRE and FOXP3 may produce changes in thymic function and therefore may influence the systemic immune responses [2, 15, 16]. The thymus is a common target organ during cancer [17, 18] and infectious diseases [19–21], undergoing relevant changes in function and morphology. Viral infections can invade and spread into thymus influencing positive and negative selection, facilitating the appearance of potentially autoreactive thymus-derived immature CD4+CD8+ T cells [21]. In genetically predisposed individuals, viruses can also trigger events that may promote the onset of autoimmune disorders with an overstimulation of the immune system, which may allow molecular mimicry between pathogen associated-antigens and host antigens [22].
The Yellow Fever Virus (YFV) is a member of the Flaviviridae family, exhibiting a positive-sense single-stranded RNA, which infects humans and other vertebrates [23]. This virus is mainly transmitted by bites of female mosquitoes (mostly from genus Aedes), occurring in endemic areas of tropical regions [24], causing a form of hemorrhagic fever characterized by headache, high fever, nausea, myalgia, jaundice, cardiovascular complications, and multiple organ failure in severe cases [23, 25, 26]. There are no antiviral therapies against YFV; however, since 1945, a live virus attenuated vaccine (YFV17D) is available in two substrain forms: a 17DD and 17D204 [23, 27].
YFV17D immunization is considered to be a safe and effective method for the protection against Yellow Fever Virus infection [28]. However, severe adverse effects related to YFV17D immunization as the Yellow Fever Virus Associated Viscerotropic Disease (YEL–AVD) and the Yellow Fever Virus Associated Neurotropic Disease (YEL–AND) have been described [23]. Interestingly, YEL-AVD cases have been associated with deaths in patients with thymic dysfunction caused by thymomas, thymectomy, myasthenia gravis, or DiGeorge syndrome [29]. Thymic disorders are one of the most relevant risk factors associated with deaths after YFV17D immunization, highlighting the involvement of thymus-derived cells in controlling the viral immune responses after immunization [29, 30].
The thymus is considered to be a primary site for expression of immunomodulatory molecules such as Aire and FOXP3 [2, 11]. Interestingly, the constitutive expression of nonclassical class I major histocompatibility complex (MHC Ib) molecules has been reported in several subpopulations of thymic cells [31–33]. Human MHC Ib molecules, such as HLA-G and HLA-E, are associated with the regulation of immune responses in the periphery by interacting with inhibitory receptors [34, 35]. These molecules exhibit restricted peripheral tissue distribution; they are not involved in antigen presentation and do not show high rate of polymorphisms [34, 35].
In mice, two molecules have been described to be functional homologues of HLA-G and HLA-E, that is, Qa-2 and Qa-1, respectively [31, 36]. Similarly to humans, Qa-2 and Qa-1 molecules are involved in the regulation of immune responses, by inhibiting maternal NK cell-mediated lysis [35–37], suppression of CD4+ T cell and NK cell responses through a preferential interaction of Qa-1 with inhibitory CD94/NKG2A receptors [38, 39], and induction of CD8+ regulatory T cells restricted to antigens presented by Qa-1 molecules [38, 39]. Qa-2 and Qa-1 are encoded by H2-Q7/H2-Q9 and H2-T23 genes, respectively, both located at the histocompatibility complex-2 in chromosome 17 [40, 41].
Due to the immunomodulatory properties of MHC Ib molecules and due to the relevance of Aire and FOXP3 in self-tolerance, it is hypothesized that changes in expression of these molecules in thymus during antiviral responses could be related to (i) alterations in thymic function, resulting in a defective PGE and negative selection, (ii) modifications in the regulatory T cell repertoire, and (iii) appearance of autoimmune disorders [7, 16, 19]. In addition, it has been reported that live virus attenuated vaccines are associated with autoimmune diseases [42–44]. Particularly, immunization with YFV17D has been related to several cases of longitudinal myelitis and Kawasaki disease [45, 46].
Based on these evidences, we hypothesized that the YFV17D immunization could promote alterations in thymic function, influencing the transcriptional profile status of immunomodulatory genes such as AIRE, FOXP3, H2-Q7 (Qa-2/HLA-G), and H2-T23 (Qa-1/HLA-E) in thymus and in relevant peripheral organs such as the spleen and liver. Also, we hypothesized that the genetic background of different murine strains (C57BL/6 and BALB/c) could contribute to differential modulation in gene expression, therefore influencing the patterns of susceptibility/resistance during immune response to YFV17D infection.
2. Material and Methods
2.1. Animals
Young adult C57BL/6 and BALB/c mice were obtained from the local animal facility and maintained in isolated cages, provided with 0.45 μm pore size air filter. Experimental procedures followed ethical guidelines under strict guidance and approval by the University of São Paulo Ethics Committee for Animal Experimental Research (Protocol number 180/2009).
2.2. Virus
The Yellow Fever Virus vaccine substrain 17DD (YFV17D) used for mouse infection was produced in the Virology Research Center, University of São Paulo, Ribeirão Preto, Brazil. Briefly, virus stocks were obtained from brains of intracerebrally infected newborn mice. Mouse brains were macerated in phosphate buffered saline (PBS) (dilution 1 : 20 wt/vol) and centrifuged at 10000 RPM for 10 min at 4°C, and supernatants were stored at −80°C.
2.3. Infection of Mice by YFV17D and Negative Controls
Fifty 8-week-old females of wild-type young adult C57BL/6 and BALB/c mice were infected intraperitoneally (IP) with 10,000 LD50 of YFV17D in a volume of 200 μL/PBS 1x pH 7.4 solution at Day 0. The animals were monitored daily and most of them developed encephalitis signs such as hind limb paralysis, tremors, muscle weakness, ruffled pile, and difficulty to feed. The animals were killed at Days 1, 5, 7, 15, and 30 after inoculation (a.i.). Negative control mouse groups (N = 50) were inoculated IP with PBS 1x pH 7.4 and killed at Days 1, 5, 7, 15, and 30 a.i. and their tissues were used for comparisons with those of YFV17D-infected animals. Tissue samples from thymus (both lobes), spleen, and liver (posterior extremity and right lobe, resp.) were surgically collected from at least 5 different animals for each Day a.i., washed in saline solution, and then processed without separation of hematopoietic cells and parenchyma.
2.4. RNA Extraction
After tissue isolation, total RNA was obtained by maceration of each tissue in TRizol reagent using a Potter homogenizer, according to the manufacturer's instructions (Invitrogen, Carlsbad, CA), and treated with DNAse (deoxyribonuclease I amplification grade, Invitrogen). RNA integrity was checked by the presence of the 28S and 18S bands in 1.5% denaturing agarose gel, and purity of RNA was accessed by UV-Vis spectrophotometer (NanoDrop 2000, Thermo Scientific, MA), regarding amount of protein and organic compounds in the samples. Only RNAs exhibiting 260/230 and 260/280 ratios of ~2.0 ratio were used for quantitative PCR experiments. Total RNA was reverse transcribed to cDNA using the High Capacity cDNA Transcription Kit (Applied Biosystems, Foster City, CA), following the manufacturer's instructions. cDNA amplification was initially carried out in a total volume of 25 μL, corresponding to 500 ηg of the initial RNA.
