Abstract
ARS2 is a regulator of RNA polymerase II transcript processing through its role in the maturation of distinct nuclear cap-binding complex (CBC)-controlled RNA families. In this study, we examined ARS2 domain function in transcript processing. Structural modeling based on the plant ARS2 orthologue, SERRATE, revealed 2 previously uncharacterized domains in mammalian ARS2: an N-terminal domain of unknown function (DUF3546), which is also present in SERRATE, and an RNA recognition motif (RRM) that is present in metazoan ARS2 but not in plants. Both the DUF3546 and zinc finger domain (ZnF) were required for association with microRNA and replication-dependent histone mRNA. Mutations in the ZnF disrupted interaction with FLASH, a key component in histone pre-mRNA processing. Mutations targeting the Mid domain implicated it in DROSHA interaction and microRNA biogenesis. The unstructured C terminus was required for interaction with the CBC protein CBP20, while the RRM was required for cell cycle progression and for binding to FLASH. Together, our results support a bridging model in which ARS2 plays a central role in RNA recognition and processing through multiple protein and RNA interactions.
INTRODUCTION
The generation of mature RNA in the nucleus is a highly coordinated process that requires distinct complexes for the biogenesis of different RNA families. For RNA polymerase II (RNAP II) transcripts, a critical step is the addition of a 7-methylguanosine (m7G) cap to the nascent transcript (1), which is subsequently bound by CBP20 and CBP80 of the nuclear cap-binding complex (CBC) (2). CBC-controlled transcripts include mRNA, microRNA (miRNA), replication-dependent histone (RDH) mRNA, small nucleolar RNA (snoRNA), and small nuclear RNA (snRNA), each with its own unique processing requirements. Binding of the CBC to the m7G cap of these RNAs occurs cotranscriptionally, protects the transcripts from degradation, and plays a central role in recruiting the appropriate machinery for processing different RNA families (3–8). However, exactly how distinct RNA families are differentially recognized to allow for correct processing complex formation is not fully understood. Recently, a protein called ARS2 (or SRRT in humans) has been shown to be part of the CBC, and it plays an important role in the maturation of several distinct RNA families (8, 9).
Clues to how ARS2 participates in recruiting different RNA processing machineries are beginning to emerge from biochemical and structural studies. ARS2 interacts directly with the assembled CBP20/80 cap complex to form a tertiary complex termed CBCA (9). One hypothesis is that ARS2 bridges the CBCA to the appropriate processing machinery by interacting with both protein and RNA elements. This hypothesis is based on studies of the ARS2 plant orthologue SERRATE in miRNA biogenesis. Both the Arg/Pro-rich N terminus and zinc finger (ZnF) domain of SERRATE are required for primary miRNA (pri-miRNA) binding (10, 11). Additionally, SERRATE, through its N terminus and ZnF, has been shown to bind directly to DCL-1, the plant RNase III enzyme responsible for pri-miRNA processing (10, 11). A similar process is thought to occur in metazoans for ARS2. Indeed, binding of ARS2 to DROSHA has been shown, and loss of ARS2 function leads to pri-miRNA instability and misprocessing (7, 12). However, how ARS2 mediates these interactions is not understood.
During replication-dependent histone (RDH) mRNA maturation in mammals, ARS2 interacts directly with the 3′-end processing machinery through the protein FLASH (13), which, in turn, binds to the U7 snRNP component LSM11 (14) and is required for 3′-end cleavage (15). FLASH has been shown to interact with ARS2 through a 13 amino acid sequence called FARB (FLASH ARS2 binding), which is both necessary and sufficient for interaction (13). It has also been hypothesized that ARS2 recognizes the stem-loop found in the 3′ untranslated region (UTR) of RDH transcripts (16). In the absence of ARS2, there is a reduction of properly processed histone mRNA and protein (12), which impairs cell cycle progression through S phase (13).
ARS2 appears to be involved in most CBC functions. In addition to the above-described roles, ARS2 has been shown to promote snRNA 3′-end processing and transport through PHAX (8), cap-proximal processing of the 3′-end of mRNA through CLP1 (8), RNA export through TREX (17), and RNA degradation through the exosome (9). However, little is known about the mechanistic details of ARS2's role in these processes.
In this study, we determined the importance of ARS2 and its domains in the myogenic C2C12 cell line. Depletion or overexpression of ARS2 caused cells to stall in S phase, indicating that ARS2 levels must be tightly regulated. To facilitate structure-function analysis, we modeled the structure of ARS2 based on the plant SERRATE structure and identified 2 new domains in mammalian ARS2: an N-terminal DUF3546 domain, which is also present in SERRATE, and an RRM domain that is present in metazoan ARS2 but not in plants. We then performed systematic mutagenesis to decipher which of ARS2's domains mediate interactions with the miRNA and RDH mRNA processing pathways. Our results support a scaffold or bridging model in which ARS2 plays a central role in RNA recognition and processing through both protein and RNA interactions.
MATERIALS AND METHODS
Cell culture and transfection.
Primary mouse myoblasts were isolated from P0 C57BL/6 mice as described previously (18). Primary and C2C12 myoblasts were cultured in growth medium (Dulbecco modified Eagle medium [DMEM] supplemented with 10% fetal bovine serum [FBS] and 1% penicillin-streptomycin). Transfections were performed using JetPrime reagent (VWR) as per the manufacturer's instructions. Ars2 small interfering RNAs (siRNAs) were purchased from Qiagen and had the following target sequences: Ars2 siRNA 1, CCCGTCGTGTCCGCAACATAA; Ars2 siRNA 2, CCGAAGAAGCACTTAAAGAAA; and Ars2 siRNA 3, CCCAGCTTTGCCTGAGATCAA. AllStars negative-control siRNA (Qiagen) was used as a control siRNA. Short hairpin RNA (shRNAs) were purchased from Origene and had the following targeting sequences: Ars2 sh78, ATGCAGCTGTCATTAAGATGGAAGGTGGC; Ars2 sh79, CAGGCTGAGAATGACAGTTCCAACGATGA; Ars2 sh80, GCAAGGATAAGTGGCTATGTCCTCTCAGT; FLASH sh, AACATTGTGCCAATAATGTCTGGTCACGT; Drosha sh, GTTCATTGAGCGGAAATACAGACAAGAGT; and Lsm11 sh, GGATTACCAGCAGGTATTCACTCGGCACA.
Plasmids.
miRNA 24 (miR-24) and miR-155 firefly luciferase reporters were obtained from Signosis. Renilla luciferase was obtained from Promega. For the histone DsRed reporter, the first 67 amino acids encoded by the H2A ORF were fused to the 3′ UTR of histone H2A, including the histone downstream element (HDE) but not the downstream polyadenylation signals (adapted from reference 19). This was cloned into pIRES2-DsRed-Express using EcoRI and BstX1 restriction sites to generate histone DsRed. Enhanced GFP-C1 (eGFP-C1) was used as a green fluorescent protein (GFP) expression control, and ARS2 (wild type and mutants) was synthesized and cloned into pEGFP-C1 by Bio Basic to generate eGFP-ARS2. For eGFP-FARB, the FARB peptide sequence (13) was cloned into pEGFP-C1. For scrambled FARB, a random generator scrambled the FARB sequence to EGEIESELDEDDR, which was then cloned into pEGFP-C1. 3×FLAG-ARS2 and ARS2-mychis were generated as described previously (20).
Cell cycle analysis.
For bromodeoxyuridine (BrdU) pulse-chase experiments, C2C12 cells were seeded on 6-cm plates at 6.0 × 105/plate and 18 h later were transfected with the desired plasmids. Twenty-four hours posttransfection, cells were passaged onto a 10-cm plate to allow growth. The next day, cells were processed according to the protocol in reference 21. Briefly, cells were given a 30-min BrdU pulse and fixed at intervals between 0 and 10 h. Cells were labeled with propidium iodide (PI) to measure DNA content and with mouse anti-BrdU–AF488 conjugate (Life Technologies) at 1:50. For cells transfected with eGFP vectors, cells were also labeled with chicken anti-GFP (Abcam) at 1:400 and donkey anti-chicken antibody–AF647 (Jackson ImmunoResearch) at 1:100 since HCl treatment abolished GFP fluorophore activity. A total of 1.0 × 104 GFP-positive events per sample were collected on a BD FACSCalibur flow cytometer.
Luciferase reporter assays.
C2C12 cells were seeded on 96-well plates at 1.0 × 104/well and 18 h later were cotransfected with the desired eGFP vector, firefly luciferase reporter, and Renilla luciferase vector. Twenty-four hours posttransfection, samples were analyzed for luminescence using the Dual-Glo luciferase assay system (Promega) on a PerkinElmer Victor3V 1420 multilabel plate counter.
DsRed reporter assays.
For the histone DsRed reporter assay, cells were seeded onto 12-well plates at 2.5 × 105/well and 18 h later were cotransfected with shRNA (containing a turbo GFP [tGFP] cassette) or eGFP/eGFP-ARS2 along with histone DsRed. Forty-eight hours posttransfection, 1.0 × 105 events were acquired using a BD FACSCalibur. Reporter activity was quantified as the percent double positive (DsRed and GFP)/total transfected cells.
For the let-7c DsRed reporter, C2C12 cells were seeded at 5.0 × 104/well, 18 h later were cotransfected with shRNA and let-7c DsRed, and 48 h posttransfection were analyzed as described for histone DsRed.
Apoptosis assay.
C2C12 cells were seeded on 24-well plates at 4.0 × 104/well and were transfected with either siRNA or shRNA 18 h later. Twenty-four hours posttransfection, cells were analyzed for apoptosis with a BD Biosciences annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit according to the manufacturer's instructions using a BD FACSCalibur.
