Abstract
Cdc14 is a phosphatase that controls mitotic exit and cytokinesis in budding yeast. In mammals, the two Cdc14 homologues, Cdc14A and Cdc14B, have been proposed to regulate DNA damage repair, whereas the mitotic exit and cytokinesis rely on another phosphatase, PP2A-B55α. It is unclear if the two Cdc14s work redundantly in DNA repair and which repair pathways they participate in. More importantly, their target(s) in DNA repair remains elusive. Here we report that Cdc14B knockout (Cdc14B−/−) mouse embryonic fibroblasts (MEFs) showed defects in repairing ionizing radiation (IR)-induced DNA double-strand breaks (DSBs), which occurred only at late passages when Cdc14A levels were low. This repair defect could occur at early passages if Cdc14A levels were also compromised. These results indicate redundancy between Cdc14B and Cdc14A in DSB repair. Further, we found that Cdc14B deficiency impaired both homologous recombination (HR) and nonhomologous end joining (NHEJ), the two major DSB repair pathways. We also provide evidence that Cdh1 is a downstream target of Cdc14B in DSB repair.
INTRODUCTION
Genome stability of mammalian cells is constantly challenged by DNA damage resulting from DNA replication errors and attacks by cellular metabolites, radiation, and other environmental hazards. If not repaired, DNA damage can lead to gene mutations and even chromosome aberrations. Thus, dealing with damaged DNA is the utmost priority of a cell. Cells respond to DNA damage by evoking DNA damage checkpoints which rely on a number of protein kinases to transduce the damage signal (1). A major function of the checkpoints is to activate DNA damage repair mechanisms (2). It has become apparent that dynamic phosphorylation of DNA repair proteins regulated by opposing kinases and phosphatases plays an important role in the timely response to and repair of DNA damage. Cdc14 phosphatases have been shown to guard the genome stability, probably by reversing Cdk phosphorylation (3, 4).
Cdc14 phosphatases are highly conserved across species, especially with a conserved N-terminal domain for substrate recognition and catalysis (3, 5). It was first identified as an essential cell cycle regulator in budding yeast by counteracting Cdk activity to allow mitotic exit (6). However, in higher eukaryotes, Cdc14 seems to play roles other than cell cycle control. Recent functional studies in zebrafish showed redundant roles of the two vertebrate Cdc14 homologues, Cdc14A and Cdc14B, in ciliogenesis. Cdc14A- or Cdc14B-deficient zebrafish embryos displayed shorter cilia in Kupffer's vesicle, and the cilium length defects could be rescued by injection of Cdc14B or Cdc14A mRNA, respectively (7). In mammals, Cdc14A and Cdc14B appear to play roles in DNA repair (3, 8–10). Knockout of Cdc14A or Cdc14B in chicken or human cell lines was shown to cause DNA repair defects (9). To study the physiological functions of Cdc14B, we previously generated Cdc14B knockout (Cdc14B−/−) mice and also showed DNA repair defects in its absence (10). Consistent with a role in DNA damage repair, Cdc14B−/− mice developed early-onset aging phenotypes, including cataracts and kyphosis. Interestingly, the repair defects in Cdc14B−/− mouse embryonic fibroblasts (MEFs) appeared only in late-passage cells. It is unclear why that is the case. Furthermore, the identity of the target of Cdc14B in DNA damage repair remains elusive.
Here we report that Cdc14A and Cdc14B are redundant in DNA damage repair. We show that these two phosphatases are required for both homologous recombination (HR)- and nonhomologous end joining (NHEJ)-mediated double-strand break (DSB) repair. Further, we provide evidence that Cdh1/Fzr1 is a downstream target of Cdc14B in DSB repair.
MATERIALS AND METHODS
Cell culture.