2.5. Detection of the Viral Load by Quantitative Real-Time PCR
The detection of the viral load and quantification was performed by quantitative real-time PCR (qPCR) method, using specific primers (forward: 5-GTGACGGCTCTGACCAT-3; reverse: 5-ATGCAGTGAGCTGAGTA-3) for the viral envelope gene (E) protein. In brief, about 150 ηm of each primer; 5 μL SYBR Green PCR Kit (Qiagen, Valencia, CA); and 3 μL of cDNA and nuclease-free water (Sigma-Aldrich, St. Louis, MO) in sufficient quantity for a final volume of 10 μL per reaction were used. The number of genome copies was obtained by absolute quantification using a serial dilution of envelope (E) protein gene, previously cloned in a plasmid (InsTAclone, Fermentas, Burlington, ON), to generate a standard curve to be used as reference values.
2.6. Analysis of Expression by Quantitative Real-Time PCR
The expression of H2-Q7 (Qa-2/HLA-G), H2-T23 (Qa-1/HLA-E), H2-Q10 (Qa10), H2-K1, AIRE, and FOXP3 genes was assessed by qPCR, using TaqMan Probe-Based Gene Expression Analysis (Applied Biosystems) in a total volume of 10 μL, containing 75 ηg total RNA, 5 μL TaqMan PCR Universal Master Mix (Applied Biosystems), and 0.5 μL TaqMan Gene Expression Assays. An ABI System Sequence Detector 7500 (Applied Biosystems) was used with the following regimen of thermal cycling: stage 1: one cycle for 2 minutes at 50°C; stage 2: one cycle for 10 minutes at 95°C; stage 3: forty cycles for 15 seconds at 95°C, followed by the last cycle for 1 minute at 60° and 25 seconds at 72°C. Gene expression was normalized relatively to the TaqMan endogenous controls (Applied Biosystems), using geometric mean glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin genes. The relative quantification of transcript levels was performed by the comparative 2−ΔΔCt method. Each sample was tested in triplicate. The TaqMan Inventoried Assays and TaqMan endogenous control references are listed as follows: AIRE: Mm00477461_m1; FOXP3: Mm00475162_m1; H2-Q7 (Qa-2/HLA-G): Mm00843895_m1; H2-T23 (Qa-1/HLA-E): Mm00439246_m1; H2-Q10 (Qa10): Mm01275264_m; H2-K1: Mm02342236_gH; GAPDH: 4352339E; ACTB: 4352341E.
Statistical analysis was performed using nonparametric tests according to the number of studied groups: (i) two groups: Mann-Whitney two-tailed; (ii) three or more groups: Kruskal-Wallis followed by Dunn's multiple comparison. Values of P < 0.05 were considered to be statistically significant. All statistical tests were performed with the GraphPad Prism software version 5.0 (GraphPad Software Inc., San Diego, CA).
3. Results and Discussion
3.1. Detection of YFV17D Viral Load in Harvested Tissues
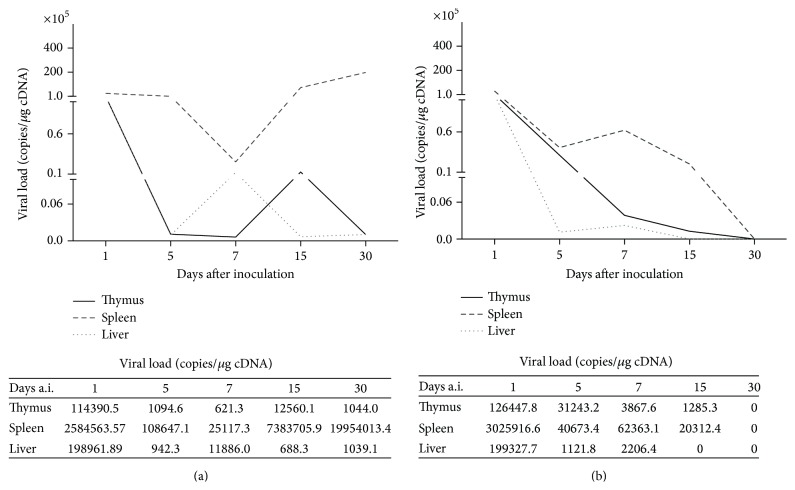
First, we evaluated if the primary and secondary immune system organs could be targeted during YFV17D immunization. Samples of thymus, liver, and spleen harvested at Days 1, 5, 7, 15, and 30 a.i. were tested by qPCR for virus detection and quantification of viral load. In both mice strains, it was observed that the YFV17D could infect the thymus and replicate in this organ until Day 15 a.i. (Figures 1(a) and 1(b)). Increased amount of viral copies in the spleen of C57BL/6 animals was observed throughout infection (Figure 1(a)). Compared with BALB/c mice, the C57BL/6 strain showed increased viral load in all tissues analyzed and was not able to eliminate the infection in spleen at Day 30 a.i. (Figure 1(a)).
Figure 1.
Detection of YFV17D viral load in harvested tissues of (a) C57BL/6 and (b) BALB/c mice. Representative of geometric mean values of viral load (copies/1 μg cDNA) measured by qPCR.
It has been reported that YFV17D infects and replicates in immunocompetent cells, such as dendritic cells (DCs), monocytes, and macrophages [23, 47]. In this study, the virus was able to infect the thymus, the liver, and the spleen. It is hypothesized that the interaction of the viral envelope (E) protein with the specific intercellular adhesion molecule-3-grabbing nonintegrin (SIGN molecules) present on the surface of DCs (CD209) and lymphocytes (CD209L) may allow virus internalization and cell infection [47]. Like other infectious agents, including HIV (Human Immunodeficiency Virus), protozoan parasites, and fungi, which have been reported to infect the thymus [19], we also reported that despite the existence of the blood-thymic barrier, the intraperitoneal inoculation of 10,000 LD50 of YFV17D in C57BL/6 and BALB/c mice infected the thymus in both strains.
Despite the absence of studies involving the characterization of immune response against YFV17D in C57BL/6 and BALB/c strains, we hypothesized that the differences in the genetic background of these two murine strains could influence the immune response pattern during viral infection. This hypothesis is supported by the fact that the signs of infection, such as hind limb paralysis, tremors, muscle weakness, ruffled pile, and difficulty to feed observed from Days 7 to 30 a.i. in both mice strains, were more pronounced in BALB/c when compared to C57BL/6 mice (data not shown). Additionally, the morphological analysis of internal organs in immunized animals revealed liver and spleen hypertrophy at Day 7 a.i in BALB/c mice compared to C57BL/6 and to control groups (see Supplemental data I in Supplementary Materials available online at http://dx.doi.org/10.1155/2015/503087). These results suggest that BALB/c mice could perform a more robust antiviral response against YFV17D that could reflect in less pronounced morbidity and in a better control of the viral load.