RNA IP.
Immunoprecipitation (IP) was performed using EZview M2 anti-FLAG beads (Sigma-Aldrich) as described in reference 8. Briefly, cells were extracted in HNTG buffer (20 mM HEPES, pH 7.9, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM MgCl2, 1 mM EGTA, and protease inhibitors) for 30 min at 4°C, and cellular debris was removed by centrifugation (20 min at 20,000 × g). Extracts were incubated with beads for 2 h at 4°C. Beads were washed twice with HNTG buffer and three times in PBS. For samples treated with a high-salt wash, beads were washed once in low-salt buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% Triton X-100, 0.5% NP-40), once in high-salt buffer (50 mM Tris-HCl [pH 7.5], 450 mM NaCl, 0.5% Triton X-100, 0.5% NP-40), and again in low-salt buffer. Beads were resuspended in TRIzol (Life Technologies), RNA was isolated using Direct-zol RNA MiniPrep (Zymo Research), and cDNA was synthesized using random primers with the high-capacity cDNA reverse transcription kit (Life Technologies).
Reverse transcription-quantitative PCR (qPCR).
Histone transcripts were amplified using Ssofast EvaGreen Supermix (Bio-Rad) on a Stratagene MX3000P qPCR system. HIST3H2A-Tot was amplified using the primers 5′-AAGCTCGTGCAAAAGCGAAG-3′ and 5′-TATTCTAGCACAGCCGCCAG-3′. HIST3H2A-NC was amplified using the primers 5′-GCTACTGCCCAAGAAGACCG-3′ and 5′-GTAACTGGCGACAGGCTCAG-3′. HIST1H2BE-NC was amplified using the primers 5′-ACACCAGCTCCAAGTGAGTT-3′ and 5′-AGGTGGTACTGCGGGTACT-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified using the primers 5′-ACGACCCCTTCATTGACCTC-3′ and 5′-GTCTCGCTCCTGGAAGATGG-3′. pri-miRNAs were amplified using TaqMan Universal PCR master mix, no AmpErase UNG (Life Technologies), and TaqMan primers. pri-miR-24 was amplified using mmu-mir-24-2, and pri-miR-155 was amplified using mmu-mir-155 (Life Technologies). For samples sorted for high GFP, cells were sorted on a BD Influx, and relative quantification of RNA was analyzed using the threshold cycle (ΔΔCT) method. For RNA IPs, fold enrichment was quantified as 2(CT untransfected − CT ARS2/ARS2 mutant). Equal pulldown efficiency was verified by Western blotting.
IP.
Cells cotransfected with DROSHA-FLAG or 3×FLAG-ARS2 and eGFP-ARS2 or eGFP-FARB were lysed in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100 [pH 7.5]) for 30 min at 4°C, and debris was removed by centrifugation (20 min at 20, 000 × g). For samples treated with RNase, RNase A was added to the lysis buffer at a concentration of 100 μg/ml. Extracts were incubated with anti-FLAG beads for 2 h at 4°C. Beads were washed three times with lysis buffer-PBS at 1:4 and eluted with 3×FLAG peptide (Sigma-Aldrich). For samples treated with a high-salt wash, beads were washed once in low-salt buffer, once in high-salt buffer, and again in low-salt buffer before elution.
Western blotting.
Cell lysates were resuspended in 2× Laemmli sample buffer, resolved by SDS-PAGE (10%), and transferred to polyvinylidene difluoride (PVDF) membranes. The membrane was blocked in 5% dehydrated milk in Tris-buffered saline–Tween 20 (0.5%) (TBST) for 1 h. Antibodies were diluted in TBST–1% dehydrated milk at the following concentrations: 1:4,000 for mouse antiactin (Sigma), 1:2,000 for rabbit anti-ARS2 (XL12.2), 1:1,000 for mouse anti-FLAG M2 (Agilent Technologies), 1:1,000 for mouse anti-GFP (Roche), and 1:500 rabbit anti-CBP20 (Novus Biologicals).
Immunofluorescence.
To measure histone H3 levels, cells were fixed for 15 min with 1% paraformaldehyde in PBS at 4°C, washed, and incubated in 70% ethanol for ≥1 h. Samples were washed and incubated in rabbit anti-histone H3 (Abcam) at 1:2,000 overnight at 4°C. Samples were then washed and incubated in goat anti-rabbit antibody–AF647 (Life Technologies) at 1:100 for 2 h at 4°C, resuspended in PBS-BSA, and analyzed on a BD FACSCalibur. For fluorescence microscopy, cells were transfected on 6-well dishes with the indicated eGFP-ARS2 constructs. Twenty-four hours posttransfection, cells were live imaged at a magnification of ×40 using a Leica DMIRE2 inverted fluorescence microscope.
Silver stain.
Cell lysates were resuspended in 2× Laemmli sample buffer and resolved by SDS-PAGE (8%). The gel was washed in H2O, fixed for 1 h (40% ethanol, 10% acetic acid, 50% H2O), and washed overnight in H2O on a shaker. The gel was sensitized for 1 min (0.02% Na2S2O3), washed three times in H2O, and incubated in 0.1% AgNO3 for 20 min at 4°C. The gel then washed three times in H2O, developed in 3% Na2CO3 with 0.05% formaldehyde, and, once developed, washed once in H2O, incubated with 5% acetic acid for 5 min, and washed and incubated in gel drying solution (30% methanol, 5% glycerol). The gel was dried overnight in cellophane.
RESULTS
ARS2 is required for cell cycle progression.
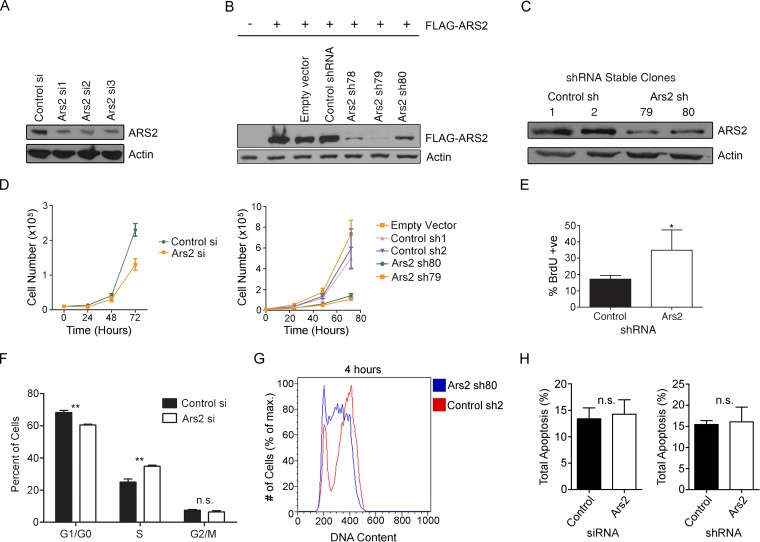
To investigate the role of Ars2 in progenitor cell proliferation, we used the C2C12 myoblast progenitor cell line (22). This cell line was chosen because it is diploid and has progenitor-like properties, including the ability to differentiate in vitro. Given the important role of Ars2 in stem and progenitor cells (20, 23–26), we felt it was important to examine Ars2 function in this context. Knockdown of ARS2 with multiple siRNAs or shRNAs (Fig. 1A to C) reduced the growth of both primary and C2C12 myoblasts (Fig. 1D). ARS2 knockdown increased the accumulation of cells in S phase 48 h posttransfection, as measured by an increased proportion of BrdU-positive cells (Fig. 1E) and by propidium iodide (PI) incorporation using flow cytometry (Fig. 1F). Additionally, C2C12 stable clones expressing Ars2 shRNA accumulated in S phase relative to nontargeting control shRNA-expressing stable clones (Fig. 1G). To demonstrate that the decrease in growth was the consequence of improper cell cycling and not due to an increase in cell death, we also examined apoptosis following ARS2 knockdown using both siRNA and shRNA. As shown in Fig. 1H, decreased expression of ARS2 in C2C12 cells did not increase apoptosis. ARS2-deficient cells do eventually progress through the cell cycle (data not shown), which is consistent with the fact that we were able to maintain stable knockdown cell lines and we did not observe an increase in apoptosis. Taken together, these results indicate ARS2-deficient cells display slower cell cycle kinetics as a result of an S phase delay.
FIG 1.
ARS2 is required for cell cycle progression. (A to C) Western blots show knockdown of endogenous ARS2 in C2C12 myoblasts using three different siRNAs (A), knockdown of FLAG-ARS2 using three different shRNAs (lanes 5 to 7) (B), and knockdown of endogenous ARS2 in C2C12 clones stably expressing Ars2 shRNA79 or shRNA80 (C). (D) ARS2 knockdown decreased cell number in primary myoblasts using siRNA (left) and in C2C12 stable shRNA clones (right). (E) ARS2 knockdown increased the proportion of BrdU-positive cells as measured by immunocytochemistry and fluorescence microscopy 48 h posttransfection. (F) Cell cycle analysis using propidium iodide (PI) and flow cytometry in primary mouse myoblasts 48 h after siRNA transfection shows that ARS2 knockdown increases the proportion of cells in S phase. An unpaired-sample t test at each phase was used to compare means in control and knockdown groups. A Bonferroni correction method was used to modify the P value cutoff to 0.017 (i.e., 0.05/3) to reflect error due to multiple comparisons. For the S phase comparison, t = 8.27 and P < 0.017. (G) C2C12 stable shRNA clones were pulsed with BrdU for 30 min and 4 h later were fixed, labeled with PI and BrdU antibody, and analyzed using flow cytometry. The histogram shows BrdU-positive events. (H) Knockdown of ARS2 with either siRNA or shRNA did not increase levels of apoptosis 24 h posttransfection as measured by PI and annexin V-FITC using flow cytometry. A two-tailed unpaired t test was used to determine whether there was a significant difference between control and Ars2 knockdown in panels E (n = 3, P < 0.05) and H (n = 3, P > 0.05). *, P < 0.05; **, P < 0.01. n.s., not significant. Error bars represent standard deviations (SD).
