Abstract
Notch1 is an evolutionarily conserved transmembrane receptor involved in melanoma growth. Notch1 is first cleaved by furin in the Golgi apparatus to produce the biologically active heterodimer. Following ligand binding, Notch1 is cleaved at the cell membrane by proteases such as ADAM10 and -17 and membrane type 1 matrix metalloproteinase (MT1-MMP), the latter of which we recently identified as a novel protease involved in Notch1 processing. The final cleavage is γ-secretase dependent and releases the active Notch intracellular domain (NIC). We now demonstrate that Notch1 directly regulates furin expression. Aside from activating Notch1, furin cleaves and activates several proteases, including MT1-MMP, ADAM10, and ADAM17. By chromatin immunoprecipitation and a reporter assay, we demonstrate that Notch1 binds at position −1236 of the furin promoter and drives furin expression. The Notch1-dependent enhancement of furin expression increases the activities of MT1-MMP and ADAM10 but not that of ADAM17, as demonstrated by short hairpin RNA (shRNA) knockdown of furin, and promotes the cleavage of Notch1 itself. These data highlight a novel positive-feedback loop whereby Notch1-dependent furin expression can induce Notch1 signaling by increasing Notch1 processing and by potentiating the activity of the proteases responsible for Notch1 activation. This leads to Notch1 signal amplification, which can promote melanoma tumor growth and progression, as demonstrated by the inhibition of cell migration and invasion upon furin inhibition downstream of Notch1. Disruption of such feedback signaling might represent an avenue for the treatment of melanoma.
INTRODUCTION
Melanoma is the deadliest form of skin cancer, causing approximately 50,000 deaths a year, despite promising new therapies (1–3). It is imperative to understand the biology of melanoma in order to find novel therapeutic targets.
Notch proteins are transmembrane receptors of approximately 300 kDa. In humans, there are four Notch receptors, Notch1 to Notch4, which share the same basic structure: (i) an extracellular domain, containing an epidermal growth factor-like repeat domain (EGF repeats), a LIN12-Notch repeat (LNR) domain, and a heterodimerization (HD) domain, and (ii) an intracellular domain, containing an RBP-JK (recombination signal-binding protein 1 for Jκ)-associated module (RAM) domain, an ankyrin repeat (ANK) domain, a transactivation domain (TAD), and a PEST (proline-glutamate-serine-threonine) domain (4). Full-length Notch is first cleaved by furin in the trans-Golgi network. Such cleavage is necessary for the proper positioning of Notch1 at the plasma membrane (5, 6). Once Notch1 is at the membrane, after ligand binding, a second cleavage occurs that is canonically driven by ADAM (a disintegrin and metalloproteinase) proteases (ADAM10 and -17) (7–10). This produces an unstable fragment that is immediately cleaved by γ-secretase to release the Notch intracellular domain (NIC) (11). The NIC translocates into the nucleus and binds with transcription factors CSL (C-promoter binding factor 1) and MAML (Mastermind-like) to regulate the transcription of multiple downstream targets, including HES (hairy and enhancer of split) family members, c-Myc, and cyclin D3 (12–20).
The Notch pathway is an evolutionarily conserved signaling cascade that plays essential roles in embryogenesis and in cell renewal in the adult by participating in the maintenance of stem cell pluripotency in a variety of tissues (21–23). Previous studies have shown that Notch1 plays essential roles in melanocyte stem and precursor cell homeostasis (24–27). Furthermore, Notch1 is highly expressed in melanomas. Overexpression of active Notch1 (NIC1) transforms primary human melanocytes in vitro and confers metastatic properties on primary melanoma cells (28–31). Notch1 has also been shown to be required for melanoma cell growth and survival (31). However, no Notch1-activating mutations have been observed in melanoma so far, suggesting that other mechanisms are responsible for the high activity of Notch1 in melanoma cells.
One potential mechanism is an increase in processing. We recently demonstrated that membrane type 1 matrix metalloproteinase (MT1-MMP), a membrane-tethered zinc-dependent MMP, acts as a protease involved in the second cleavage of Notch1 (32). MT1-MMP is very abundant in melanomas and is associated with disease progression and poor patient outcome (32, 33). MT1-MMP is synthesized as a preproenzyme of 64 kDa that is also cleaved by furin prior to its transport to the plasma membrane as an active 55-kDa enzyme (34). MT1-MMP is one of the most important MMPs that promote cell migration and invasion in cancer. We have demonstrated that MT1-MMP mediates melanoma growth and metastasis via various mechanisms, including Notch1 activation (32).
ADAM10 and ADAM17 belong to the ADAM family of zinc-dependent metalloproteinases and are considered the canonical proteases involved in the cleavage of Notch1 at the S2 site (7). Structurally similar to MT1-MMP, they also are activated in the Golgi apparatus by furin and then arrive at the membrane as active enzymes (34). Dysregulated expression of ADAM proteins has been reported in multiple tumors. These proteases are implicated as positive regulators in tumor cell proliferation, angiogenesis, and metastasis (35). Importantly, both ADAM10 and ADAM17 have been shown to be overexpressed in melanoma (36–39).
Furin cleaves many substrates, including MT1-MMP, Notch1, ADAM10, and ADAM17 (6, 34, 40–42). Furin is a calcium-dependent serine protease that belongs to the family of proprotein convertases (PCs). The expression levels of furin differ for different cell types and degrees of cell differentiation (43–45, 47). Furin has been reported to regulate tumor growth and malignant tumor phenotypes (48–50). Importantly, the expression of furin has been shown to be coordinated with that of its substrates, such as transforming growth factor (TGF), bone morphogenetic protein (BMP), and insulin-like growth factor (51–53).
Our present data establish that furin is regulated by its own substrate Notch1. In turn, Notch1-induced furin further activates MT1-MMP and ADAM10, which can then enhance the cleavage of Notch1. Hence, we have identified a novel positive-feedback loop whereby Notch1 signaling self-activates by regulating not only its own processing but also that of its regulatory proteases. This novel feedback signaling might account for the high activity of Notch1 in melanoma cells.
MATERIALS AND METHODS
Cell lines.
Primary and metastatic melanoma cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA) or were gifts from Marianne Broome Powell (Stanford University) (54). The use of the cells was approved by the Case Cancer Institutional Review Board (IRB). The cell lines used in this study were V2387, WM115, WM266-4, and SKMel2. These are human melanoma cells derived from primary melanoma (WM115) and from metastases to the skin (WM266-4 and SKMel2) and lymph nodes (V2387). All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 1% glutamine, and 1% penicillin-streptomycin.
shRNAs and expression plasmids.
Short hairpin RNAs (shRNAs) against human Notch1 (TRCN0000003359) and furin (TRCN000075238 and TRCN000075239) were purchased from Sigma. The Notch1-NIC lentiviral expression vector was constructed by inserting the cDNA sequence corresponding to human Notch1-NIC (bp 5278 to 7668 of full-length human Notch1) into the pLM-CMV-Ha-puro-PL3 lentiviral plasmid (55), between XbaI and XhoI. The lentiviral plasmid iDuet101a-DN-MAML (with dominant negative MAML) was kindly provided by Eli Bar (Case Western Reserve University). Viral particles were produced in 293FT cells by using the X-tremeGENE 9 reagent (Roche, Mannheim, Germany) according to the manufacturer's instructions. The packaging plasmids used were pMD2.G and psPAX2, which were purchased from Addgene (Cambridge, MA).
Western blot analysis.