3.2. Influence of YFV17D Immunization in the Gene Expression of Immunomodulatory Molecules in Thymus
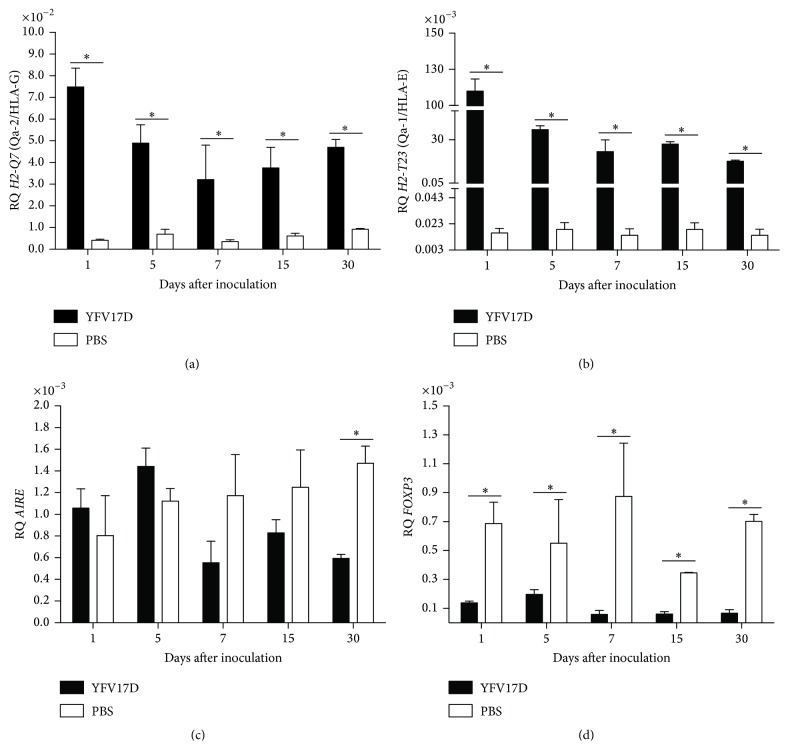
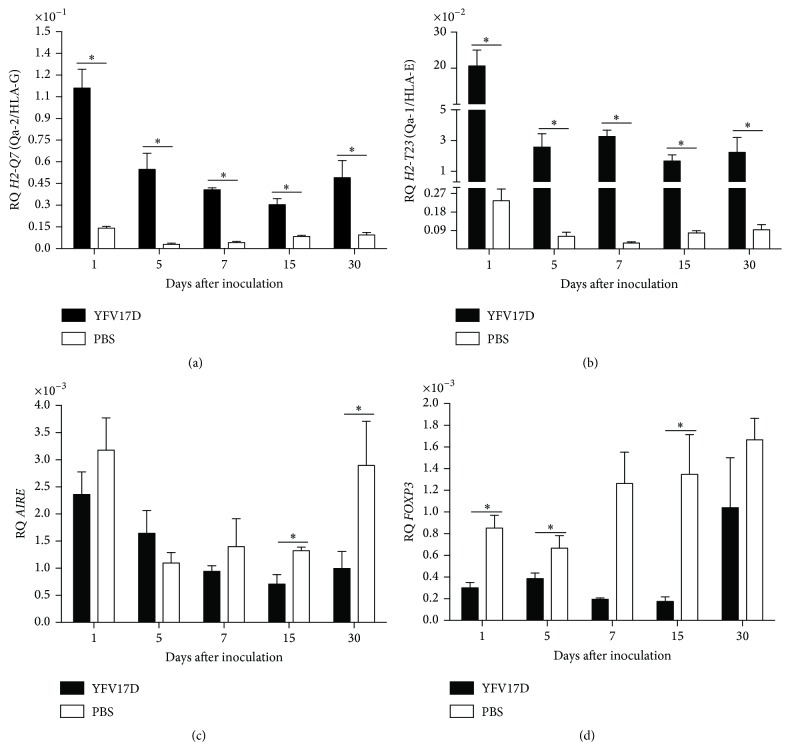
Since YFV17D could be detected in thymus, we evaluated the influence of YFV17D immunization on the transcription of immunomodulatory genes in thymi of C57BL/6 and BALB/c mice. The expression of immunomodulatory genes in samples of thymus of YFV17D-infected mice was significantly modified in both strains compared to mice control group. In thymi of YFV17D-treated C57BL/6 mice a transcriptional activation of MHC class Ib genes was observed. Transcripts for H2-Q7 (Qa-2/HLA-G), H2-T23 (Qa-1/HLA-E) (Figures 2(a) and 2(b), resp.), and H2-Q10 (Qa10) (Supplemental data II) were significantly increased from Day 1 a.i. and at all analyzed days compared to PBS-treated mice control group (P < 0.05). Differently, a significant transcriptional repression was observed for AIRE and FOXP3 genes in thymi of YFV17D-treated mice compared to control group (P < 0.05) (Figures 2(c) and 2(d), resp.). YFV17D-treated BALB/C mice showed similar transcriptional profiles observed for C57BL/6, with significant increased transcripts levels for H2-Q7 (Qa-2/HLA-G) and H2-T23 (Qa-1/HLA-E) (Figures 3(a) and 3(b), resp.) and downregulation in AIRE and FOXP3 transcription compared to PBS-treated mice control group (P < 0.05) (Figures 3(c) and 3(d), resp.). No differences in H2-Q10 (Qa10) transcription were observed (Supplemental data II).
Figure 2.
Gene expression of immunomodulatory molecules: (a) H2-Q7 (Qa-2/HLA-G); (b) H2-T23 (Qa-1/HLA-E); (c) AIRE; and (d) FOXP3 in thymus of C57BL/6 mice inoculated with 10.000 LD50 of YFV17D or PBS. RQ = relative quantification representative of geometric mean values of 2−ΔΔCt. Tissue samples were obtained in triplicate from different animals for each treatment. Each experiment was independently performed at least three times. Statistical analysis was performed using the nonparametric test Mann-Whitney two-tailed. Values close to the level of significance (P < 0.05) are marked with ∗.
Figure 3.
Gene expression of immunomodulatory molecules: (a) H2-Q7 (Qa-2/HLA-G); (b) H2-T23 (Qa-1/HLA-E); (c) AIRE; and (d) FOXP3 in thymus of BALB/c mice inoculated with 10.000 LD50 of YFV17D or PBS. RQ = relative quantification representative of geometric mean values of 2−ΔΔCt. Tissue samples were obtained in triplicate from different animals for each treatment. Each experiment was independently performed at least three times. Statistical analysis was performed using the nonparametric test Mann-Whitney two-tailed. Values close to the level of significance (P < 0.05) are marked with ∗.
These results agree with evidences showing that several pathogens can infect and replicate in thymus influencing its function [19]. It is known that thymus could act as a site of infection during immune responses, where mature T cells are recruited back to thymus in response to intrathymic cytokine (IFN-γ) and chemokine (CXCL9, CXCL10, and CXCR3) production during infection [48]. Additionally, several infectious agents are able to modulate the expression of nonclassical MHC class I molecules, promoting the escape of immunosurveillance in peripheral tissues [35, 49–51].