ARS2 overexpression arrests cells in early S phase.
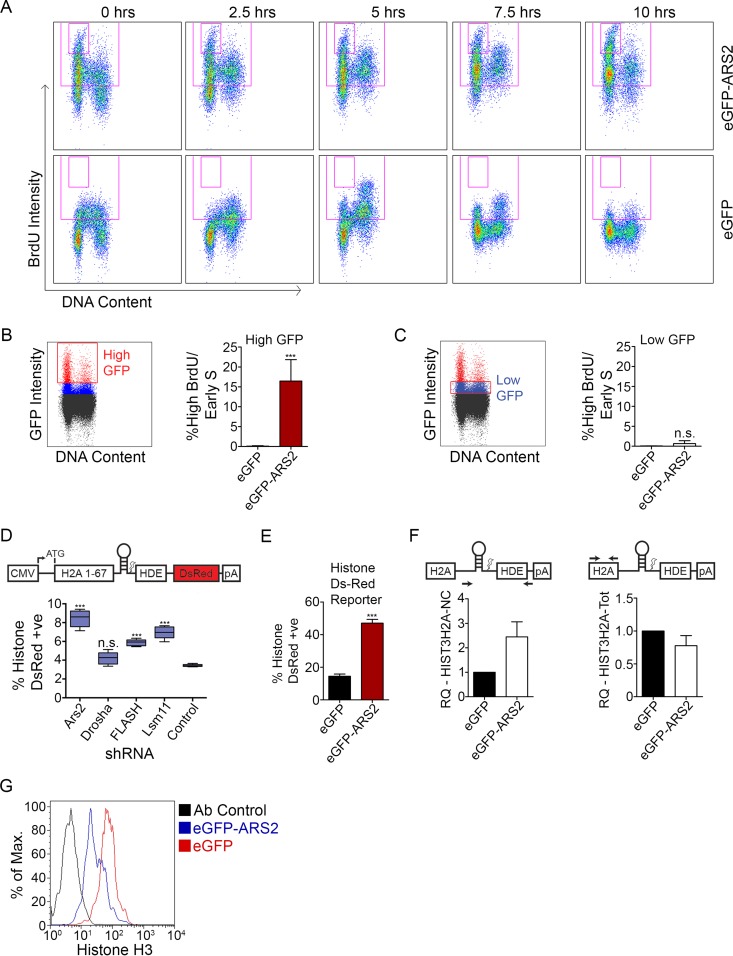
Since ARS2 is a critical component of several multiprotein/RNA complexes, we reasoned that overexpression of ARS2 may generate a dominant negative phenotype. Such dominant negative effects through overexpression are common in genes encoding components of multiprotein complexes, where misexpression of one component can disrupt complex stoichiometry (27). To determine if ARS2 overexpression conferred a dominant negative phenotype, myoblasts were transfected with GFP-ARS2 and pulsed with BrdU for 30 min to identify the cells in S phase, and the DNA content and BrdU incorporation were determined for the GFP population at various time points. As shown in Fig. 2A, overexpression of GFP-ARS2, but not GFP alone, generated a high-intensity BrdU population. This high-BrdU population accumulated in early S phase immediately postpulse (0 h) and failed to progress through the cell cycle in the subsequent 10 h. This effect was evident in the high-level GFP-expressing cells, which presumably also expressed the highest levels of ARS2 (Fig. 2B), whereas low GFP/ARS2 levels did not show this high-BrdU/early-S-phase accumulation (Fig. 2C). Thus, while reducing the levels of ARS2 slows cell cycle progression, the high-level overexpression of ARS2 results in an early S phase cell cycle arrest through a dominant negative effect.
FIG 2.
ARS2 dominant negative affects histone mRNA processing and expression. (A) C2C12 myoblasts transfected with eGFP-ARS2 and eGFP were given a 30-min BrdU pulse, fixed every 2.5 h, and labeled with PI to measure DNA content, and BrdU-positive cells were tracked over time. Cells were gated for GFP; total BrdU-positive events are in the large gate, and high-BrdU/early-S phase cells are in the small inset gate. (B) Cells treated as for panel A were gated for high GFP intensity (red gate). The graph shows the percentage of high-GFP gated cells (average percentage of each time point) contributing to the high-BrdU/early-S-phase-arrested population. (C) Cells treated as for panel A were gated for low GFP intensity (red gate). (D) Cells were cotransfected with shRNA containing a TurboGFP (tGFP) cassette and a histone DsRed reporter that expresses DsRed upon disrupted processing (8, 19). The percent double positive (DsRed and GFP) is shown relative to total transfected cells. (E) Cells were cotransfected with eGFP or eGFP-ARS2 and the histone DsRed reporter and the percent double positive was quantified as for panel D. (F) Noncleaved HIST3H2A (left) and HIST3H2A-Total (Tot) mRNAs (right) were quantified using qPCR with the ΔΔCT method from cells sorted for high GFP intensity. GAPDH was used as a control RNA, and samples were normalized to eGFP samples. (G) Cells were cotransfected with eGFP or eGFP-ARS2 and gated for high GFP intensity, and histone H3 levels were measured using flow cytometry. A two-tailed unpaired t test was used to determine whether there was a significant difference between eGFP and eGFP-ARS2 in panel B (n = 5; P < 0.001), panel C (n = 5; P > 0.05), and panel E (n = 5; P < 0.001). A one-way analysis of variance (ANOVA) (F[4, 20] = 56.79; P < 0.001) followed by Tukey's multiple-comparison post hoc test was used to determine whether there were significant differences between groups in panel D. ***, P < 0.001. n.s., not significant. Error bars represent SD.
ARS2 is required for histone processing and expression.
The high BrdU population associated with ARS2 overexpression is intriguing, since arrest in early S phase should decrease the amount of BrdU incorporated per cell. However, the increase in BrdU labeling suggested a likely mechanism for the arrest. Since ARS2 is required for RDH expression (12, 13), we reasoned that the increase in BrdU signal we observe may simply reflect an increase in the accessibility of antibody to BrdU-DNA due to a histone deficiency and under packaged chromatin. To examine this, we knocked down ARS2, or components of the RDH mRNA processing machinery, FLASH and LSM11, and assessed histone mRNA processing using a reporter in which DsRed expression is dependent on improper histone 3′-UTR processing (8) (Fig. 2D). This assay revealed elevated levels of reporter expression following ARS2, FLASH, and LSM11 knockdowns but not in DROSHA knockdowns (Fig. 2D), confirming that DROSHA is not involved in RDH mRNA processing. Overexpression of ARS2 similarly increased expression of the reporter, suggesting improper processing of histone transcripts (Fig. 2E). To further evaluate this, we isolated the high-GFP populations and assessed the levels of endogenous noncleaved histone H2A transcripts using qPCR with primers that amplify across the 3′-UTR cleavage site. Overexpression of GFP-ARS2 increased the levels of misprocessed H2A transcripts (Fig. 2F). Consistent with previous reports, the levels of total H2A transcripts declined slightly (Fig. 2F) (12, 13). In addition, defective RDH transcript processing led to a decrease in histone H3 protein levels following ARS2 overexpression (Fig. 2G). Although we cannot rule out contributions to the cell cycle phenotype from other types of RNA processed by ARS2 with known roles in the cell cycle (28–30), histone deficiency during S phase and the inability to package chromatin caused by improper processing of histone RNAs are likely major contributors to the S phase arrest associated with high levels of ARS2 overexpression.
Predicted structures of ARS2 and RRM domains.
To help understand how SERRATE functions, Machida et al. determined its crystal structure (11), revealing a unique “walking man-like” topology of 3 domains: N-terminal, Mid, and zinc finger (ZnF) domains, which form the leading leg, body, and lagging leg, respectively (11) (Fig. 3A). Both the N and C termini of the protein are unstructured. The recent crystal structure of Arabidopsis SERRATE created an opportunity to model the mouse ARS2 sequence and reexamine the conserved sequences of ARS2 for additional functional groups. Although the overall identity between mouse ARS2 and Arabidopsis SERRATE is relatively low (∼28%), bioinformatic analysis showed that the DUF3546, Mid, and ZnF domains are conserved (Fig. 3D). However, ARS2 diverges considerably from SERRATE at the unstructured C terminus and, interestingly, contains an insert within the Mid domain that is not present in SERRATE (Fig. 3D). Alignment of this insert within metazoans shows that it contains an unstructured glutamate-rich region that is followed by an RNA recognition motif (RRM). Homology modeling of the mouse ARS2 RRM predicted a fold similar to the RRM domain of splicing factor 3B subunit 4 (SF3B4) (Z score of 14.9; root mean square deviation [RMSD] of 0.1 Å; 73 Cα) (Fig. 3B).
FIG 3.