Total protein for all assays was extracted with urea lysis buffer (9 M urea, 75 mM Tris-HCl [pH 7.5], and 100 mM 2-mercaptoethanol), and 20 μg/sample was separated by 8%-to-10% SDS-PAGE and was transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were probed with antibodies against Notch1 (C20; Santa Cruz Biotechnology, Santa Cruz, CA), full-length and cleaved Notch1 (mN1A; Novus Biologicals LLC, Littleton, CO), the Notch1 NIC cleaved at Val-1744 (Cell Signaling Technology, Beverly, MA), MT1-MMP (clone LEM-2/15.8; Millipore, Billerica, MA), ADAM10 (Abcam, Cambridge, MA), tumor necrosis factor alpha-converting enzyme (TACE; also called ADAM17) (eBioscience, San Diego, CA), and furin (Abcam, Cambridge, MA). Bands were detected using the SuperSignal detection reagent (Thermo Scientific). Loading was normalized with anti-β-actin (Santa Cruz Biotechnology). Densitometric quantifications (by ImageJ) of band intensity were normalized to the intensity of the respective β-actin band.
Real-time PCR analysis.
cDNA was synthesized from total RNA using the SuperScript first-strand synthesis system for reverse transcription-PCR (RT-PCR) (Invitrogen). Then the cDNA was used for PCR amplification with SYBR green PCR master mix (Roche). The following primer sets were used to amplify specific target genes: human ADAM10 forward (5′-CAAAGTCTGAGAAGTGTCGGG-3′) and reverse (5′-CTGCACATTGCCCATTAATG-3′) primers, human ADAM17 forward (5′-ACCTGAAGAGCTTGTTCATCGAG-3′) and reverse (5′-CCATGAAGTGTTCCGATAGATGTC-3′) primers, human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward (5′-CGCTCTCTGCTCCTCCTGTT-3′) and reverse (5′-CCATGGTGTCTGAGCGATGT-3′) primers, human MT1-MMP forward (5′-CTCCCTCGGCTCGGCCCAAA-3′) and reverse (5′-CGCCTCATGGCCTTCATGGTGTCT-3′) primers, human furin forward (5′-GAAGCAGCAGCGGCCAGGAT-3′) and reverse (5′-CGAAGATCTGGCCCAGGTTGAGG-3′) primers, human PC5 forward (5′-TGCCAGGGACCAACCCAGGA-3′) and reverse (5′-TGCTCTCGGCCATAGTTGTCTGC-3′) primers, and human PC7 forward (5′-CACCAGCACGGTTTCGGCCT-3′) and reverse (5′-AGCCGTTGGGATCCGAGTCCAT-3′) primers. The relative quantification of mRNA expression levels was normalized by GAPDH.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed using ChIP assay kits (Upstate Biotechnology) according to the manufacturer's recommendations. The following primer sequences were used: for the furin promoter at B1 (bp −3556), forward primer 5′-ATCAGGAGGGTCACCTATGGTGC-3′ and reverse primer 5′-CAGAGGTGCTGGGATTACAGGC-3′; for the furin promoter at B2 (bp −1236), forward primer 5′-CTCAGAGCTTAGTTCCCAGCAGAC-3′ and reverse primer 5′-GATCTCCAGGTATTGCCAAATGTC-3′; for the furin promoter at B3 (bp +960), forward primer 5′-ATGCAGATTGAAGAGGCAAGCTG-3′ and reverse primer 5′-GCAATACTTGCTGCTTCAGGTGC-3′; for the HES1 promoter, forward primer 5′-CGTGTCTCCTCCTCCCATTG-3′ and reverse primer 5′-CCAGGACCAAGGAGAGAGGT-3′. The antibody employed for the immunoprecipitation has been described previously (56).
Luciferase assays.
The Hes1-reporter plasmid was a kind gift from Ryoichiro Kageyama (Kyoto University, Kyoto, Japan) (57). The human furin promoter-luciferase constructs pGL2-Basic and pGL2-SacI were generously provided by Claire M. Dubois (University of Sherbrooke, Sherbrooke, Quebec, Canada) (43). The mutations within the pGL2-SacI reporter were generated by PCR using primers 5′-CCTGTGACGTCACAGCTCCTCATTCTGCGACAGTGG-3′ and 5′-GAATGAGGAGCTGTGACGTCACAGGATGGTGGTTAG-3′, which replace the sequence 5′-TTCCCAC-3′ with 5′-TCACAGC-3′. V2387, WM115, or SKMel2 cells (5 ×104/well, in 24-well plates) were transfected by using the X-tremeGENE high-performance (HP) reagent (Roche Applied Science) according to the manufacturer's instructions. After 48 h, the cells were lysed in 100 μl of lysis buffer (Promega, Madison, WI). A Renilla luciferase reporter plasmid driven by a cytomegalovirus (CMV) promoter was cotransfected with the reporter constructs at a 1:20 ratio in order to assess transfection efficiency. Firefly and Renilla luciferase activities were assessed by the Dual-Luciferase assay system (Promega), and light production was measured for 10 s in a Monolight 2010 luminometer (Molecular Devices, Sunnyvale, CA).
MT1-MMP and ADAM10 activity assay.
Membrane proteins were extracted from 106 cells by three freeze-thaw cycles in dry-ice–ethanol baths at 37°C. The lysates were sonicated for 3 s, and the membranes were pelleted by centrifugation (30 min, 13,000 × g, 4°C) and were resuspended in phosphate-buffered saline (PBS). Equal protein amounts per sample were incubated with a fluorogenic MT1-MMP substrate [Mca-PLGL-Dap (Dnp)-AR-NH2] or the ADAM10 substrate 5-carboxyfluorescein (5-FAM)–SLGRKIQIQ-K (QXL 520)–NH2 provided by the manufacturer (AnaSpec, Fremont, CA). Fluorescence intensity was measured at an excitation wavelength of 490 + 20 nm and an emission wavelength of 520 + 20 nm by using a SpectraMax M2 enzyme-linked immunosorbent assay (ELISA) reader (Molecular Devices, Sunnyvale, CA).
Notch ligand stimulation assay.
Notch signaling was induced by plating cells (32,000/cm2) on dishes displaying an immobilized fragment crystallizable region (Fc) or Fc-JAGGED1 ligand anchored to protein A. Plasmids expressing Fc and Fc-JAGGED1 (58, 59) were kindly provided by Aaron Proweller (Case Western Reserve University, Cleveland, OH).
Migration and invasion assays.
SKMel2 cells were plated in a confluent monolayer in duplicate. A scratch was produced using a pipette tip, and the detached cells were gently washed away with PBS. Plates were incubated under a time lapse microscope for 24 h. Pictures were taken every hour for the duration of the experiment. Frames were aligned, and the distance from one of the edges of the wound in the first frame (considered time zero) to the migration front was calculated for the time points indicated in Fig. 9B. For the transwell invasion assay, a suspension of 25 × 104 cells was added to an 8-μm-pore-size insert, either uncoated (control) or coated with a Matrigel growth factor reduced basement membrane matrix (BD Biosciences, MA). DMEM containing 5% fetal bovine serum (FBS) was added to the lower chamber (24-well plate) as a chemoattractant. After a 24-h incubation, cells that had migrated through the control inserts or Matrigel and were collected on the bottom membrane, fixed with 4% formaldehyde, and stained with Coomassie blue. Each treatment was carried out in triplicate. Membranes were incubated in 10% acetic acid to extract the Coomassie stain, and the color intensity was quantified at 295 nm. The percentage of invasion was calculated as (mean reads of Matrigel)/(mean reads of control insert) × 100.