In humans, YFV17D induces the transcription of sets of genes related to innate immunity, including several Toll-like receptors (TLRs), type I IFNs, KIR2DL3, and KIR2DL4 [52]. Additionally, YFV17D promotes the maturation and activation of DCs by the recognition of TLR2, TLR7, TLR8, and TLR9, resulting in a dose-dependent response of proinflammatory cytokines, such as IL-6, IL-1β, TNF-α, MCP-1, IP-10, and INF-α and INF-β [53, 54]. YFV17D also stimulates the differentiation of YFV17D-specific CD8+ T and CD4+ T cells [23] that can be found from Day 7 a.i.; from Day 15 these cells begin to express the receptor for IL-10 and to secrete IFN-γ [55].
The transcriptional activation of nonclassical MHC class I H2-Q7 (Qa-2/HLA-G) and H2-T23 (Qa-1/HLA-E) in thymus during YFV17D immunization may be attributed to the increased production of proinflammatory cytokines IL-6, IL-1β, and TNF-α and also INF-α and INF-β released in thymus or present in bloodstream, during the early events of innate immune response, and persisting at high levels during the adaptive immune response. Additionally, production of IL-10 and TGF-β is increased after Day 15 a.i. in YFV17D-immunized patients [55]. Reportedly, these cytokines are related to the induction of nonclassical MHC molecules, such as HLA-G [34, 35].
Considering that the thymus (i) is the primary site of expression of MHC Ib molecules, related to functional maturation of medullary thymocytes, as described for Qa-2 [33, 56], and (ii) plays a relevant role on positive selection of subpopulations of CD8+ T cells and regulatory CD8+ T cells restricted to recognition of antigens presented by Qa-1 [57, 58] and HLA-E [59], we believe that the transcriptional activation of MHC Ib genes after YFV17D infection could influence the thymic function by altering thymocyte differentiation, possibly by increasing the rate of medullary Qa-2+ thymocyte emigration and Qa-1-restricted T cells selection, thus influencing the antiviral response in peripheral tissues.
Additionally, since MHC Ib are related to the modulation of immune system by interaction with inhibitory receptors, such as CD94/NKG2A for HLA-E/Qa-1 [58, 60] and ILT-2/ILT-4 for HLA-G [34, 35], the upregulation of MHC Ib expression in thymus by YFV17D could be related to an increased thymic immunoregulatory network to maintain thymic microenvironment and functionality during antiviral response. Particularly, the upregulation of Qa-1 expression could enhance regulatory mechanisms by the interaction with CD94/NKG2A receptor, promoting inhibition of CD8+ T and NK cells cytotoxicity and increasing the regulatory activity and anti-inflammatory cytokine production by Qa-1-restricted suppressor CD8+ T cells [61, 62].
The transcriptional repression observed for AIRE and FOXP3 genes in thymus after YFV17D immunization was an interesting finding. Although there are no evidences relating that viral infections can modulate the expression of AIRE in human or murine thymus, we hypothesize that the inhibition of this gene during the YFV17D immunization could contribute to the onset of autoimmune manifestations associated with vaccination. We raised this hypothesis due to several evidences suggesting the onset of autoimmune diseases as a result of viral infections and immunizations [44, 46] and, more particularly, due to the important role of AIRE in the TRA expression regulation during thymic selection [2, 7]. Decreased levels of AIRE may imply a defective negative selection, therefore releasing autoreactive clones of T cells to peripheral tissues [2, 7].
FOXP3 expression decreases during acute infection caused by herpes simplex 1/2 [63] and by West Nile viruses [64], being associated with the aggravation of infection, increasing the mortality rate and increasing viral load in infectious site. Similarly, the ratio between the frequency of effector T cells and regulatory T cells plays an important role on the immunopathogenesis of infections caused by flaviviruses, as reported for dengue [65]. We speculate that the transcriptional repression of FOXP3 in thymus after administration of YFV17D could contribute to higher severity pathogenic effects associated with the vaccine, as observed for BALB/c mice in the present study.
Aire regulates the induction and frequency of the repertoire of subpopulations of natural regulatory T cells in thymus by unknown mechanisms [66, 67]. We believe that the decreased AIRE expression after YFV17D immunization could influence the expression of FOXP3 and consequently the frequency of nTregs. The diminished frequency of nTreg cells associated with a defective negative selection, due to low levels of AIRE, could be an additional evidence for the onset of autoimmunity after immunization with YFV17D.
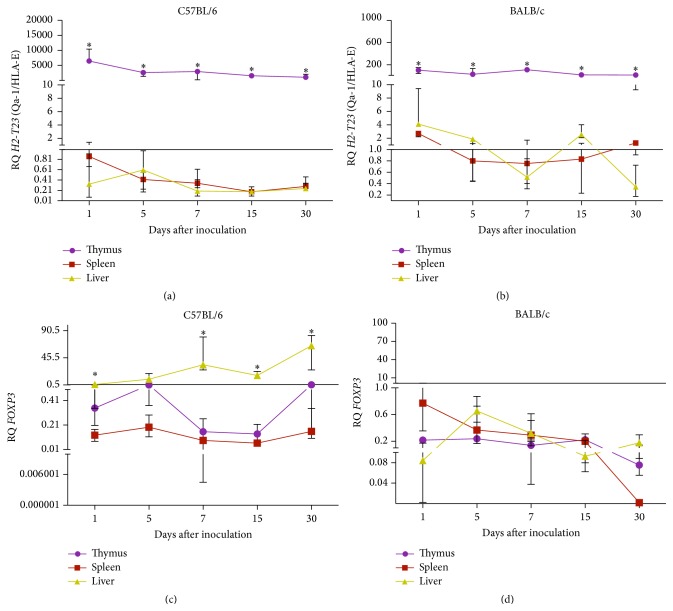
3.3. Transcriptional Profiles of Immunomodulatory Genes during YFV17D Immunization
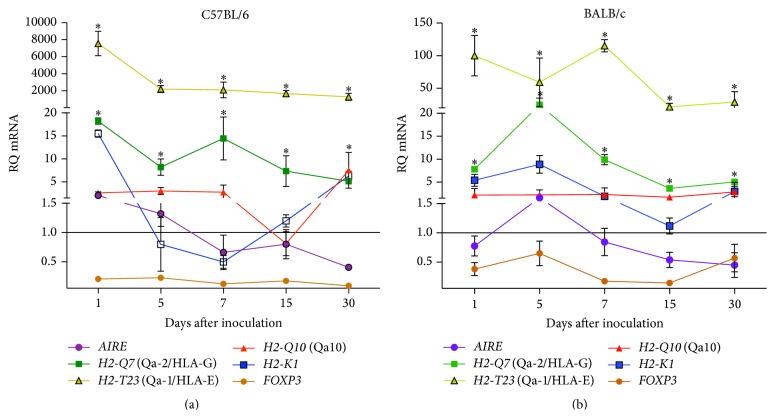
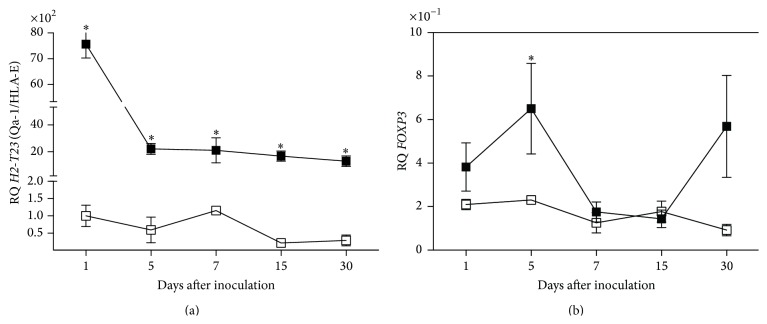
To understand the effect of YFV17D immunization in the transcription of immunomodulatory genes, we characterized the transcriptional profiles of these genes in thymus and peripheral tissues (liver and spleen) of YFV17D-treated mice, comparing the profiles between C57BL/6 and BALB/c strains. After YFV17D immunization, the thymic expression of MHC class Ib transcripts, particularly H2-T23 (Qa-1/HLA-E) and H2-Q7 (Qa-2/HLA-G), was increased when compared to MHC class Ia, H2-K1 transcripts (P < 0.05, for each comparison) for both mouse strains. Decreased expression of AIRE and FOXP3 was also observed compared to all genes analyzed (Figures 4(a) and 4(b)). H2-T23 (Qa-1/HLA-E) was the most widely expressed gene in thymus throughout the immunization, and the thymus was the tissue which most expressed this gene, compared to liver and spleen, in both strains analyzed (P < 0.05) (Figures 5(a) and 5(b)). No significant differences were observed for other genes in all studied tissues.