Predicted structures of ARS2 and RRM domains. (A) Crystal structure of Arabidopsis thaliana SERRATE (11) (PDB accession code 3AX1) highlighting the DUF3546 domain (green), Mid domain (blue), and ZnF domain (purple). (B) Mus musculus ARS2 RRM domain (blue-green) was modeled using SF3B4 as a reference (PDB accession code 1X5U, shown in white) (37). The mutated residues of ARS2 RRM are highlighted. (C) The Mus musculus ARS2 amino acid sequence (without the glutamate-rich and RRM domain insert) was modeled using Arabidopsis SERRATE as a reference (37). Domains are highlighted as in panel A with the exception of the disordered region colored in red, which is where the glutamate-rich region and RRM domain are predicted to be inserted. Also highlighted are the residues chosen to be mutated to alanines, in addition to the residues predicted to coordinate zinc. (D) Bioinformatic alignment of SERRATE/ARS2 using Clustal Omega (38) and manual alignment. Asterisks indicate amino acids that were mutated, arrowheads indicate residues predicted to coordinate Zinc, and the arrow indicates the C-terminal region that was deleted in ARS2-ΔC. The regions that correspond to structural motifs or domains are outlined with boxes and labeled above the alignment. NLS, predicted nuclear localization sequence. PXXP motifs are underlined below the alignment.
We next modeled the regions that were conserved between mouse ARS2 (minus the unstructured N and C termini, and glutamate-rich and RRM domains) and SERRATE. The computational model conforms very well to the previously described SERRATE structure (Z-score of 30.1; RMSD of 0.7 Å; 279 Cα) (Fig. 3A and C). Importantly, the glutamate-rich and RRM domains are expected to insert into the region of ARS2 between the predicted α-helices 4 and 5 (Fig. 3C and D). The orientation of the RRM domain relative to the core ARS2 structure is unknown. C terminal to the ZnF in metazoans is a proline-rich region. Eight PXXP motifs are present within the unstructured C terminus of mouse ARS2 (amino acids 763 to 875) (Fig. 3D). These motifs often mediate protein interactions, suggesting that the C terminus may perform this role. In summary, the core of metazoan ARS2 consists of 4 domains: a DUF3546 domain, a newly identified RRM domain, a Mid domain, and a ZnF domain.
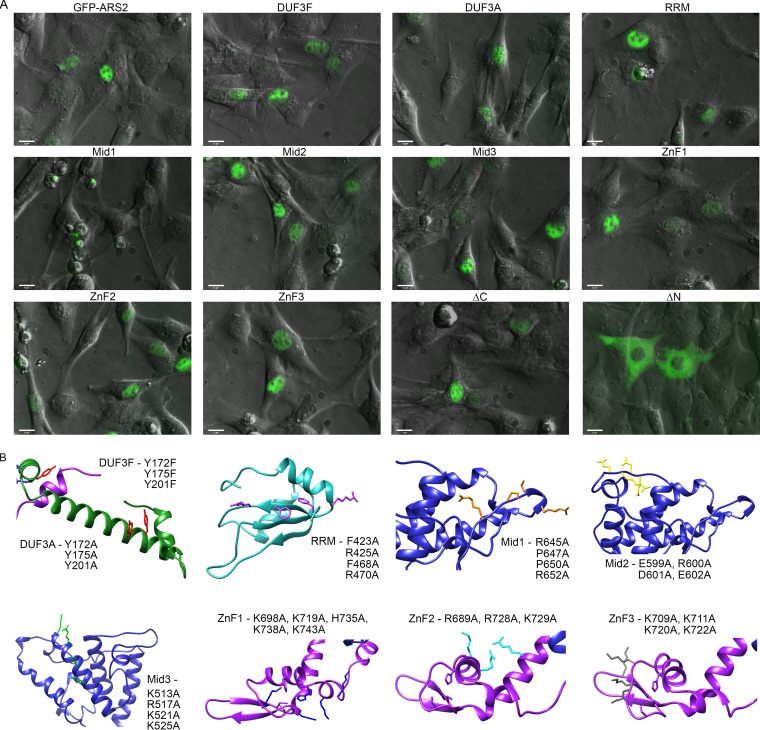
The strong dominant negative phenotype associated with ARS2 overexpression created an opportunity for screening mutations in ARS2 functional domains. Specifically, we asked if conserved amino acids in these regions could abrogate or exacerbate the ARS2 overexpression phenotype. If so, these mutations would reveal the importance of the different ARS2 domains. Since deletions beyond the unstructured N and C termini affect the overall fold of the protein (11), we decided to mutate several conserved motifs within the DUF3546, RRM, Mid, and ZnF domains. We also generated deletions of the N and C termini. Mutation sites, shown in Fig. 3D and 4B, were selected based on their conservation and predicted surface exposure. Because of the large size of the protein and paucity of interaction data, we mutated 3 or 4 residues at a time to ensure that mutations would abolish function. Each mutant was expressed as a GFP-ARS2 fusion and its localization was observed (Fig. 4A). Only mutants that exhibited wild-type localization and expression level and retained some ability to associate with protein and/or RNA were included in our phenotypic analysis. Deletion of the N-terminal residues (amino acids 1 to 227) resulted in cytoplasmic localization (Fig. 4A), confirming the presence of a nuclear localization signal within this region (20). We focused our analysis on mutants with nuclear expression and their effects on the RDH and miRNA biogenesis pathways, as well as the CBC, since ARS2 has been shown to form direct protein interactions with these complexes.
FIG 4.
ARS2 mutant localization. (A) C2C12 cells transfected with eGFP-ARS2 or the indicated mutants were live imaged at a magnification of ×40. Shown is an overlay of GFP and phase contrast. Bars, 15 μm. (B) Using the predicted model structures of ARS2 from Fig. 3B and C, the positions of the mutated amino acids are highlighted using the same color scheme as in Fig. 3D, and the names of the specific mutations are shown adjacent to their respective locations within the structure.
The ZnF domain mediates interactions with FLASH and RNA.
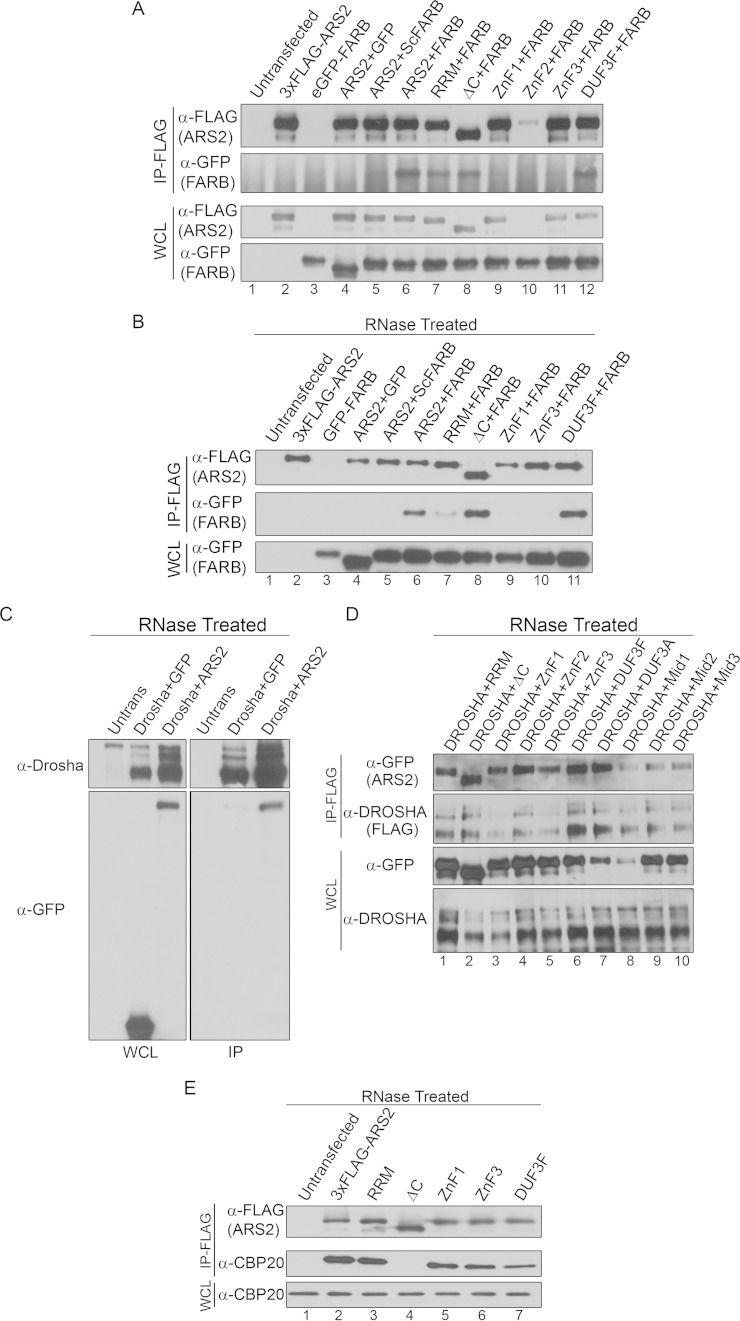
Amino acids 550 to 876 of ARS2 (ZnF and proline-rich region) have previously been shown to be important for RNA and FLASH/FARB binding (13). To refine the region of ARS2 required for FLASH interaction, we tested the ability of ARS2 ZnF mutants to interact with FLASH. Three groups of mutations were generated within the ZnF, either in the predicted double-stranded RNA (dsRNA) interface (ZnF1) or in basic regions (ZnF2 and ZnF3) that might associate with the highly acidic FARB peptide (DELEEGEIRSDDE) (Fig. 3C and D and 4B). As endogenous FLASH protein was difficult to detect in C2C12 cells, we used FARB peptide to map the ARS2 region required for FLASH interaction. As shown in Fig. 5A and B, ZnF1 and ZnF3 mutants lost the ability of ARS2 to coimmunoprecipitate (co-IP) with FARB. RNase treatment demonstrates that this effect is not mediated by RNA (Fig. 5A and B). Since the ZnF2 did not express well (Fig. 5A, lane 10), it was not pursued further. In contrast, ZnF1 and ZnF3 maintained the ability to bind to DROSHA (Fig. 5C and D) and CBP20 (Fig. 5E), confirming that these proteins remain folded. Thus, FLASH interaction with ARS2 requires these conserved features of the ZnF domain.