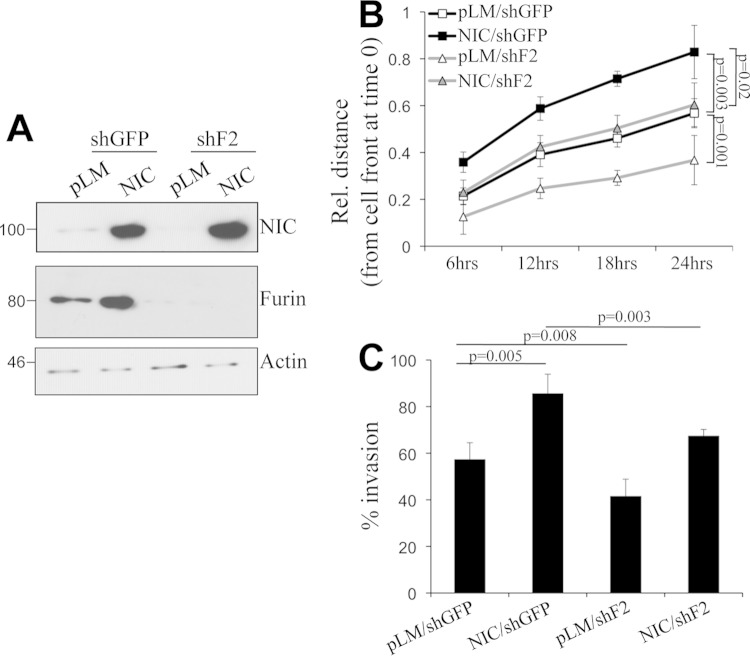
FIG 9.
Furin inhibition reduces migration and invasion by melanoma cells. (A) Expression levels of Notch1-NIC and furin in cells expressing active Notch1 and furin shRNA. The same cells were used for all panels. (B) Migration of the cells (determined by a scratch assay) over 24 h. (C) Invasion of the cells through Matrigel over a 24-h period. The reductions in the levels of migration and invasion in shFurin-expressing cells are statistically significant (by Student's t test).
RESULTS
Notch1 modulates MT1-MMP activity.
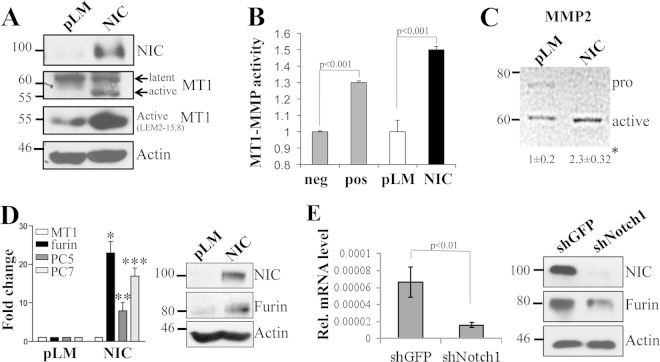
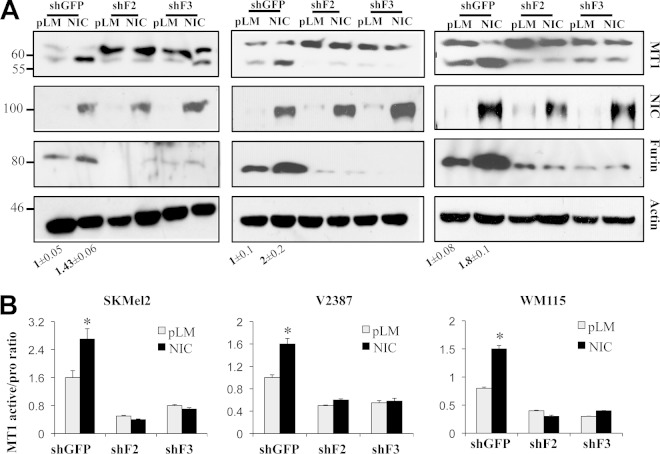
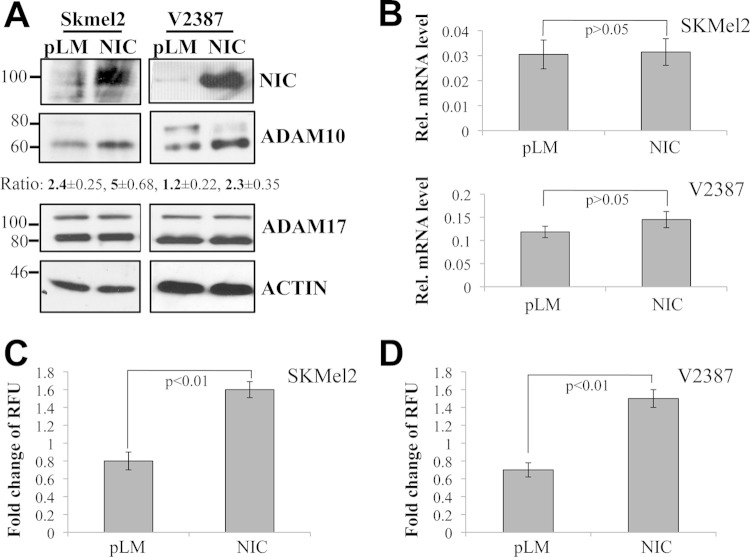
Our previous data showed that MT1-MMP is involved in the cleavage of Notch1 and that it is a critical modulator of melanoma metastasis and melanoma cell growth (32, 33). Thus, we decided to explore the mechanisms of MT1-MMP regulation. Interestingly, we observed that Notch1 promotes MT1-MMP activity. Expression of active Notch1 (NIC) in melanoma cells increases the 55-kDa fraction of MT1-MMP (Fig. 1A), the catalytically active enzyme, as assessed by two different antibodies, one of which recognizes the active MT1-MMP peptide specifically. This protein fraction is indeed the active protein, since it leads to an increase in MT1-MMP activity as measured by an in vitro activity assay (Fig. 1B) (61, 62). Furthermore, expression of active Notch1 leads to an increase in the processing of MMP2 (Fig. 1C), a known target of MT1-MMP (63). Given that MT1-MMP activation requires the cleavage of the inhibitory propeptide by furin or furin-like convertases, and given that Notch1 expression did not change MT1-MMP transcript levels in the cells (Fig. 1D), we investigated if Notch1 could regulate the expression of the convertases. Indeed, we found that Notch1-NIC increased both mRNA and protein levels of furin (Fig. 1D). The transcripts of the furin-like convertases PC5 and PC7, also involved in MT1-MMP activation, were increased as well (Fig. 1D). Conversely, inhibition of Notch1 by a specific shRNA resulted in decreases in furin mRNA and protein levels. In a parallel experiment, cleavage of endogenous Notch1 by stimulation with the recombinant JAGGED1 ligand led to an increase in the level of furin and a corresponding increase in MT1-MMP processing (Fig. 2), further suggesting that Notch1 activates MT1-MMP via furin expression.
FIG 1.
Notch1 promotes MT1-MMP activity. (A) The active MT1-MMP fraction (55 kDa) is increased in Notch1-NIC-expressing cells. β-Actin is used as a loading control. (B) Notch1-NIC increases MT1-MMP activity in vitro. neg, negative control (buffer); pos, positive control (0.5 μg/ml recombinant active MT1-MMP). (C) Zymogram showing MMP2 processing in pLM- or NIC-expressing cells. Numbers below the gel represent the fold induction (±standard deviation) of the active band over the control (pLM). pro, inactive form. Results are averages for three repeats. The asterisk indicates a significant difference (P < 0.05) by Student's t test. (D) Notch1 increases both mRNA and protein levels of furin but does not affect the expression of MT1-MMP. The same cells used for panel A were used in these experiments. Quantitative RT-PCR results (left) and a Western blot (right) show changes in the expression of furin and furin-like convertases. The differences in expression from the expression of the control are significant (P < 0.001) by Student's t test. *, **, and *** refer to the statistical significance calculated between the control and NIC for three different targets. (E) Inhibition of Notch1 reduces both mRNA and protein levels of furin. Rel., relative.
FIG 2.