Figure 4.
Transcriptional profiles of immunomodulatory genes in thymus of (a) C57BL/6 and (b) BALB/c mice inoculated with 10.000 LD50 of YFV17D. RQ = relative quantification representative of geometric mean values of 2−ΔΔCt. RQ values expressed as fold change relative to control group values (baseline = 1). Statistical analysis was performed using the nonparametric test Kruskal-Wallis Dunn's multiple comparison. Values close to the level of significance (P < 0.05) are marked with ∗.
Figure 5.
Transcriptional profiles of H2-T23 (Qa-1/HLA-E) ((a) and (b)) and FOXP3 ((c) and (d)) in harvested tissues of C56BL/6 and BALB/c mice inoculated with 10.000 LD50 of YFV17D. RQ = relative quantification representative of geometric mean values of 2−ΔΔCt. RQ values expressed as fold change relative to control group values (baseline = 1). Statistical analysis was performed using the nonparametric test Kruskal-Wallis Dunn's multiple comparison. Values close to the level of significance (P < 0.05) are marked with ∗.
The role of Qa-1 in antiviral immunity has been demonstrated in several viral infections. Increased levels of Qa-1 have been reported in mice infected with Herpes Simplex Virus [62] and cytomegalovirus, when compared to classical MHC class I (H2-D) molecules in peripheral tissues [68]. Particularly, viral infections caused by other flaviviruses, as observed for Japanese Encephalitis Virus, promoted the upregulation of Qa-1 expression in a type 1 IFN-dependent manner, independently of NF-kB pathway [69].
We demonstrated that YFV17D immunization promotes a significant upregulation of Qa-1 in thymus in higher levels than in other peripheral tissues, as liver and spleen, evidencing this central lymphoid organ as a primary expression site of MHC Ib during YFV17D infection. Interestingly, during YFV17D immunization in both strains we observed decreased levels of classical MHC class I gene (H2-K1), typically involved in antiviral responses [70, 71], compared to MHC Ib genes (Figures 4(a) and 4(b)). We believe that low levels of H2-K1 transcripts, as observed in this study, could be related to a viral escape mechanism of immunosurveillance acting in a transcriptional regulation level, contributing to a reduced antigen processing and presentation, as described at molecular level [72].
It is known that the blockade of Qa-1 during acute and chronic antiviral responses enhances the cytolytic activity of effector CD8+ T cells, the repression of PD-1 (programmed cell death-1), and the upregulation of activating receptor NKG2D [61]. We speculate that this molecule and its human counterpart HLA-E could play a relevant role during antiviral immunity against YFV and other flaviviruses.
Interestingly, we observed an increased expression FOXP3 in liver of YFV17D-treated C57BL/6 mice compared to thymus and spleen (P < 0.05) (Figure 5(c)). No differences in FOXP3 transcript levels among tissues were observed in BALB/c mice (Figure 5(d)). Increased transcript levels of H2-Q10 (Q10) were observed in the liver of YFV17D-treated C57BL/6 mice compared to thymus and spleen (P < 0.05) (Supplemental data III). Noteworthy, H2-Q10 is a murine MHC Ib gene encoding the Q10 molecule, expressed mainly in the liver and related to the regulation of immune responses [73, 74]. Finally, compared to BALB/c mice, the C57BL/6 thymus showed significant higher transcript levels for H2-T23 (Qa-1/HLA-E) in all harvested days a.i. (P < 0.05) and FOXP3 at Day 5 a.i. (Figures 6(a) and 6(b)). No differences were found for the other genes between both strains.
Figure 6.
Comparison of transcriptional profiles of (a) H2-T23 (Qa-1/HLA-E) and (b) FOXP3 in thymus of C57BL/6 and BALB/c mice inoculated with 10.000 LD50 of YFV17D. RQ = relative quantification representative of geometric mean values of 2−ΔΔCt. RQ values expressed as fold change relative to control group values (baseline = 1). Statistical analysis was performed using the nonparametric test Mann-Whitney two-tailed. Values close to the level of significance (P < 0.05) are marked with ∗.
We speculate that the increased expression of H2-T23 (Qa-1/HLA-E), H2-Q10 (Q10), and FOXP3 in YFV17D-immunized C57BL/6 mice may contribute to the establishment of a regulatory network that protects the YFV17D-infected cells from immune system, since both molecules are clearly involved in the modulation of the immune system and the generation of regulatory T cells, thus, reflecting in an inability of viral load control and in a greater susceptibility of this strain to YFV17D infection. In BALB/c mice, on the other hand, the observation of liver and spleen hypertrophy, more pronounced signs of infection, which may be indicative of more robust antiviral immune response, better control of viral load, together with lower transcription of immunomodulatory genes in thymus and peripheral tissues in YFV17D-immunized mice compared to C57BL/6, may contribute to a better resistance to YFV17D infection.
Among the several factors involved in the onset of autoimmunity, not only viral infections but also live virus attenuated vaccines are potentially relevant to development of autoimmune disorders [42–44]. The thymus is the primary organ involved in induction and maintenance of self-tolerance in T lymphocytes, particularly due to Aire activity and generation of natural regulatory T cells [2, 66]. Moreover, the expression of immunomodulatory molecules, such as those from MHC Ib, is essential to ensure maintenance of self-tolerance in potentially autoreactive T lymphocyte clones in peripheral tissues [11, 16, 35, 38]. Alterations in thymic function and in MHC Ib expression could change the regulation of immune responses permitting the onset of autoimmune disorders.