FIG 5.
Mapping of ARS2 protein interactions. (A) Cells were cotransfected with 3×FLAG-ARS2 (or 3×FLAG-ARS2 mutants) and either eGFP control (GFP), eGFP-ScrambledFARB (ScFARB), or eGFP-FARB. IP was carried out using 3×FLAG beads. (B) Cells were transfected and immunoprecipitated as for panel A but were treated with 100 μg/ml of RNase A. (C and D) Cells cotransfected with DROSHA-FLAG and eGFP, eGFP-ARS2, or eGFP-ARS2 mutants were treated with 100 μg/ml of RNase A and immunoprecipitated using 3×FLAG beads. Untrans, untransfected. (E) C2C12 cells transfected with 3×FLAG-ARS2 or the indicated mutants were treated with 100 μg/ml of RNase A and immunoprecipitated using FLAG beads.
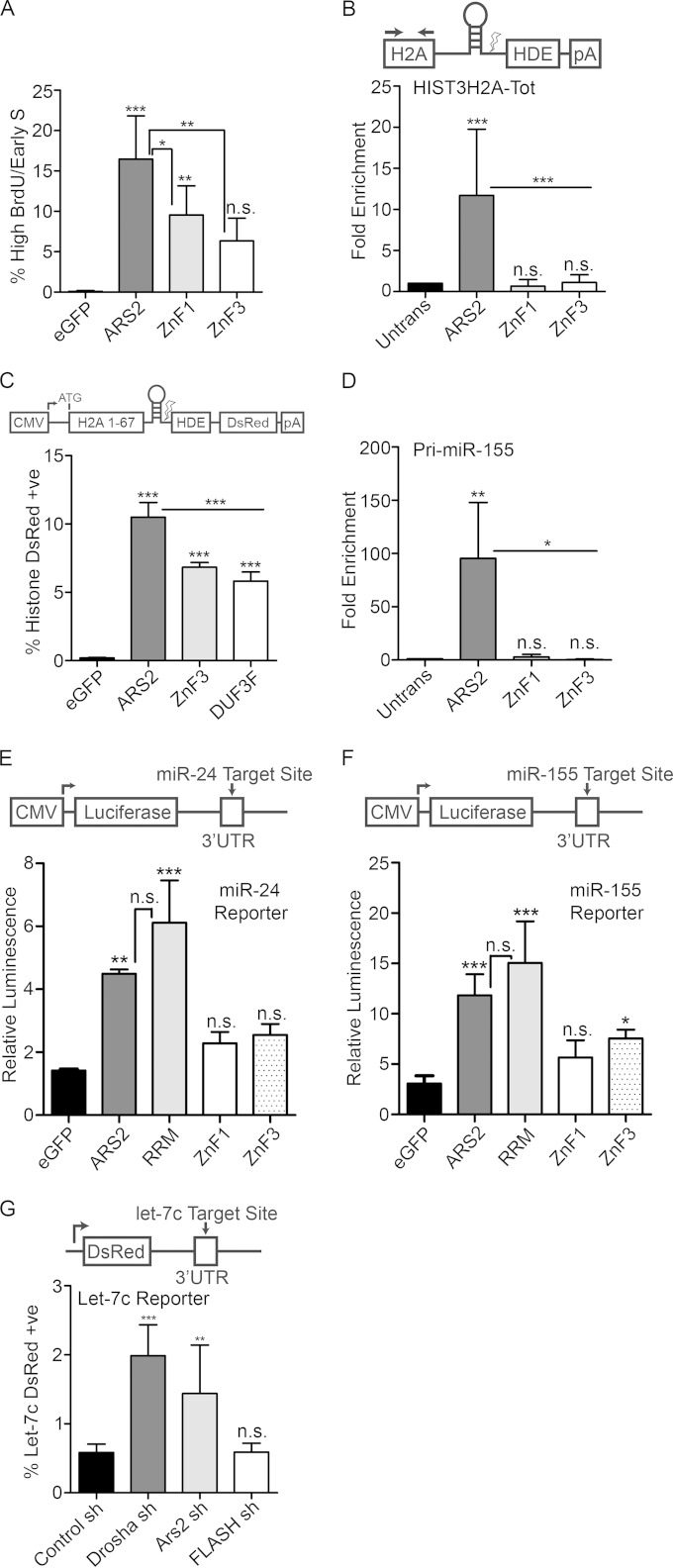
We also examined whether the ZnF mutants were able to induce ARS2 dominant negative phenotypes. Consistent with the ZnF participating in RDH mRNA processing and FLASH interaction, ZnF1 and ZnF3 were less effective at inducing the ARS2 S phase arrest (Fig. 6A). We next determined whether the ZnF was important for RDH mRNA recognition. Interestingly, mutation of the ZnF domain reduced the ability of ARS2 to IP endogenous histone mRNA (Fig. 6B) and partially rescued RDH transcript processing, as measured by the DsRed reporter assay (Fig. 6C). Thus, the ARS2 ZnF plays a critical role in the recognition, and possibly processing, of RDH mRNA.
FIG 6.
The zinc finger domain mediates interaction with RNA. (A) Cells transfected with eGFP or eGFP-ARS2, -ZnF1 or -ZnF3 were pulsed with BrdU, fixed every 2.5 h, and gated for high GFP. Shown is the percentage of gated cells in the high-BrdU/early-S-phase-arrested population (average percent for each time point). (B) Cells transfected with 3×FLAG-ARS2 or the indicated mutants were immunoprecipitated, and the fold enrichment of RNA pulldown was quantified relative to that for untransfected samples using qPCR for HIST3H2A-Tot and primers upstream of the cleavage site. (C) Cells cotransfected with eGFP or eGFP-ARS2, -DUF3F, or -ZnF3 and histone DsRed were quantified as in Fig. 2E. (D) Samples were treated as for panel B and amplified using primers for pri-miR-155. (E and F) Cells were transfected with eGFP and eGFP-ARS2, -RRM, -ZnF1, or -ZnF3 along with the indicated firefly luciferase reporter and Renilla luciferase. Each firefly luciferase reporter contains an miRNA target site of perfect complementarity in its 3′ UTR. Firefly luminescence was normalized to Renilla luminescence. (G) Cells were cotransfected with shRNA containing a tGFP cassette and a let-7c DsRed reporter which contains a let-7c binding site in the 3′ UTR. The percent double positive (DsRed and GFP) is shown relative to total transfected cells. A one-way ANOVA was performed for panels A to G, with results as follows: F(3, 16) = 18.66 and P < 0.001 (A), F(3, 18) = 11.28 and P < 0.001 (B), F(3, 16) = 210.4 and P < 0.001 (C), F(3, 8) = 9.65 and P< 0.01 (D), F (4, 10) = 26.18 and P < 0.001 (E), F(4, 25) = 26.90 and P < 0.001 (F), and F(3, 20) = 22.84 and P < 0.001 (G), followed by Tukey's multiple-comparison post hoc test to determine whether there were significant differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant. Error bars represent SD.
The ZnF of SERRATE has been implicated in miRNA biogenesis in plants (10, 11). We examined whether the ZnF domain was required for miRNA biogenesis in mammalian ARS2. Mutations in the ZnF abrogated miRNA interaction (Fig. 6D) and overexpression of these mutants did not elicit a miRNA biogenesis defect (Fig. 6E and F), indicating an additional role for this region in miRNA biogenesis. Since FLASH potentially binds to the same domain of ARS2 as miRNA, we next asked whether FLASH was required for miRNA biogenesis using a let-7c reporter. While knockdown of DROSHA or ARS2 resulted in the expected decrease in mature miRNA, knockdown of FLASH did not affect these miRNA levels (Fig. 6G). Together, these results indicate that the ARS2 ZnF mediates interactions with miRNA, RDH mRNA, and FLASH.
The DUF3546 domain is required for miRNA and histone mRNA pulldown.
The DUF3546 domain forms the leading leg of the protein and consists of a helix-turn-helix-turn-helix motif. Notably, it contains 3 invariant tyrosine residues in helices α2 and α3 (Tyr residues 172, 175, and 201), which we mutated to alanine or phenylalanine (Fig. 3C and D and 4B). Mutating these residues to alanine resulted in poor expression and protein mislocalization, with ARS2 forming nuclear aggregates indicative of protein misfolding (Fig. 4A). However, mutating these tyrosines to phenylalanine yielded an ARS2 (3F) mutant with distribution and expression levels similar to those of wild-type GFP-ARS2 (Fig. 4A).