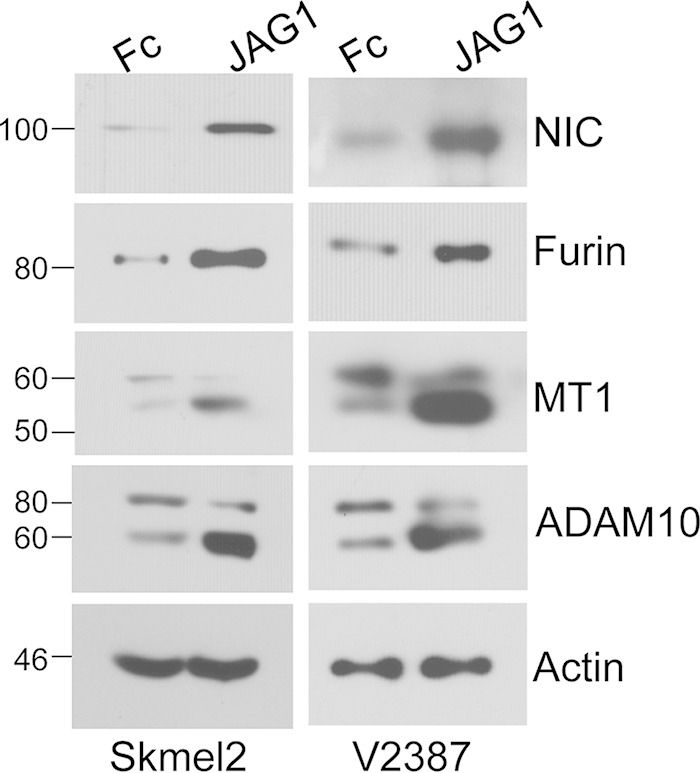
Stimulation of endogenous Notch1. JAGGED1-dependent activation of endogenous Notch1 increases furin levels and promotes the processing of MT1-MMP and ADAM10 in two melanoma cell lines.
Notch1 affects furin expression through binding to the furin promoter.
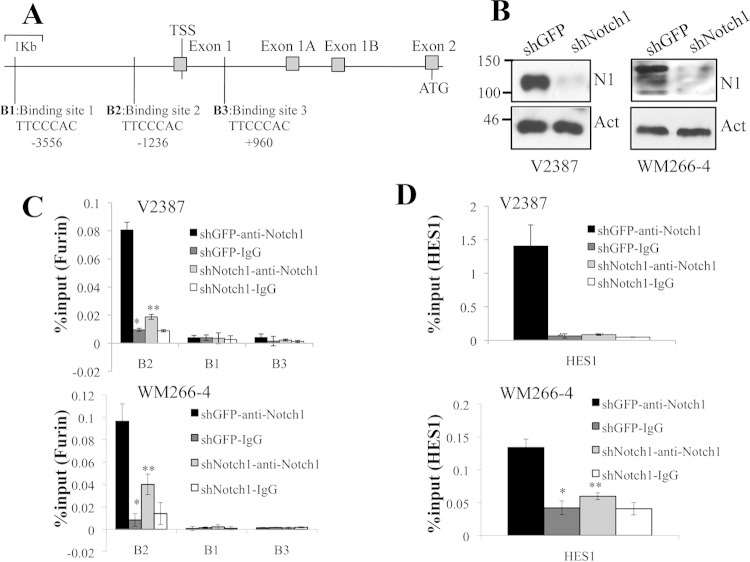
Based on the previous data, we hypothesized that Notch1 affects the activity of its protease, MT1-MMP, by increasing the levels of furin available to cleave the inhibitory propeptide. We therefore sought to determine whether Notch1 might regulate furin by directly driving its expression. Active Notch1, though not capable of binding directly to DNA sequences, functions in a transcription complex together with CBF1 and Mastermind-like (MAML). Sequence analysis of the furin promoter through Motif Search (http://www.genome.jp/tools/motif/) revealed three putative CBF1/Notch-binding sequences (TTCCCAC) within a 5-kb sequence encompassing 4 kb upstream and 1 kb downstream of the furin transcription start site: one at position −3556, one at −1236, and one at +960 (Fig. 3A). The sites were then tested by a chromatin immunoprecipitation (ChIP) assay for Notch1 occupancy in V2387 and WM266-4 cells expressing either a control shRNA (green fluorescent protein shRNA [shGFP]) or Notch1 shRNA (shNotch1) (Fig. 3B). We observed enrichment in shGFP-expressing cells at the B2 site (position −1236), but not at positions −3556 and +960, that was inhibited in shNotch1-expressing cells (Fig. 3C). The binding of Notch1 to the HES1 promoter was tested as a positive control; this assay also showed enrichment in shGFP-expressing cells over shNotch1-expressing cells (Fig. 3D).
FIG 3.
Furin is a direct target of Notch1. (A) Schematic representation of the furin promoter. There are three putative CSL binding sequences (TTCCCAC), located at bp −3556 (B1), −1236 (B2), and +960 (B3). TSS, transcription start site. (B) Western blot analysis of V2387 and WM266-4 cells showing Notch1 protein levels in control and shNotch1-expressing cells. (C) ChIP analysis showing the association of Notch1 with the binding site at bp −1236 (B2) in both V2387 and WM266-4 cells. Asterisks indicate significant differences (P < 0.01) by Student's t test. (D) The HES1 promoter was used as a positive control for CSL/Notch-binding activity. Asterisks indicate significant differences (P < 0.01) by Student's t test. All data are representative of the results of three independent experiments.
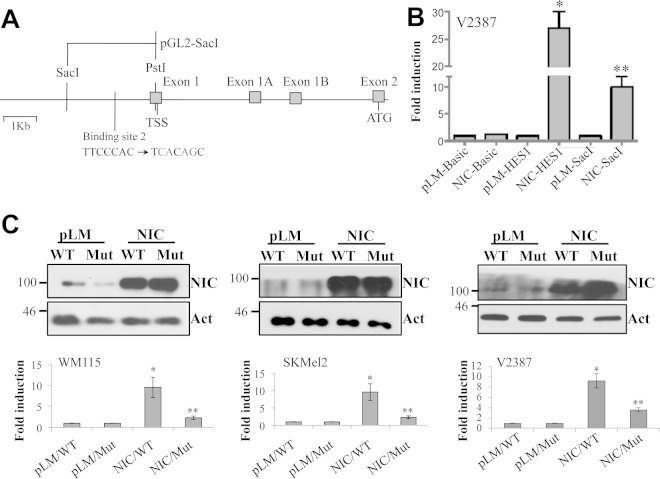
To further confirm that the B2 site is indeed the location where Notch1 binds and drives furin transcription, we performed a series of luciferase reporter assays (Fig. 4A). In V2387 cells, the SacI reporter construct, containing the wild-type (WT) B2 binding site, showed a 10-fold induction of luciferase activity in cells expressing the active form of Notch1 (NIC) over activity in cells expressing the control (pLM) (Fig. 4B). The HES1 reporter construct was used as a positive control. Mutation of the B2 binding site from TTCCCAC to TCACAGC significantly reduced the induction of luciferase activity in NIC-expressing cells from that with the WT sequence in three different melanoma cell lines (Fig. 4C). The upper section of Fig. 4C shows the expression levels of Notch1-NIC in each cell line tested. These data identify furin as a novel direct target of Notch1.
FIG 4.