Besides the fact that the YFV17D vaccine is the only preventive measure against the hemorrhagic fever caused by YFV, studies involving the biological effects after immunization with YFV17D are relevant since several evidences show the emergence of autoimmune disorders in patients after YFV17D immunization [45, 46]. Additionally, the contraindication of YFV17D vaccine in individuals with thymic diseases [29], immunosuppressed, or with rheumatic diseases, such as lupus erythematosus, rheumatoid arthritis, and polymyalgia rheumatica [75], leads to speculation about the influence of YFV17D immunization in thymus, particularly in the immunoregulatory mechanisms of tolerance.
4. Conclusions
The present study showed that YFV17D immunization influences the transcriptional profiles of immunomodulatory genes in thymus by promoting the induction of MHC Ib genes: H2-Q7 (Qa-2/HLA-G) and H2-T23 (Qa-1/HLA-E), and the repression of AIRE and FOXP3, indicating a possible influence on thymic function. Also, we highlighted the thymus as a target organ for YFV17D virus and as a main site of expression of MHC Ib during YFV17D infection. Similarly, we demonstrated the relevance of the genetic background of C57BL/6 and BALB/c mice strains in tolerance mechanisms during YFV17D infection, suggesting that increased expression of H2-T23 (Qa-1/HLA-E) and FOXP3 transcripts in thymus and liver of C57BL/6 mice, together with a defective control of viral load, might be indicative of a higher susceptibility of this strain to YFV17D infection. The present work may contribute to further studies involving the thymus and its immunomodulatory molecules in the maintenance of central and peripheral tolerance during antiviral immune responses.
Supplementary Material
The morphological analysis of internal organs in YFV17D - immunized animals revealed liver and spleen hypertrophy at Day 7 a.i in BALB/c mice compared to C57BL/6 and compared to control groups. (Supplemental data I). The gene expression analysis of H2-Q10(Qa10) in thymus of C57BL/6 and BALB/c mice inoculated with 10.000 LD50 of YFV17D or PBS showed no differences in H2-Q10(Qa10) transcription compared to the control group. (Supplemental data II). Increased transcript levels of H2-Q10(Q10) were observed in the liver of YFV17D-treated C57BL/6 mice compared to thymus and spleen (P<0.05). (Supplemental data III).
Acknowledgments
The authors would like to thank Sandra Rodrigues, Flávia Tremmeschin, Soraya Badra, and Vinicius Reis for technical support during the study. This work was supported by grants from the binational collaborative CAPES/COFECB program (no. 653/09).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Rioux J. D., Abbas A. K. Paths to understanding the genetic basis of autoimmune disease. Nature. 2005;435(7042):584–589. doi: 10.1038/nature03723. [DOI] [PubMed] [Google Scholar]
- 2.Mathis D., Benoist C. Aire. Annual Review of Immunology. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 3.Owen J. A. P., Punt J., Stranford S. A., Jones P. P. Kuby Immunology. 7th. W. H. Freeman and Company; 2013. [Google Scholar]
- 4.Anderson G., Jenkinson W. E., Jones T., et al. Establishment and functioning of intrathymic microenvironments. Immunological Reviews. 2006;209:10–27. doi: 10.1111/j.0105-2896.2006.00347.x. [DOI] [PubMed] [Google Scholar]
- 5.Itoi M., Kawamoto H., Katsura Y., Amagai T. Two distinct steps of immigration of hematopoietic progenitors into the early thymus anlage. International Immunology. 2001;13(9):1203–1211. doi: 10.1093/intimm/13.9.1203. [DOI] [PubMed] [Google Scholar]
- 6.Kyewski B., Derbinski J. Self-representation in the thymus: an extended view. Nature Reviews Immunology. 2004;4(9):688–698. doi: 10.1038/nri1436. [DOI] [PubMed] [Google Scholar]
- 7.Zumer K., Saksela K., Peterlin B. M. The mechanism of tissue-restricted antigen gene expression by AIRE. Journal of Immunology. 2013;190(6):2479–2482. doi: 10.4049/jimmunol.1203210. [DOI] [PubMed] [Google Scholar]
- 8.Gotter J., Brors B., Hergenhahn M., Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. The Journal of Experimental Medicine. 2004;199(2):155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontenot J. D., Rudensky A. Y. A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nature Immunology. 2005;6(4):331–337. doi: 10.1038/ni1179. [DOI] [PubMed] [Google Scholar]
- 10.Cabarrocas J., Cassan C., Magnusson F., et al. Foxp3+ CD25+ regulatory T cells specific for a neo-self-antigen develop at the double-positive thymic stage. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(22):8453–8458. doi: 10.1073/pnas.0603086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller D. L. Mechanisms maintaining peripheral tolerance. Nature Immunology. 2010;11(1):21–27. doi: 10.1038/ni.1817. [DOI] [PubMed] [Google Scholar]
- 12.Ramsdell F., Ziegler S. F. Transcription factors in autoimmunity. Current Opinion in Immunology. 2003;15(6):718–724. doi: 10.1016/j.coi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Nagamine K., Peterson P., Scott H. S., et al. Positional cloning of the APECED gene. Nature Genetics. 1997;17(4):393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 14.Haiqi H., Yong Z., Yi L. Transcriptional regulation of Foxp3 in regulatory T cells. Immunobiology. 2011;216(6):678–685. doi: 10.1016/j.imbio.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Guerau-de-Arellano M., Mathis D., Benoist C. Transcriptional impact of Aire varies with cell type. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(37):14011–14016. doi: 10.1073/pnas.0806616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Fuente H., Cibrián D., Sánchez-Madrid F. Immunoregulatory molecules are master regulators of inflammation during the immune response. FEBS Letters. 2012;586(18):2897–2905. doi: 10.1016/j.febslet.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrio R., Lopez D. M. Insights into thymic involution in tumor-bearing mice. Immunologic Research. 2013;57(1–3):106–114. doi: 10.1007/s12026-013-8446-3. [DOI] [PubMed] [Google Scholar]
- 18.Kaiserlian D., Savino W., Hassid J., Dardenne M. Studies of the thymus in mice bearing the Lewis lung carcinoma. III. Possible mechanisms of tumor-induced thymic atrophy. Clinical Immunology and Immunopathology. 1984;32(3):316–325. doi: 10.1016/0090-1229(84)90275-7. [DOI] [PubMed] [Google Scholar]
- 19.Savino W. The thymus is a common target organ in infectious diseases. PLoS Pathogens. 2006;2(6, article e62) doi: 10.1371/journal.ppat.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro P., Torres B., López A., et al. Effects of different antigenic stimuli on thymic function and interleukin-7/CD127 system in patients with chronic HIV infection. Journal of Acquired Immune Deficiency Syndromes. 2014;66(5):466–472. doi: 10.1097/qai.0000000000000207. [DOI] [PubMed] [Google Scholar]