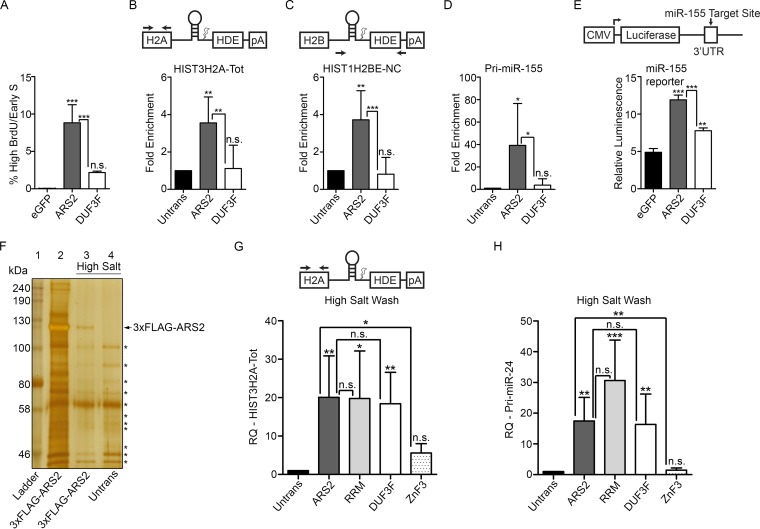
To assess the function of the DUF3546 domain's conserved tyrosines, we first examined the phenotypic consequences of the DUF3F mutant in cell cycle progression. Upon overexpression, the DUF3F mutation was not able to induce the cell cycle arrest associated with wild-type ARS2 overexpression (Fig. 7A). This demonstrates that these amino acids in the DUF3546 domain are required for the dominant negative effect of ARS2 overexpression. To investigate the mechanism behind this effect, we first examined the ability of this mutant to interact with RDH mRNA. Interestingly, the 3F mutations impaired ARS2's ability to IP both total and noncleaved misprocessed RDH mRNAs (Fig. 7B and C). This mutation also partially rescued RDH mRNA processing, as measured by the DsRed reporter (Fig. 6C). We next assessed whether the 3F mutant's inability to pull down RDH mRNA was specific for this type of RNA or was impaired in miRNA interaction as well. Similar to the RDH mRNA, this mutant was impaired in its ability to IP miRNA (Fig. 7D), and as a consequence, the ability of this mutant to affect miRNA biogenesis was significantly reduced, as measured by an miR-155 luciferase reporter (Fig. 7E). In contrast, the ability of this mutant to interact with ARS2 protein binding partners—the FLASH peptide, FARB (Fig. 5A and B), DROSHA (Fig. 5C and D), or CBP20 (Fig. 5E)—was not affected, even in the presence of RNase, confirming that these interactions were RNA independent and the mutant remained folded. Together, these data indicate that the DUF3546 domain is necessary, directly or indirectly, for ARS2 interactions with miRNA and histone mRNA.
FIG 7.
The DUF3546 domain is required for miRNA and histone mRNA pulldown. (A) C2C12 Cells transfected with eGFP or eGFP-ARS2 or -DUF3F were treated as in Fig. 2A and gated as in Fig. 2B. (B to D) Cells were transfected with 3×FLAG-ARS2 or -DUF3F, protein was immunoprecipitated using 3×FLAG beads, RNA was extracted from the immunoprecipitates, and the fold enrichment of RNA pulldown was quantified relative to untransfected samples using qPCR for HIST3H2A-Tot (B), HIST1H2BE-noncleaved (NC) (C), and pri-miRNA-155 (D). For HIST3H2A-Tot, primers amplified the histone mRNA upstream of the cleavage site, and for HISTH2BE-NC, primers span the cleavage site. (E) Cells were cotransfected with eGFP, ARS2, or DUF3F, miR-155 firefly luciferase, and Renilla luciferase. Firefly luminescence was normalized to Renilla luminescence. (F) Silver stain of elution following IP of 3×FLAG-ARS2 under normal conditions (lane 2) or following a high-salt wash (lane 3). An untransfected sample treated with a high-salt wash was used as a negative control (lane 4). Asterisks indicate nonspecific bands present under both ARS2 and untransfected conditions (lanes 3 and 4). (G and H) Cells transfected with the indicated plasmids were immunoprecipitated following a high-salt wash, RNA was extracted, and HIST3H2A-Tot (G) or pri-miR-24 (H) was quantified using qPCR with the ΔΔCT method. GAPDH was used as a control RNA, and samples were normalized to the untransfected control. A one-way ANOVA was performed, with the following results: F(2, 12) = 52.73 and P < 0.001 (A), F(2, 15) = 10.82 and P < 0.01 (B), F(2, 15) = 14.81 and P < 0.001 (C), F(2, 15) = 5.75 and P < 0.01 (D), F(2, 6) = 142.1 and P < 0.001 (E), F(4, 22) = 7.89 and P < 0.001 (G), and F(4, 22) = 13.33 and P < 0.001 (H), followed by Tukey's multiple-comparision post hoc test to determine whether there were significant differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant. Error bars represent SD.
To assess whether ARS2 interacts directly with histone mRNA and miRNA, IPs were subject to a high-salt wash, which effectively removed all detectable ARS2-interacting proteins as determined by silver staining (Fig. 7F). Wild-type ARS2 remained associated with miRNA and histone mRNA after a high-salt wash (Fig. 7G and H). Consistent with its inability to IP RDH and miRNAs under milder conditions (Fig. 6B and D), the high-salt-washed ZnF mutant did not associate with RDH mRNA or miRNA (Fig. 7G and H). However, we found that when copurifying factors were removed from ARS2, the DUF3F mutant retained both miRNA and RDH mRNA binding ability (Fig. 7G and H). The contrasting results between the low- and high-salt washes with the DUF3F mutant suggest that this domain may facilitate ARS2-RNA interactions in vivo but is not a necessary component of the protein-RNA interface. Regardless, since purified ARS2 captures RNA as efficiently as ARS2-multiprotein complexes, it is likely that ARS2 binds directly to miRNA and RDH mRNA and that this interaction requires the ZnF domain.
The RRM domain is required for cell cycle progression and is involved in FLASH interaction.
During our bioinformatics analysis, we identified a previously undescribed RRM domain within metazoan ARS2. Canonical RRM domains have two conserved sequence motifs: RNP1 ([RK]-[G]-[FY]-[GA}-[FY]-[ILV]-[X]-[FY]) and RNP2 ([ILV]-[FY]-[ILV]-X-N-L) (31). Critical aromatic residues (in bold) of RNP1 and RNP2 form stacked pi interactions with RNA bases, and the arginine or lysine of RNP1 forms multiple hydrogen bonds with nitrogenous bases (32). Noncanonical RRMs diverge from the consensus as they utilize other interfaces to interact with RNA or protein (33). Analysis of ARS2 indicates that it contains a conserved RNP2-like motif (LFMRNI). The RNP1 (RGWVTFDR) deviates from the consensus (Fig. 3D), and it is therefore unclear whether the ARS2 RRM mediates RNA binding.
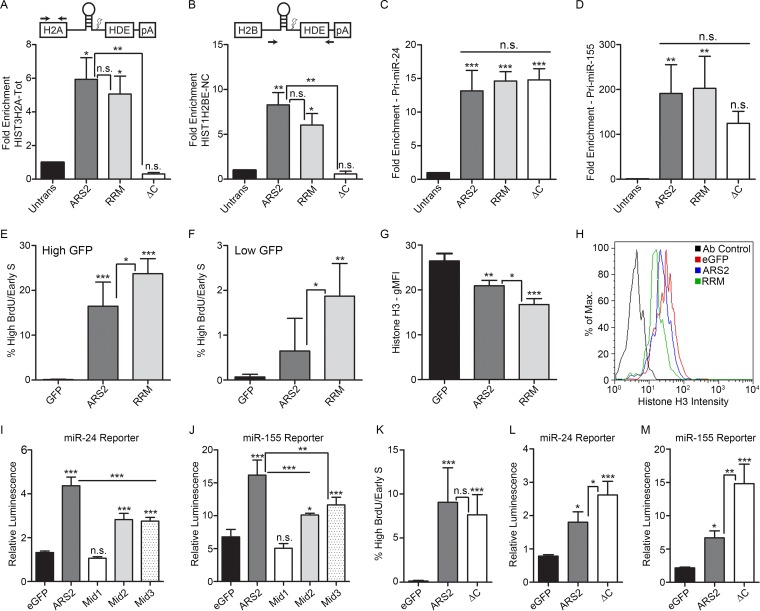
Based on the modeled RRM structure, we mutated conserved residues within RNP1 (F468A and R470A) and RNP2 (F423A and R425A) (Fig. 3B and D and 4B). This mutant expressed at wild-type levels and showed normal localization (Fig. 4A). However, these mutations had no effect on histone mRNA or miRNA pulldown under low- or high-salt wash conditions (Fig. 7G and H and 8A to D).
FIG 8.
The RRM domain is required for cell cycle progression, and the Mid domain and C terminus are important for miRNA biogenesis. (A to D) Cells transfected with 3×FLAG-ARS2, -RRM Mut (RRM), and -ΔC were immunoprecipitated, and the fold enrichment of RNA pulldown was quantified as in Fig. 6B for HIST3H2A-Tot (A), HIST1H2BE-NC (B), pri-miR-24 (C), and pri-miR-155 (D). (E and F) Cells transfected with eGFP, eGFP-ARS2, or eGFP-RRM were treated as in Fig. 2A. Cells were gated for either high GFP (E) or low GFP intensity (F), and the percentage of gated cells in the high-BrdU/early-S-phase-arrested population is shown (average percent for each time point). (G) Cells were transfected as for panel E, and histone H3 levels were analyzed using flow cytometry. The geometric mean fluorescence intensity (gMFI) is shown for the high-GFP gated population. (H) Representative samples from panel G are shown as a histogram overlay for histone H3 fluorescence intensity. (I and J) Cells were cotransfected with eGFP, eGFP-ARS2, or eGFP-Mid mutants along with the indicated firefly luciferase reporter and Renilla luciferase. Firefly luminescence was normalized to Renilla luminescence. (K) Cells transfected with the indicated plasmids were treated as in Fig. 2A, gated, and quantified as in Fig. 2B. (L and M) Cells were transfected with eGFP or eGFP-ARS2 or -ΔC along with the indicated firefly luciferase reporter and Renilla luciferase. A one-way ANOVA was performed, with the following results: F(3, 8) = 16.00 and P < 0.01 (A), F(3, 8) = 11.48 and P < 0.01 (B), F(3, 8) = 37.96 and P < 0.001 (C), F(3, 8) = 10.33 and P < 0.01 (D), F(2, 12) = 54.86 and P < 0.001 (E), F(2, 12) = 11.97 and P < 0.01 (F), F(2, 6) = 36.23 and P < 0.001 (G), F(4, 25) = 192.9 and P < 0.001 (I), F(4, 14) = 44.05 and P < 0.001 (J), F(2, 12) = 16.78 and P < 0.001 (K), F(2, 6) = 28.62 and P < 0.001 (L), and F(2, 6) = 38.14 and P <0.001 (M), followed by Tukey's multiple-comparison post hoc test to determine whether there were significant differences between groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001. n.s., not significant. Error bars represent SD.