Notch1 affects furin expression through binding to the furin promoter. (A) Schematic representation of luciferase reporter constructs containing the WT (TTCCCAC) or mutated (TCACAGC) binding site 2 sequence. The pGL2-SacI construct contains the sequence from PstI at the transcription start site (TSS) to SacI. (B) Luciferase reporter assay with V2387 cells expressing the control (pLM) or Notch1-NIC. The pGL2-Basic luciferase construct, without the promoter region, represents the negative control for the luciferase assay; the HES1 construct, which contains the promoter region of the Notch1 target HES1, represents the positive control. Significant induction of luciferase activity was observed in NIC-expressing cells when the WT SacI construct was used. Asterisks indicate significant differences (P < 0.01) by Student's t test. (C) (Bottom) Luciferase induction is seen in NIC-expressing cells with the WT SacI construct (NIC/WT) and is significantly reduced when the SacI construct is mutated (NIC/Mut). (Top) Notch1-NIC protein levels in pLM- and NIC-expressing cells. Experiments were carried out in three cell lines: WM115, SKMel2, and V2387. Data are averages for three independent experiments. Act, actin.
Notch1 affects MT1-MMP activity through furin.
Since MT1-MMP activity is dependent on the cleavage of the inhibitory propeptide by furin, once we established that Notch1 modulates furin expression, we next evaluated whether Notch1 plays a role in the conversion of MT1-MMP from the inactive (64-kDa) to the active (55-kDa) form through furin. We tested whether inhibition of furin by specific shRNA sequences would impair MT1-MMP activation downstream of Notch1. Since furin also processes Notch1, we used cell lines expressing an exogenous, active Notch1-NIC that is independent of furin cleavage. Western blot analysis showed that the ratio of active to inactive MT1-MMP increases in NIC-expressing cells but decreases in furin knockdown cells (Fig. 5). Importantly, inhibition of furin expression abolishes the ability of active Notch1 to increase the active MT1-MMP fraction (Fig. 5), indicating that Notch1 affects MT1-MMP activation through furin.
FIG 5.
Notch1 induces MT1-MMP activation through furin. (A) Western blot analysis showing that the active fraction of MT1-MMP increases in NIC-expressing cells but decreases in furin knockdown cells. Experiments were carried out in three cell lines: SKMel2, V2387, and WM115. The average quantifications of furin bands (relative fold changes with respect to the band density of the control [pLM] band) for three separate experiments ± standard deviations are given below the blots. (B) Ratio of active to inactive MT1-MMP bands normalized to the expression of the corresponding β-actin band. Data are averages from three independent experiments. In all three cell lines, differences between control cells and furin knockdown cells are significant (P < 0.01) by Student's t test.
Notch1 affects ADAM10 activity.
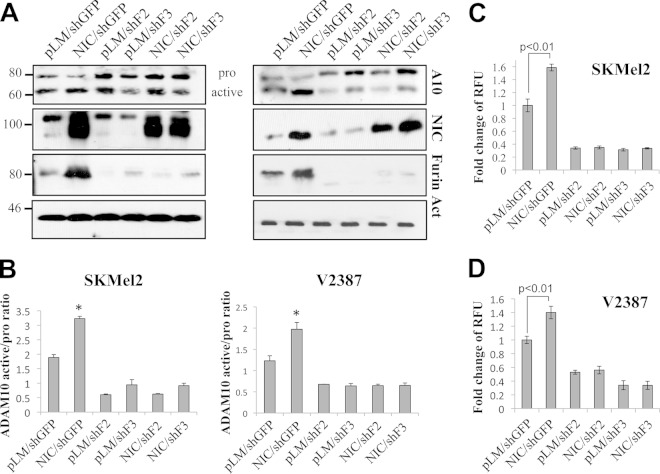
ADAM10 and ADAM17 are generally accepted as proteases responsible for Notch1 cleavage following ligand binding. Like MT1-MMP, ADAM10 and -17 are also activated by furin prior to reaching the plasma membrane as fully active enzymes (7, 34, 40). Previous data from our group showed that melanoma tumors and cells do express these proteases, although not at the same levels as MT1-MMP (32). We therefore wanted to determine whether ADAM10 and -17 were also affected by Notch1 through furin induction. As shown in Fig. 6A, the intensity of the active band of ADAM10 (60 kDa) increases in NIC-expressing cells, indicating an increase in processing. Similarly, in cells stimulated by recombinant JAGGED1, ADAM10 processing is increased (Fig. 2). This is not true for ADAM17, however. We further measured the activity of ADAM10 in vitro by using a commercially available activity assay (AnaSpec) and found that overexpression of NIC significantly increased ADAM10 activity in both SKMel2 and V2387 cells (Fig. 6C and D). Of note, Notch1 did not affect ADAM10 gene expression (Fig. 6B). We next evaluated whether Notch1 plays a role in the conversion of ADAM10 through furin. As Fig. 7 shows, the intensity of the active band of ADAM10 increases in NIC-expressing cells but decreases in furin knockdown cells. Again, inhibition of furin abolished the ability of Notch1-NIC to process ADAM10, as seen above for MT1-MMP. The in vitro activity assay mirrored the Western blot data, showing activation in the presence of NIC and a lack of activation by Notch1 when furin is inhibited (Fig. 7C and D).
FIG 6.
Notch1 promotes ADAM10 activity. (A) In SKMel2 and V2387 cells, the active ADAM10 fraction (60 kDa) is increased in Notch1-NIC-expressing cells. The level of ADAM17 does not change. The ratio of active to inactive ADAM10 normalized to the corresponding β-actin level is given. Values are averages ± standard deviations for three independent experiments. (B) Quantitative RT-PCR of ADAM10 in pLM- or NIC-expressing cells. Data are ADAM10 mRNA levels normalized to GAPDH mRNA levels. (C and D) In vitro activity assay for ADAM10 in pLM- or NIC-expressing cells. Fold changes in relative fluorescence units (RFU) in SKMel2 and V2387 cells were determined. Statistical significance (by Student's t test) is indicated.
FIG 7.
Notch1 increases the active fraction of ADAM10 through furin. (A) Western blot analysis showing the expression of the propeptide and active forms of ADAM10 and the expression of Notch1-NIC in NIC-expressing SKMel2 (left) and V2387 (right) cells in the presence of shGFP or furin shRNA (shFurin) (shF2 and shF3). (B) Ratio of active to inactive ADAM10 in the lanes in panel A (normalized to the β-actin level). Asterisks indicate significant differences (P < 0.01) by Student's t test between pLM/shGFP and NIC/shGFP. Data are averages for four independent experiments. (C and D) In vitro activity assay for ADAM10 in SKMel2 and V2387 cells expressing the empty vector (pLM) or active Notch1 (NIC) in the presence of shGFP or shFurin. Statistical significance (by Student's t test) is indicated. Results are averages for four independent experiments.
Inhibition of furin diminishes Notch1 processing.