- 21.Takamura S., Kajiwara E., Tsuji-Kawahara S., et al. Infection of adult thymus with murine retrovirus induces virus-specific central tolerance that prevents functional memory CD8+ T cell differentiation. PLoS Pathogens. 2014;10(3) doi: 10.1371/journal.ppat.1003937.e1003937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooke A., Ferraccioli G. F., Herrmann M., et al. Induction and protection of autoimmune rheumatic diseases. The role of infections. Clinical and Experimental Rheumatology. 2008;26(1) supplement 48:S1–S7. [PubMed] [Google Scholar]
- 23.Bethke N. Characterization of the immune response after Yellow Fever vaccination [Ph.D. thesis] Berlin, Germany: Berlin-Brandenburg Zentrum für Regenerative Therapien der Medizinischen Fakultät Charité, Universitätsmedizin; 2011. [Google Scholar]
- 24.Finlay C. The mosquito hypothetically considered as an agent of yellow fever transmission. 1881. Salud Pública de México. 1992;34(4):474–483. [PubMed] [Google Scholar]
- 25.WHO. Meeting of Global Advisory Committee on Vaccine Safety, 18-19 June 2008. The Weekly Epidemiological Record. 2008;83(32):287–292. [PubMed] [Google Scholar]
- 26.Robertson S. E., Hull B. P., Tomori O., Bele O., LeDuc J. W., Esteves K. Yellow fever: a decade of reemergence. The Journal of the American Medical Association. 1996;276(14):1157–1162. doi: 10.1001/jama.276.14.1157. [DOI] [PubMed] [Google Scholar]
- 27.Hayes E. B. Is it time for a new yellow fever vaccine? Vaccine. 2010;28(51):8073–8076. doi: 10.1016/j.vaccine.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Monath T. P. Yellow fever: an update. The Lancet Infectious Diseases. 2001;1(1):11–20. doi: 10.1016/s1473-3099(01)00016-0. [DOI] [PubMed] [Google Scholar]
- 29.Eidex R. B. History of thymoma and yellow fever vaccination. The Lancet. 2004;364(9438):p. 936. doi: 10.1016/s0140-6736(04)17017-7. [DOI] [PubMed] [Google Scholar]
- 30.Barrett A. D., Teuwen D. E. Yellow fever vaccine—how does it work and why do rare cases of serious adverse events take place? Current Opinion in Immunology. 2009;21(3):308–313. doi: 10.1016/j.coi.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Melo-Lima B. L., Evangelista A. F., de Magalhães D. A. R., Passos G. A., Moreau P., Donadi E. A. Differential transcript profiles of MHC class Ib(Qa-1, Qa-2, and Qa-10) and aire genes during the ontogeny of thymus and other tissues. Journal of Immunology Research. 2014;2014:12. doi: 10.1155/2014/159247.159247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crisa L., McMaster M. T., Ishii J. K., Fisher S. J., Salomon D. R. Identification of a thymic epithelial cell subset sharing expression of the class Ib HLA-G molecule with fetal trophoblasts. The Journal of Experimental Medicine. 1997;186(2):289–298. doi: 10.1084/jem.186.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ge Q., Chen W.-F. Effect of murine thymic epithelial cell line (MTEC1) on the functional expression of CD4+CD8− thymocyte subgroups. International Immunology. 2000;12(8):1127–1133. doi: 10.1093/intimm/12.8.1127. [DOI] [PubMed] [Google Scholar]
- 34.Donadi E. A., Castelli E. C., Arnaiz-Villena A., Roger M., Rey D., Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cellular and Molecular Life Sciences. 2011;68(3):369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carosella E. D., Moreau P., LeMaoult J., Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends in Immunology. 2008;29(3):125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Comiskey M., Goldstein C. Y., De Fazio S. R., Mammolenti M., Newmark J. A., Warner C. M. Evidence that HLA-G is the functional homolog of mouse Qa-2, the Ped gene product. Human Immunology. 2003;64(11):999–1004. doi: 10.1016/j.humimm.2003.08.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson P. J., Millrain M., Antoniou J., Simpson E., Mellor A. L. A glycophospholipid anchor is required for Qa-2-mediated T cell activation. Nature. 1989;342(6245):85–87. doi: 10.1038/342085a0. [DOI] [PubMed] [Google Scholar]
- 38.Lu L., Cantor H. Generation and regulation of CD8+ regulatory T cells. Cellular and Molecular Immunology. 2008;5(6):401–406. doi: 10.1038/cmi.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith T. R. F., Kumar V. Revival of CD8+ Treg-mediated suppression. Trends in Immunology. 2008;29(7):337–342. doi: 10.1016/j.it.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Stroynowski I., Tabaczewski P. Multiple products of class Ib Qa-2 genes which ones are functional? Research in Immunology. 1996;147(5):290–301. doi: 10.1016/0923-2494(96)89642-8. [DOI] [PubMed] [Google Scholar]
- 41.Klein J., Figueroa F., David C. S. H-2 haplotypes, genes and antigens: second listing. II. The H-2 complex. Immunogenetics. 1983;17(6):553–596. doi: 10.1007/bf00366126. [DOI] [PubMed] [Google Scholar]
- 42.Oliveira A. C. V., Maria Henrique Da Mota L., Dos Santos-Neto L. L., et al. Occurrence of autoimmune diseases related to the vaccine against yellow Fever. Autoimmune Diseases. 2014;2014:8. doi: 10.1155/2014/473170.473170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gout O. Vaccinations and multiple sclerosis. Neurological Sciences. 2001;22(2):151–154. doi: 10.1007/s100720170014. [DOI] [PubMed] [Google Scholar]
- 44.Tishler M., Shoenfeld Y. Vaccination may be associated with autoimmune diseases. Israel Medical Association Journal. 2004;6(7):430–432. [PubMed] [Google Scholar]
- 45.Chaves M., Riccio P., Patrucco L., Rojas J., Cristiano E. Longitudinal myelitis associated with yellow fever vaccination. Journal of NeuroVirology. 2009;15(4):348–350. doi: 10.1080/13550280903062805. [DOI] [PubMed] [Google Scholar]
- 46.Schmöeller D., Keiserman M. W., Staub H. L., Velho F. P., de Fátima Grohe M. Yellow fever vaccination and Kawasaki disease. Pediatric Infectious Disease Journal. 2009;28(11):1037–1038. doi: 10.1097/inf.0b013e3181bbc571. [DOI] [PubMed] [Google Scholar]
- 47.Barrett A. D. T., Monath T. P. Epidemiology and ecology of yellow fever virus. Advances in Virus Research. 2003;61:291–315. doi: 10.1016/S0065-3527(03)61007-9. [DOI] [PubMed] [Google Scholar]
- 48.Nobrega C., Cláudio Nunes-Alves, Cerqueira-Rodrigues B., et al. T cells home to the thymus and control infection. Journal of Immunology. 2013;190(4):1646–1658. doi: 10.4049/jimmunol.1202412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C., Toth I., Schulze zur Wiesch J., et al. Functional characterization of HLA-G+ regulatory T cells in HIV-1 infection. PLoS Pathogens. 2013;9(1) doi: 10.1371/journal.ppat.1003140.e1003140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Söderberg-Nauclér C., Nelson J. A. Human cytomegalovirus latency and reactivation—a delicate balance between the virus and its host's immune system. Intervirology. 2000;42(5-6):314–321. doi: 10.1159/000053966. [DOI] [PubMed] [Google Scholar]