We next asked if the RRM is required for the ARS2 cell cycle progression phenotype. Expression of the GFP-ARS2 RRM mutant resulted in an increase in the early-S-phase-arrested/high-BrdU population compared to wild-type GFP-ARS2 (Fig. 8E). With this mutant, even low-level GFP-ARS2-expressing populations resulted in high BrdU incorporation and cell cycle arrest in early S phase (Fig. 8F). Additionally, the RRM mutant resulted in decreased levels of histone H3 protein relative to those with wild-type ARS2 (Fig. 8G and H), in agreement with histone deficiency being a major contributor to the cell cycle phenotype.
Consistent with the strong effect of this mutation on cell cycle progression and histone expression, the ability of the RRM mutation to interact with FLASH/FARB was reduced, but not lost, under RNase treatment conditions (Fig. 5B, lane 7). In contrast, the interactions with DROSHA (Fig. 5D, lane 1) and CBP20 (Fig. 5E, lane 3) were not affected, indicating that global destabilization of the protein as a result of the RRM mutations did not occur. In addition, two independent miRNA biogenesis reporters did not show a significant change as a result of this mutation, suggesting that this domain is not required for miRNA biogenesis (Fig. 6E and F). Finally, the reduced binding of the RRM mutant to FARB suggests that this domain and the ZnF domain are each important for FLASH interaction. Further mutagenesis is required to determine the site of interaction and how these two domains mediate this interaction.
The Mid domain is important for miRNA biogenesis.
In Arabidopsis SERRATE, the Mid domain comprises a 3-helix bundle (α5 to α7) orientated orthogonally against a long fourth alpha helix (α4) (11). The long unstructured loop regions between each of these three helices were predicted to mediate protein-protein interactions (11). Within the Mid domain, we generated three groups of mutations that are each based on well-conserved residues: Mid1 (R645A, P647A, P650A, and R652A), Mid2 (E599A, R600A, D601A, and E602A), and Mid3 (K513A, R517A, K521A, and K525A) (Fig. 3C and D and 4B). DROSHA specifically coimmunoprecipitates wild-type GFP-ARS2 in the presence of RNase, confirming that this interaction is not RNA dependent (7) (Fig. 5C). Under the same conditions, all Mid domain mutations reduced, but did not abolish, DROSHA binding (Fig. 5D, lanes 8 to 10), suggesting that these conserved residues may contribute to the DROSHA interaction surface. The Mid1 mutant did not express well and displayed mislocalization (Fig. 4A and 5D, lane 8), likely explaining its reduced binding to DROSHA and rescue of the miR-24 and miR-155 reporters (Fig. 8I and J). However, Mid2 and Mid3 mutants expressed and localized normally (Fig. 4A and 5D), suggesting that their ability to interact with DROSHA is compromised. Consistent with this interpretation, Mid2 and Mid3 mutants had a markedly reduced RNA-misprocessing phenotype (Fig. 8I and J), i.e., an impaired ability to elicit a dominant negative effect on this pathway. Taken together, these results suggest that the DROSHA binding site likely lies within the Mid domain. Further mutagenesis will be required to determine the precise binding site.
The ARS2 C terminus is required for CBP20 interaction.
Hallais et al. recently showed that C-terminal amino acids 502 to 871 of mammalian ARS2 are sufficient for binding to the CBC (8). Using a deletion spanning amino acids 763 to 875, we refined this region and found that the unstructured C-terminal proline-rich region is necessary for CBP20 co-IP (Fig. 5E, lane 4). We next utilized the cell cycle defect associated with ARS2 overexpression to assay the involvement of the C terminus in this phenotype. Perhaps surprisingly, deletion of the proline-rich C terminus did not affect the cell cycle arrest, as overexpression of the deletion mutant blocked cell cycle progression to the same degree as wild-type ARS2 (Fig. 8K). This was despite the C-terminal deletion abolishing the ability of ARS2 to IP histone mRNA (Fig. 8A and B). Notably, these data indicate that sequestration of RDH mRNA by ARS2 is not the major mechanism contributing to the dominant negative cell cycle phenotype. This suggests that while the C terminus is required for RDH mRNA interaction, the intact domains in ARS2-ΔC (i.e., the RRM, ZnF, and/or DUF3546 domain) may sequester proteins necessary for RDH processing and/or expression. Our results with the RRM and ZnF mutants strongly implicate these domains. Furthermore, while the ZnF and DUF3546 domains are necessary for RDH mRNA pulldown, they are not sufficient, as ARS2 requires an interaction with the CBC to tightly associate with this RNA class (Fig. 8A and B).
We next examined whether the C-terminal proline-rich region (amino acids 763 to 875) was required for miRNA biogenesis. As shown in Fig. 8L and M, ARS2 requires this region for miRNA biogenesis, since deletion of the C terminus results in a more severe dominant negative. However, the C-terminal deletion mutant can still interact with pri-miRNA (Fig. 8C and D). This suggests that although ARS2-ΔC retains the ability to interact with pri-miRNA, likely through the DUF3546 and ZnF domains, the C terminus of ARS2 and, presumably, the interaction with the CBC are required for pri-miRNA processing.
DISCUSSION
ARS2 knockdown has been shown to disrupt RDH mRNA processing and cause a reduction in histone expression, leading to disruption of the cell cycle (12, 13). Consistent with this, we found that myogenic cells deficient in ARS2 cycle more slowly and spend an increased time in S phase. The more severe S phase arrest phenotype associated with ARS2 overexpression reflects the ability of the dominant negative to more severely compromise ARS2 function versus RNA interference (RNAi). Many types of RNAP II transcripts controlled by ARS2 likely contribute to the cell cycle phenotype of altering ARS2 levels. Indeed, microRNA (28), mRNA splicing (29, 30), and replication-dependent histones are all important for cell cycle progression (13, 34). However, the distinctive phenotype of the high-BrdU-containing cells in early S phase points to defective chromatin packaging as a consequence of histone deficiency as a major contributor for this unique population within the overexpressing cells.
We have generated the first models of mammalian ARS2 and its RRM domain and designed mutations that disrupt the function of each of ARS2's domains. These models, coupled with the finding that ARS2 overexpression generates a dominant negative, allowed us to screen these mutations for the ability to disrupt known protein interactions, or to change the magnitude of the dominant negative effect of ARS2 overexpression on miRNA and RDH mRNA biogenesis, RNA binding, and cell cycle progression. Through this process, we identified two types of mutations: those that exacerbate the dominant negative effect by forming nonfunctional complexes better than the wild type and those that lose the ability to evoke a dominant negative effect because they lose the ability to sequester key interacting partners. Both of these types of mutations are expected to represent regions in ARS2 that mediate protein or RNA interactions. Although our models proved useful for our experimental rational design, a crystal structure of mammalian ARS2 is needed to gain insight into how the RRM is oriented relative to the core structure.
Our data show that the DUF3546 domain of ARS2 indirectly contributes to RDH mRNA and miRNA binding. Although the DUF3546 was required for ARS2 to associate with these classes of RNA under low-salt conditions where ARS2 complexes were intact, this domain was not required when ARS2 was purified under high-salt wash conditions. This suggests that the DUF3546 domain likely facilitates protein interactions, or conformations of ARS2, that permit RNA interaction with the ZnF domain. A similar requirement for both the DUF3546 and ZnF of SERRATE for miRNA interaction was found in plants (10, 11).
The ZnF domain of ARS2 is critical for both miRNA and RDH mRNA interaction. The mutations in this domain obviated the ARS2 dominant negative in miRNA and RDH mRNA biogenesis, as well as the high-BrdU/cell cycle phenotypes. This domain was necessary for miRNA and RDH mRNA interaction under both low- and high-salt wash conditions. Thus, these RNA interactions are not dependent on other interacting proteins, which is evidence that the ZnF domain is the principal interface for these classes of RNA. Further work with recombinant ZnF domain is required to determine the mechanism of RNA binding and confirm a direct interaction.
The ARS2 complexes involved in processing the different RNA classes are thought to be mutually exclusive (16). We speculate that differences in the manner in which the different classes of RNA interact with ARS2 may induce conformational changes. Interestingly, the ZnF domain was also found to be necessary for binding the FLASH peptide FARB. Since it is unlikely that FLASH binds to the same interface of the ZnF at the same time as histone mRNA, one possibility that accounts for the loss of FLASH binding with the ZnF mutations is that histone mRNA binding is a prerequisite for FLASH binding at another site, possibly within the RRM domain, or another site within the ZnF.