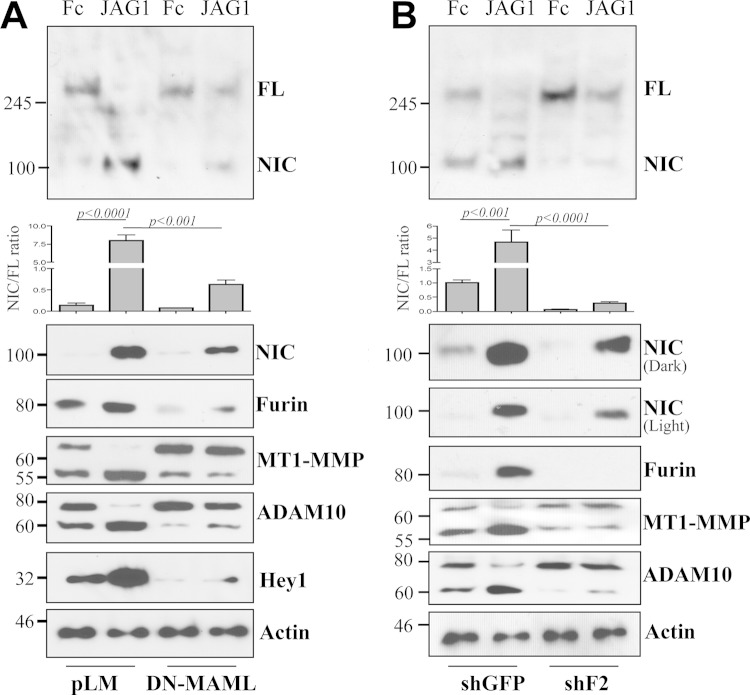
Given that furin promotes the first cleavage of Notch1 and that such cleavage is required for proper Notch positioning at the plasma membrane and access to the ligand (5, 6), we wanted first to determine whether interruption of Notch1 signaling at the DNA binding level would affect furin and consequently Notch1-NIC release. For this purpose, cells were transduced with a dominant negative form of MAML (DN-MAML). Expression of DN-MAML inhibited the expression of furin and HEY1, a known direct Notch transcription target, and resulted in diminished processing of full-length Notch1 to cleaved Notch1, as shown by an antibody that recognizes both full-length and processed Notch1 (Fig. 8A, top and center) and by an antibody that specifically recognizes Notch1 cleaved by γ-secretase at valine 1744 (Fig. 8A, bottom, NIC). Inhibition of furin by a specific shRNA reduced Notch processing in a similar manner (Fig. 8B). These data indicate that Notch1 can promote its own activation through a signaling loop involving furin and the proteases ADAM10 and MT1-MMP.
FIG 8.
Inhibition of furin decreases Notch1 processing. (A) Expression of full-length (FL) and intracellular (NIC) Notch1 (probed with antibody mN1A), Notch-NIC (probed with an antibody that recognizes Notch1 NIC cleaved at Val-1744), MT1-MMP, ADAM10, and furin in cells stimulated with recombinant JAGGED1 and expressing either an empty vector (pLM) or dominant negative MAML (DN-MAML). (B) Expression of FL and intracellular (NIC) Notch1 (probed with antibody mN1A), Notch-NIC (probed with an antibody that recognizes Notch1 NIC cleaved at Val-1744), MT1-MMP, ADAM10, and furin in cells stimulated with recombinant JAGGED1 and expressing either a specific furin shRNA (shF2) or a control shRNA (shGFP). Each graph shows the mean ratio (±standard deviation) of NIC to FL Notch1 for two independent experiments. P values were calculated by the Student t test.
Inhibition of furin affects migration and invasion by melanoma cells.
We have shown previously that MT1-MMP is required for melanoma cell migration and invasion (33). Given that Notch1-dependent furin expression promotes MT1-MMP activation, we next examined whether furin inhibition could affect migration and invasion by cells downstream of Notch1. Furin was downregulated in SKMel2 cells expressing constitutive Notch1-NIC (Fig. 9A). Then a scratch assay and a Matrigel invasion assay were performed. Cells expressing Notch1-NIC migrated faster than controls; however, a 30% reduction in migration (on average) was observed when furin was inhibited in both control (pLM) and NIC-expressing cells (Fig. 9B; see also Fig. S1 in the supplemental material). Similarly, cells expressing active Notch1 were more invasive than control cells, but inhibition of furin reduced these invasion-promoting effects (Fig. 9C).
DISCUSSION
Notch processing is an unexplored and potentially therapeutically targetable mechanism of increased Notch signaling in cancer. We have established that elevated Notch levels play a critical role in the pathogenesis of melanoma, and we describe here a novel circuit by which Notch drives an autoregulatory positive-feedback loop initiating with the elevated expression of furin. The full-length precursor of Notch1 is cleaved by furin in the trans-Golgi network, prior to arriving as a heterodimer at the plasma membrane, where it can be activated by interaction with Delta-like and Jagged ligands expressed on the surfaces of adjacent cells (6, 9, 64, 65). The cleavage by furin is required for the proper surface presentation of Notch1 and for proper ligand binding (64). In fact, studies show that furin-resistant Notch1 receptors exhibit decreased surface expression and ligand-mediated receptor activation (5). Although furin can be regulated by a variety of factors, including Sox9 (66), TGF-β (67), and BMPs (68), here we find that furin is a novel direct transcriptional target of its own substrate Notch1. Furthermore, we show that the activities of two major proteases involved in Notch1 cleavage following ligand binding are induced by active Notch1 through the modulation of furin.
Both MT1-MMP and ADAM10 are synthesized as inactive zymogens whose activation requires the cleavage of an inhibitory prodomain sequence (40, 69). Furin is an essential activator of pro-MT1-MMP that controls the level of active MT1-MMP on the cell surface (34). On the other hand, previous studies have demonstrated that overexpression of furin and the proprotein convertase PC7 increased the levels of active ADAM10 and that mutation of the convertase-dependent cleavage sites blocked the processing of ADAM10 to its active form (40). Indeed, we demonstrate here that Notch1 promotes the processing of MT1-MMP and ADAM10 to their active forms through furin, since downregulation of furin expression is sufficient to abolish the ability of Notch1 to promote the processing of the proteases and to increase their activity. On the basis of these data, we suggest that Notch1, by regulating the expression of furin, establishes a positive-feedback loop that can promote not only the processing of Notch1 but also its activation by modulating the activities of MT1-MMP and ADAM10.
MT1-MMP belongs to a family of membrane-tethered matrix metalloproteinases and is considered one of the most important MMPs in promoting cancer cell migration and invasion (33). Recently, we demonstrated that MT1-MMP operates as a protease involved in Notch1 activation and that MT1-MMP-dependent Notch1 activation mediates part of the protumorigenic properties of MT1-MMP (32). Melanomas express high levels of active Notch1, and its expression is correlated with that of MT1-MMP (32, 54, 70, 71). Hence, considering the results presented here, it is likely that in tumors where the two proteins are present, they functionally influence each other by promoting their reciprocal activation. The decrease in cell migration and invasion levels downstream of active Notch1 upon furin inhibition would substantiate this notion.
On the other hand, although ADAM10 and ADAM17 are both capable of processing Notch1 at the S2 site (10, 72), more and more evidence points to ADAM10 as the main enzyme responsible for such activity. First, the defects found in ADAM10-deficient mice phenocopy those of Notch1-deficient mice (73–75); second, van Tetering et al. (10) have demonstrated that Notch1 cleavage is severely diminished only in ADAM10 knockout fibroblasts, not in ADAM17 knockout cells. ADAM10 expression has been shown to be significantly higher in melanoma metastases than in primary tumors (37, 38). Knockdown of ADAM10 inhibits melanoma cell growth and migration, while overexpression of ADAM10 increases the migration of melanoma cells (38). Our study shows that overexpression of Notch1 increases the activity of ADAM10 through the modulation of furin. In contrast, Notch1 did not promote the processing of ADAM17. These data suggest that ADAM17, at least in melanoma cells, is not under the influence of Notch1 signaling.