- 51.Ely K. H., Matsuoka M., DeBerge M. P., et al. Tissue-protective effects of NKG2A in immune-mediated clearance of virus infection. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0108385.e108385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaucher D., Therrien R., Kettaf N., et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. Journal of Experimental Medicine. 2008;205(13):3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Querec T. D., Akondy R. S., Lee E. K., et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature Immunology. 2009;10(1):116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer D. R., Fernandez S., Bisbing J., et al. Restricted replication and lysosomal trafficking of yellow fever 17D vaccine virus in human dendritic cells. Journal of General Virology. 2007;88, part 1:148–156. doi: 10.1099/vir.0.82272-0. [DOI] [PubMed] [Google Scholar]
- 55.Martins M. Â., Silva M. L., Marciano A. P. V., et al. Activation/modulation of adaptive immunity emerges simultaneously after 17DD yellow fever first-time vaccination: is this the key to prevent severe adverse reactions following immunization? Clinical and Experimental Immunology. 2007;148(1):90–100. doi: 10.1111/j.1365-2249.2006.03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ge Q., Chen W. F. Phenotypic identification of the subgroups of murine T-cell receptor alphabeta+ CD4+ CD8- thymocytes and its implication in the late stage of thymocyte development. Immunology. 1999;97(4):665–671. doi: 10.1046/j.1365-2567.1999.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarantopoulos S., Lu L., Cantor H. Qa-1 restriction of CD8+ suppressor T cells. The Journal of Clinical Investigation. 2004;114(9):1218–1221. doi: 10.1172/jci200423152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang H., Chess L. Qa-1/HLA-E-restricted regulatory CD8+ T cells and self-nonself discrimination: an essay on peripheral T-cell regulation. Human Immunology. 2008;69(11):721–727. doi: 10.1016/j.humimm.2008.08.279. [DOI] [PubMed] [Google Scholar]
- 59.O'Callaghan C. A., Bell J. I. Structure and function of the human MHC class Ib molecules HLA-E, HLA-F and HLA-G. Immunological Reviews. 1998;163:129–138. doi: 10.1111/j.1600-065x.1998.tb01192.x. [DOI] [PubMed] [Google Scholar]
- 60.Lu L., Werneck M. B. F., Cantor H. The immunoregulatory effects of Qa-1. Immunological Reviews. 2006;212:51–59. doi: 10.1111/j.0105-2896.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- 61.Holderried T. A. W., Lang P. A., Kim H.-J., Cantor H. Genetic disruption of CD8+ Treg activity enhances the immune response to viral infection. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(52):21089–21094. doi: 10.1073/pnas.1320999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suvas S., Azkur A. K., Rouse B. T. Qa-1b and CD94-NKG2a interaction regulate cytolytic activity of herpes simplex virus-specific memory CD8+ T cells in the latently infected trigeminal ganglia. The Journal of Immunology. 2006;176(3):1703–1711. doi: 10.4049/jimmunol.176.3.1703. [DOI] [PubMed] [Google Scholar]
- 63.Lund J. M., Hsing L., Pham T. T., Rudensky A. Y. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320(5880):1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanteri M. C., O'Brien K. M., Purtha W. E., et al. Tregs control the development of symptomatic West Nile virus infection in humans and mice. Journal of Clinical Investigation. 2009;119(11):3266–3277. doi: 10.1172/jci39387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lühn K., Simmons C. P., Moran E., et al. Increased frequencies of CD4+CD25high regulatory T cells in acute dengue infection. The Journal of Experimental Medicine. 2007;204(5):979–985. doi: 10.1084/jem.20061381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aschenbrenner K., D'Cruz L. M., Vollmann E. H., et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nature Immunology. 2007;8(4):351–358. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 67.Hinterberger M., Aichinger M., da Costa O. P., Voehringer D., Hoffmann R., Klein L. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nature Immunology. 2010;11(6):512–519. doi: 10.1038/ni.1874. [DOI] [PubMed] [Google Scholar]
- 68.Cavanaugh V. J., Raulet D. H., Campbell A. E. Upregulation of CD94/NKG2A receptors and Qa-1b ligand during murine cytomegalovirus infection of salivary glands. Journal of General Virology. 2007;88(5):1440–1445. doi: 10.1099/vir.0.82444-0. [DOI] [PubMed] [Google Scholar]
- 69.Sojan A., Shwetank V., Gaurav K., Ramanathapuram M. Japanese encephalitis virus: innate and adaptive immunity. In: Ruzek D., editor. Flavivirus Encephalitis. InTech; 2011. [Google Scholar]
- 70.Fodil-Cornu N., Loredo-Osti J. C., Vidal S. M. Nk cell receptor/H2-Dk-dependent host resistance to viral infection is quantitatively modulated by H2q inhibitory signals. PLoS Genetics. 2011;7(4) doi: 10.1371/journal.pgen.1001368.e1001368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie X., Stadnisky M. D., Brown M. G. MHC class I Dk locus and Ly49G2+ NK cells confer H-2k resistance to murine cytomegalovirus. Journal of Immunology. 2009;182(11):7163–7171. doi: 10.4049/jimmunol.0803933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boname J. M., de Lima B. D., Lehner P. J., Stevenson P. G. Viral degradation of the MHC class I peptide loading complex. Immunity. 2004;20(3):305–317. doi: 10.1016/s1074-7613(04)00047-0. [DOI] [PubMed] [Google Scholar]
- 73.Lew A. M., Maloy W. L., Coligan J. E. Characteristics of the expression of the murine soluble class I molecule (Q10) Journal of Immunology. 1986;136(1):254–258. [PubMed] [Google Scholar]
- 74.Wieties K., Hammer R. E., Jones-Youngblood S., Forman J. Peripheral tolerance in mice expressing a liver-specific class I molecule: inactivation/deletion of a T-cell subpopulation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(17):6604–6608. doi: 10.1073/pnas.87.17.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oliveira A. C. V., Da Mota L. M. H., Dos Santos-Neto L. L., Tauil P. L. What a rheumatologist needs to know about yellow fever vaccine. Revista Brasileira de Reumatologia. 2013;53(2):206–210. doi: 10.1590/S0482-50042013000200008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The morphological analysis of internal organs in YFV17D - immunized animals revealed liver and spleen hypertrophy at Day 7 a.i in BALB/c mice compared to C57BL/6 and compared to control groups. (Supplemental data I). The gene expression analysis of H2-Q10(Qa10) in thymus of C57BL/6 and BALB/c mice inoculated with 10.000 LD50 of YFV17D or PBS showed no differences in H2-Q10(Qa10) transcription compared to the control group. (Supplemental data II). Increased transcript levels of H2-Q10(Q10) were observed in the liver of YFV17D-treated C57BL/6 mice compared to thymus and spleen (P<0.05). (Supplemental data III).