The proline-rich C terminus of ARS2 is required for CBC interaction (8). We refined the region involved to amino acids 743 to 875 of ARS2. Although deletion of the C terminus impaired RDH mRNA pulldown, the overexpression of ARS2-ΔC still generated the dominant negative high-BrdU cell cycle phenotype. This suggests that ARS2-ΔC remains capable of sequestering key proteins involved in RDH processing through its other intact regions. Conversely, the ARS2 C terminus was not required to IP miRNA but was involved in miRNA biogenesis, as ARS2-ΔC exacerbated this phenotype, suggesting that ARS2 interaction with the CBC is critical for proper miRNA biogenesis.
The RRM domain is found only in metazoan ARS2, implying that it confers unique functionality to ARS2 not found in the plant orthologue SERRATE. We showed that the RRM mutant exacerbated the high-BrdU cell cycle phenotype and further reduced histone H3 levels relative to those with wild-type ARS2, suggesting that this domain may be specifically required for RDH mRNA processing. Consistent with this interpretation, we found no observable effects of this mutation on the miRNA biogenesis phenotypes. Our data suggest that the RRM domain is involved in interactions with histone mRNA 3′-end processing machinery, since the FARB peptide showed reduced binding to the RRM mutant in the presence of RNase. We found no effect of the RRM mutant on binding to RDH mRNA or miRNA, nor does it appear that the RRM-RNA interaction is masked by binding to an interacting partner, since the high-salt wash also showed no effect on ARS2 interaction with RNA. Since we mutated the canonical RNA interface, we cannot rule out that the RRM interacts with these RNA through another interface. However, the lack of conservation of RNP1 and the effect on FARB binding suggests that the RRM domain likely mediates protein interactions. In this manner, the ARS2 RRM bears similarity with the Y14 RRM, which specifically interacts with MAGOH protein, and the UPF3 RRM, which interacts with UPF2 but not RNA (35, 36).
The Mid domain of ARS2 is the largest part of the protein and was predicted to be a platform for protein-protein interactions (11). We found that this domain was important for miRNA biogenesis, as the mutations we made partially negated the dominant negative effect of ARS2 on miR-24 and miR-155 levels. One possible mechanism to account for this finding is that this domain is involved in DROSHA interaction. None of the mutations completely disrupted the binding to DROSHA, but we found reduced binding in the presence of RNase with two mutations affecting different regions of the protein. One potential explanation for this result is that these mutations locally disrupt the structure of the Mid domain and thereby diminish the interaction with DROSHA.
We have shown that ARS2 can interact with at least two different types of RNA, likely through its ZnF domain. Additionally, ARS2 interacts with at least two RNA processing complexes, via the ZnF, RRM, and Mid domains (Fig. 9). The ability to recognize different RNA and processing complexes suggests that ARS2 has a key role in linking the nuclear cap to the appropriate RNA biogenesis machinery. A key function of the CBC is thought to be the coupling of RNA processing steps (16). ARS2 appears to be uniquely situated to do this. For example, we found that RDH mRNA pulldown, and presumably processing, requires both CBC-ARS2 interaction through the C terminus of ARS2, as well as additional interactions through the DUF3546 and ZnF domains. Similarly, miRNA biogenesis requires both CBC-ARS2 interaction through the C terminus and miRNA binding, likely through the ARS2 ZnF/DUF3546. Since cap assembly on nascent transcripts is thought to occur cotranscriptionally within the first 20 nucleotides, it is likely that ARS2 binds first to the CBP20/80-m7G Cap complex through its proline-rich C terminus. Next, the type of RNA may be recognized through the ZnF domain. We speculate that the binding of either miRNA or histone mRNA alters the conformation of ARS2 to allow for either FLASH binding or DROSHA binding to the ZnF/RRM or Mid domain, respectively. The interaction of ARS2 with the 5′ cap and 3′ processing site thereby ensures that only capped RNAs are processed. Taken together, these results show that ARS2 is a multifunctional platform for protein and RNA interactions suited for directing primary RNA transcripts to the appropriate processing pathways.
FIG 9.
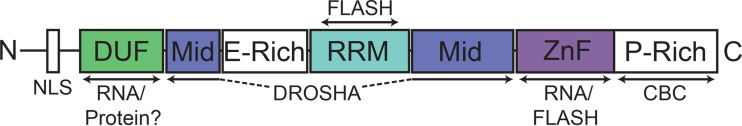
Domain architecture of ARS2 with mapping of protein and RNA interaction sites. The DUF3546 domain likely contributes to RNA interaction indirectly, while the ZnF is likely the principal interaction site for miRNA and RDH mRNA. The Mid domain is involved in interaction with DROSHA, the RRM and ZnF domains are required for interaction with FLASH, and the proline-rich (P-rich) C terminus is required for interaction with the CBC. NLS, nuclear localization sequence.
ACKNOWLEDGMENTS
This work was supported by an NSERC Discovery Grant and a Foundation Fighting Blindness operating grant awarded to P.L.H. and an NSERC Discovery Grant to R.L.C.
We thank Chris Nelson for critically reviewing the manuscript and for antibodies and reagents and help with experiments. We also thank Stephen Evans and Omid Haji-Ghassemi for providing assistance and advice on homology modeling. The let-7c DsRed sensor was a gift from Gregory Hannon, and pCK-Drosha-FLAG was a gift from V. Narry Kim. Anti-ARS2 (XL12.2 and LX186.3) was generously provided by the Ludwig Institute for Cancer Research.
REFERENCES
- 1.Lewis JD, Izaurralde E. 1997. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem 247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 2.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol 133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci U S A 94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narita T, Yung TMC, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. 2008. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci U S A 105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, Cherry S, Ihle JN, Dreyfuss G, Thompson CB. 2009. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche NEH, Gostan T, Vandermoere F, Robert MC, Cusack S, Verheggen C, Jensen TH, Bertrand E. 2013. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- 9.Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, Sandberg R, Hyman A, LaCava J, Rout MP, Andersen JS, Bertrand E, Jensen TH. 2013. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata Y, Takahashi M, Fedoroff NV, Hamdan SM. 2013. Dissecting the interactions of SERRATE with RNA and DICER-LIKE 1 in Arabidopsis microRNA precursor processing. Nucleic Acids Res 41:9129–9140. doi: 10.1093/nar/gkt667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida S, Chen HY, Adam Yuan Y. 2011. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res 39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. 2012. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell 45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiriyama M, Kobayashi Y, Saito M, Ishikawa F, Yonehara S. 2009. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol Cell Biol 29:4729–4741. doi: 10.1128/MCB.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang XC, Xu B, Sabath I, Kunduru L, Burch BD, Marzluff WF, Dominski Z. 2011. FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol Cell Biol 31:1492–1502. doi: 10.1128/MCB.00979-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller-McNicoll M, Neugebauer KM. 2014. Good cap/bad cap: how the cap-binding complex determines RNA fate. Nat Struct Mol Biol 21:9–12. doi: 10.1038/nsmb.2751. [DOI] [PubMed] [Google Scholar]
- 17.Chi B, Wang K, Du Y, Gui B, Chang X, Wang L, Fan J, Chen S, Wu X, Li G, Cheng H. 2014. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res 42:7305–7318. doi: 10.1093/nar/gku350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rando TA, Blau HM. 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol 125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Wilson MD, Wang D, Wagner R, Breyssens H, Gertsenstein M, Lobe C, Lu X, Nagy A, Burke RD, Koop BF, Howard PL. 2008. ARS2 is a conserved eukaryotic gene essential for early mammalian development. Mol Cell Biol 28:1503–1514. doi: 10.1128/MCB.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terry NHA, White RA. 2006. Flow cytometry after bromodeoxyuridine labeling to measure S and G2+M phase durations plus doubling times in vitro and in vivo. Nat Protoc 1:859–869. doi: 10.1038/nprot.2006.113. [DOI] [PubMed] [Google Scholar]
- 22.Silberstein L, Webster SG, Travis M, Blau HM. 1986. Developmental progression of myosin gene expression in cultured muscle cells. Cell 46:1075–1081. doi: 10.1016/0092-8674(86)90707-5. [DOI] [PubMed] [Google Scholar]
- 23.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. 2006. SERRATE: a new player on the plant microRNA scene. EMBO Rep 7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh SW, Kingsley T, Shin HH, Zheng Z, Chen HW, Chen X, Wang H, Ruan P, Moody M, Hou SX. 2003. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics 163:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet 31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 26.Andreu-Agullo C, Maurin T, Thompson CB, Lai EC. 2012. Ars2 maintains neural stem-cell identity through direct transcriptional activation of Sox2. Nature 481:195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veitia RA. 2007. Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19:3843–3851. doi: 10.1105/tpc.107.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco TR, Moita LF, Gomes AQ, Hacohen N, Carmo-Fonseca M. 2006. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol Biol Cell 17:4187–4199. doi: 10.1091/mbc.E06-01-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawellek A, McElroy S, Samatov T, Mitchell L, Woodland A, Ryder U, Gray D, Lührmann R, Lamond AI. 2014. Identification of small molecule inhibitors of pre-mRNA splicing. J Biol Chem 289:34683–34698. doi: 10.1074/jbc.M114.590976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerdà-Costa N, Bonet J, Fernández MR, Avilés FX, Oliva B, Villegas S. 2011. Prediction of a new class of RNA recognition motif. J Mol Model 17:1863–1875. doi: 10.1007/s00894-010-0888-0. [DOI] [PubMed] [Google Scholar]
- 32.Allain FHT, Gubser CC, Howe PWA, Nagai K, Neuhaus D, Varani G. 1996. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature 380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 33.Cléry A, Blatter M, Allain FHT. 2008. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. 2006. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A 103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fribourg S, Gatfield D, Izaurralde E, Conti E. 2003. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 36.Kadlec J, Izaurralde E, Cusack S. 2004. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 37.Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 38.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]