Together, the data presented here highlight a novel positive-feedback signaling mechanism whereby Notch1, by regulating furin expression, promotes its own cleavage in the Golgi apparatus, a process that is required for the proper localization of Notch1 on the plasma membrane, and stimulates the activation of ADAM10 and MT1-MMP, which not only are involved in the cleavage and activation of Notch1 but also contribute to tumorigenesis by modulating the processing of a variety of substrates, including extracellular matrix and surface receptors. These findings add further complexity to Notch signaling deregulation in cancer. Notch not only promotes tumorigenesis by directly regulating genes involved in tumor development and progression (76) but can also do so indirectly, by influencing the activity of protumorigenic enzymes. We therefore propose that the positive-feedback loop between Notch1 and its proteases promotes the amplification of Notch1 signaling and may represent a novel therapeutic target for the treatment of melanoma.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the Harry Lloyd Charitable Trust and by a grant from the Concern Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00116-15.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. 2012. Cancer statistics, 2012. CA Cancer J Clin 62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Weinstock MA. 2012. Reducing death from melanoma and standards of evidence. J Invest Dermatol 132:1311–1312. doi: 10.1038/jid.2012.57. [DOI] [PubMed] [Google Scholar]
- 3.Chang AE, Karnell LH, Menck HR. 1998. The National Cancer Data Base report on cutaneous and noncutaneous melanoma—a summary of 84,836 cases from the past decade. Cancer 83:1664–1678. doi:. [DOI] [PubMed] [Google Scholar]
- 4.Bray SJ. 2006. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 5.Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. 2009. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One 4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A 95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozkulak EC, Weinmaster G. 2009. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol 29:5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulis ML, Saftig P, Ferrando AA. 2011. Redundancy and specificity of the metalloprotease system mediating oncogenic NOTCH1 activation in T-ALL. Leukemia 25:1564–1569. doi: 10.1038/leu.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. 2011. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure 19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. 2009. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortini ME. 2002. γ-Secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol 3:673–684. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 12.Artavanis-Tsakonas S, Matsuno K, Fortini ME. 1995. Notch signaling. Science 268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 13.Egan SE, St-Pierre B, Leow CC. 1998. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol 228:273–324. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh JJD, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol 16:952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. 1995. Signaling downstream of activated mammalian Notch. Nature 377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 16.Kopan R, Goate A. 2000. A common enzyme connects Notch signaling and Alzheimer's disease. Genes Dev 14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 17.Oswald F, Liptay S, Adler G, Schmid RM. 1998. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol 18:2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, Sun T, Kobayashi K, Gao P, Griffin JD. 2002. Identification of a family of mastermind-like transcriptional coactivators for mammalian Notch receptors. Mol Cell Biol 22:7688–7700. doi: 10.1128/MCB.22.21.7688-7700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou SF, Fujimuro M, Hsieh JJD, Chen L, Miyamoto A, Weinmaster G, Hayward SD. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol 20:2400–2410. doi: 10.1128/MCB.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou K, Huang L, Zhou Z, Hu C, Liu W, Zhou J, Sun H. 2010. Wnt and Notch signaling pathways selectively regulating hematopoiesis. Ann Hematol 89:749–757. doi: 10.1007/s00277-010-0923-3. [DOI] [PubMed] [Google Scholar]
- 22.Brabletz S, Schmalhofer O, Brabletz T. 2009. Gastrointestinal stem cells in development and cancer. J Pathol 217:307–317. doi: 10.1002/path.2475. [DOI] [PubMed] [Google Scholar]
- 23.Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. 2010. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci 30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haass NK, Herlyn M. 2005. Normal human melanocyte homeostasis as a paradigm for understanding melanoma. J Investig Dermatol Symp Proc 10:153–163. doi: 10.1111/j.1087-0024.2005.200407.x. [DOI] [PubMed] [Google Scholar]
- 25.Kumano K, Masuda S, Sata M, Saito T, Lee SY, Sakata-Yanagimoto M, Tomita T, Iwatsubo T, Natsugari H, Kurokawa M, Ogawa S, Chiba S. 2008. Both Notch1 and Notch2 contribute to the regulation of melanocyte homeostasis. Pigment Cell Melanoma Res 21:70–78. doi: 10.1111/j.1755-148X.2007.00423.x. [DOI] [PubMed] [Google Scholar]
- 26.Schouwey K, Beermann F. 2008. The Notch pathway: hair graying and pigment cell homeostasis. Histol Histopathol 23:609–619. [DOI] [PubMed] [Google Scholar]
- 27.Osawa M, Fisher DE. 2008. Notch and melanocytes: diverse outcomes from a single signal. J Invest Dermatol 128:2571–2574. doi: 10.1038/jid.2008.289. [DOI] [PubMed] [Google Scholar]
- 28.Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, Brown EJ, Capobianco AJ, Herlyn M, Liu ZJ. 2005. Activation of Notch1 signaling is required for β-catenin-mediated human primary melanoma progression. J Clin Invest 115:3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu ZJ, Xiao M, Balint K, Smalley KSM, Brafford P, Qiu RH, Pinnix CC, Li XL, Herlyn M. 2006. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res 66:4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- 30.Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, Brafford PA, Xiao M, Himes B, Zabierowski SE, Yashiro-Ohtani Y, Nathanson KL, Bengston A, Pollock PM, Weeraratna AT, Nickoloff BJ, Pear WS, Capobianco AJ, Herlyn M. 2009. Active Notch1 confers a transformed phenotype to primary human melanocytes. Cancer Res 69:5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K, Wong P, Zhang L, Jacobs B, Borden EC, Aster JC, Bedogni B. 2012. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene 31:4609–4618. doi: 10.1038/onc.2011.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma J, Tang XY, Wong PK, Jacobs B, Borden EC, Bedogni B. 2014. Noncanonical activation of Notch1 protein by membrane type 1 matrix metalloproteinase (MT1-MMP) controls melanoma cell proliferation. J Biol Chem 289:8442–8449. doi: 10.1074/jbc.M113.516039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaverdashvili K, Wong P, Ma J, Zhang KM, Osman I, Bedogni B. 2014. MT1-MMP modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1. Pigment Cell Melanoma Res 27:287–296. doi: 10.1111/pcmr.12201. [DOI] [PubMed] [Google Scholar]
- 34.Yana I, Weiss SJ. 2000. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol Biol Cell 11:2387–2401. doi: 10.1091/mbc.11.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner SL, Blair-Zajdel ME, Bunning RA. 2009. ADAMs and ADAMTSs in cancer. Br J Biomed Sci 66:117–128. [DOI] [PubMed] [Google Scholar]
- 36.Anderegg U, Eichenberg T, Parthaune T, Haiduk C, Saalbach A, Milkova L, Ludwig A, Grosche J, Simon JC. 2008. Functions of ADAM10 in human melanoma cells. Exp Dermatol 17:275 (Abstract.) [DOI] [PubMed] [Google Scholar]
- 37.Gangemi R, Amaro A, Gino A, Barisione G, Fabbi M, Pfeffer U, Brizzolara A, Queirolo P, Salvi S, Boccardo S, Gualco M, Spagnolo F, Jager MJ, Mosci C, Rossello A, Ferrini S. 2014. ADAM10 correlates with uveal melanoma metastasis and promotes in vitro invasion. Pigment Cell Melanoma Res 27:1138–1148. doi: 10.1111/pcmr.12306. [DOI] [PubMed] [Google Scholar]
- 38.Lee SB, Schramme A, Doberstein K, Dummer R, Abdel-Bakky MS, Keller S, Altevogt P, Oh ST, Reichrath J, Oxmann D, Pfeilschifter J, Mihic-Probst D, Gutwein P. 2010. ADAM10 is upregulated in melanoma metastasis compared with primary melanoma. J Invest Dermatol 130:763–773. doi: 10.1038/jid.2009.335. [DOI] [PubMed] [Google Scholar]
- 39.Cireap N, Narita D. 2013. Molecular profiling of ADAM12 and ADAM17 genes in human malignant melanoma. Pathol Oncol Res 19:755–762. doi: 10.1007/s12253-013-9639-8. [DOI] [PubMed] [Google Scholar]
- 40.Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. 2001. Regulation of the α-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J 15:1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 41.Srour N, Lebel A, McMahon S, Fournier I, Fugere M, Day R, Dubois CM. 2003. TACE/ADAM-17 maturation and activation of sheddase activity require proprotein convertase activity. FEBS Lett 554:275–283. doi: 10.1016/S0014-5793(03)01159-1. [DOI] [PubMed] [Google Scholar]
- 42.Schlöndorff J, Becherer JD, Blobel CP. 2000. Intracellular maturation and localization of the tumour necrosis factor α convertase (TACE). Biochem J 347:131–138. doi: 10.1042/bj3470131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laprise MH, Grondin F, Cayer P, McDonald PP, Dubois CM. 2002. Furin gene (fur) regulation in differentiating human megakaryoblastic Dami cells: involvement of the proximal GATA recognition motif in the P1 promoter and impact on the maturation of furin substrates. Blood 100:3578–3587. doi: 10.1182/blood.V100.10.3578. [DOI] [PubMed] [Google Scholar]
- 44.Ayoubi TAY, Creemers JWM, Roebroek AJM, Vandeven WJM. 1994. Expression of the dibasic proprotein processing enzyme furin is directed by multiple promoters. J Biol Chem 269:9298–9303. [PubMed] [Google Scholar]
- 45.Creemers JWM, Siezen RJ, Roebroek AJM, Ayoubi TAY, Huylebroeck D, Vandeven WJM. 1993. Modulation of furin-mediated proprotein processing activity by site-directed mutagenesis. J Biol Chem 268:21826–21834. [PubMed] [Google Scholar]
- 46.Reference deleted.
- 47.Gendron FP, Mongrain S, Laprise P, McMahon S, Dubois CM, Blais M, Asselin C, Rivard N. 2006. The CDX2 transcription factor regulates furin expression during intestinal epithelial cell differentiation. Am J Physiol Gastrointest Liver Physiol 290:G310–G318. doi: 10.1152/ajpgi.00217.2005. [DOI] [PubMed] [Google Scholar]
- 48.Khatib AM, Siegfried G, Chretien M, Metrakos P, Seidah NG. 2002. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am J Pathol 160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khatib AM, Siegfried G, Prat A, Luis J, Chretien M, Metrakos P, Seidah NG. 2001. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem 276:30686–30693. doi: 10.1074/jbc.M101725200. [DOI] [PubMed] [Google Scholar]
- 50.Basak A, Chen A, Scamuffa N, Mohottalage D, Basak S, Khatib AM. 2010. Blockade of furin activity and furin-induced tumor cells malignant phenotypes by the chemically synthesized human furin prodomain. Curr Med Chem 17:2214–2221. doi: 10.2174/092986710791331040. [DOI] [PubMed] [Google Scholar]
- 51.Blanchette F, Day R, Dong W, Laprise MH, Dubois CM. 1997. TGFβ1 regulates gene expression of its own converting enzyme furin. J Clin Invest 99:1974–1983. doi: 10.1172/JCI119365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dickson MC, Slager HG, Duffie E, Mummery CL, Akhurst RJ. 1993. RNA and protein localizations of TGFβ2 in the early mouse embryo suggest an involvement in cardiac development. Development 117:625–639. [DOI] [PubMed] [Google Scholar]
- 53.Lee JE, Pintar J, Efstratiadis A. 1990. Pattern of the insulin-like growth factor II gene expression during early mouse embryogenesis. Development 110:151–159. [DOI] [PubMed] [Google Scholar]
- 54.Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. 2008. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest 118:3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Razorenova OV, Agapova LS, Budanov AV, Ivanov AV, Strunina SM, Chumakov PM. 2005. Retroviral reporter systems for assessing the activity of stress-inducible signal transduction pathways controlled by the p53, HIF-1, and HSF-1 transcription factors. Mol Biol 39:286–293. (In Russian.) [PMC free article] [PubMed] [Google Scholar]
- 56.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishimura M, Isaka F, Ishibashi M, Tomita K, Tsuda H, Nakanishi S, Kageyama R. 1998. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49:69–75. doi: 10.1006/geno.1998.5213. [DOI] [PubMed] [Google Scholar]
- 58.Proweller A, Pear WS, Parmacek MS. 2005. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J Biol Chem 280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- 59.Buas MF, Kabak S, Kadesch T. 2009. Inhibition of myogenesis by Notch: evidence for multiple pathways. J Cell Physiol 218:84–93. doi: 10.1002/jcp.21571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reference deleted.
- 61.Knäuper V, Will H, Lopez-Otin C, Smith B, Atkinson SJ, Stanton H, Hembry RM, Murphy G. 1996. Cellular mechanisms for human procollagenase-3 (MMP-13) activation. Evidence that MT1-MMP (MMP-14) and gelatinase A (MMP-2) are able to generate active enzyme. J Biol Chem 271:17124–17131. [DOI] [PubMed] [Google Scholar]
- 62.Sato H, Takino T, Okada Y, Cao J, Shinagawa A, Yamamoto E, Seiki M. 1994. A matrix metalloproteinase expressed on the surface of invasive tumour cells. Nature 370:61–65. doi: 10.1038/370061a0. [DOI] [PubMed] [Google Scholar]
- 63.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. 1995. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem 270:5331–5338. [DOI] [PubMed] [Google Scholar]
- 64.Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. 2007. DSL ligand endocytosis physically dissociates Notch 1 heterodimers before activating proteolysis can occur. J Cell Biol 176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Tetering G, Vooijs M. 2011. Proteolytic cleavage of Notch: “HIT and RUN. ” Curr Mol Med 11:255–269. doi: 10.2174/156652411795677972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guimont P, Grondin F, Dubois CM. 2007. Sox9-dependent transcriptional regulation of the proprotein convertase furin. Am J Physiol Cell Physiol 293:C172–C183. doi: 10.1152/ajpcell.00349.2006. [DOI] [PubMed] [Google Scholar]
- 67.Blanchette F, Rudd P, Grondin F, Attisano L, Dubois CM. 2001. Involvement of Smads in TGFβ1-induced furin (fur) transcription. J Cell Physiol 188:264–273. doi: 10.1002/jcp.1116. [DOI] [PubMed] [Google Scholar]
- 68.Chang HM, Cheng JC, Klausen C, Leung PC. 2015. Recombinant BMP4 and BMP7 increase activin A production by up-regulating inhibin βA subunit and furin expression in human granulosa-lutein cells. J Clin Endocrinol Metab 100:E375–E386. doi: 10.1210/jc.2014-3026. [DOI] [PubMed] [Google Scholar]
- 69.Remacle AG, Chekanov AV, Golubkov VS, Rozanov DV, Fugere M, Day R, Strongin AY. 2006. Proprotein convertases and glycosylation regulate MT1-MMP activity. Matrix Biol 25(Suppl 1):S49. doi: 10.1016/j.matbio.2006.08.136. [DOI] [Google Scholar]
- 70.Bedogni B, Powell MB. 2009. Unique transforming properties of Notch1 in human melanocytes. Pigment Cell Melanoma Res 22:702–703. doi: 10.1111/j.1755-148X.2009.00625.x. [DOI] [PubMed] [Google Scholar]
- 71.Bedogni B, Powell MB. 2009. Hypoxia, melanocytes and melanoma—survival and tumor development in the permissive microenvironment of the skin. Pigment Cell Melanoma Res 22:166–174. doi: 10.1111/j.1755-148X.2009.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell 5:207–216. doi: 10.1016/S1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 73.Hartmann D, De Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Brabant V, Luebke T, Illert AL, von Figura K, Saftig P. 2002. Deficiency for the disintegrin metalloprotease ADAM10 causes disturbed α-secretase function and a Notch deficiency-related phenotype in mice. Neurobiol Aging 23:S183. [Google Scholar]
- 74.Krebs LT, Xue YZ, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T. 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev 14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 75.Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. 2005. Essential role of endothelial Notch1 in angiogenesis. Circulation 111:1826–1832. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ntziachristos P, Lim JS, Sage J, Aifantis I. 2014. From fly wings to targeted cancer therapies: a centennial for Notch signaling. Cancer Cell 25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.