MEFs were prepared from embryos at embryonic day 13.5 (E13.5) and genotyped as reported previously (10). The mice used for MEF isolations had been backcrossed to a C57BL/6 background for more than 10 generations. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 15% fetal bovine serum (FBS). Cells were trypsinized and passaged every 3 days at a ratio of 1:5 at early passages or 1:3 at late passages. Cells from the third or fourth passage (P3 or P4) were used as early-passage MEFs, and cells from the seventh or eighth passage (P7 or P8) were used as late-passage MEFs.
293T cells were purchased from the American Type Culture Collection and cultured in DMEM with 10% FBS. The U2OS cells with a single copy of the direct-repeat–green fluorescent protein (DR-GFP) construct incorporated were kindly provided by Shiaw-Yih Lin (MD Anderson Cancer Center, Houston, TX) (11), and the cells were maintained in McCoy's 5A medium with 10% FBS and 100 μg/ml hygromycin.
Plasmids, siRNAs, and transfection.
pLKO.1 puro, pMD2.G, psPAX2, pEGFP-N1, pCAGGS, and pCSCMV:tdTomato were obtained from Addgene. Short hairpin RNAs (shRNAs) against Cdc14A, Cdc14B, and Cdh1 (shCdc14A, shCdc14B, and shCdh1, respectively) were designed with the vector pLKO.1 puro as previously described (12, 13) using oligonucleotides targeting the following sequences: mouse Cdc14A (AAGATAGTGCACTACACCTCT), human Cdc14A (AAGCACAGTAAATACCCACTA), human Cdc14B (AATATGAGAACTTCTACGCAG), and mouse Cdh1 (AACACGCTCTACAAAGGAATC). Cdh1 or Cdc14A was cloned from mouse or human cDNA to pENTR with the following primers and then subcloned to pInducer20 as previously described (14) with Gateway LR Clonase II (Invitrogen): Cdh1-F (AAGGAAAAAAGCGGCCGCATGGACTACAAGGACGATGATGACAAGGACCAGGACTATGAGCGAAGG) with Cdh1-R (ACGCGTCGACCTATCGGATCCGGGTGAAGAGGTT) and Cdc14A-F (AAGGAAAAAAGCGGCCGCATGGCAGCGGAGTCAGGGGAAC) with Cdc14A-R (ACGCGTCGACTTACTTGTCATCATCGTCCTTGTAGTCGTAATGAACATATTCAGACTG). Phosphodeficient (Cdh1-4A) and phosphomimetic (Cdh1-4D) Cdh1 mutants were generated by site-directed mutagenesis in the four putative Cdk consensus sites (S40, T121, S151, and S163, mutated to alanine in Cdh1-4A or aspartic acid in Cdh1-4D) as previously described (15).
To knock down genes in U2OS cells, the following small interfering RNAs (siRNAs) were used: siNC (siRNA universal negative control 1) (SIC001; Sigma-Aldrich), siCdc14A (SASI_Hs01_00132334; Sigma-Aldrich), siCdc14B (GAGCAGCCUUCUCCAAACUdTdT) (16), siBRCA1 (GGGAUACCAUGCAACAUAAdTdT) (17), and si53BP1 (GAAGGACGGAGUACUAAUAdTdT) (18).
Lipofectamine 2000 (Invitrogen) was used for all the plasmid and siRNA transfection except electroporation using Gene Pulser II (Bio-Rad) in the NHEJ assay. For gene knockdown with shRNAs, each shRNA was cotransfected with psPAX2 and pMD2.G in 293T cells to produce lentivirus. The lentivirus was collected to infect the target cells (MEFs and U2OS cells) as previously described (19). After puromycin selection, the infected target cells were ready for the following assays.
RNA extraction and reverse transcription-PCR (RT-PCR).
Total RNA was extracted from cells using TRIzol (Invitrogen). One microgram of total RNA was used for cDNA synthesis using SuperScript III (Invitrogen). Quantitative PCR (qPCR) was performed and analyzed with ABI Prism 7000 using SYBR green Master (Rox) (Roche). Primers for qPCR were as follows: mCdc14A-F (TGGACCTCTGAACTTGGCAAT) and mCdc14A-R (AGATGACGGCATAAGCACCTAT), m18s-F (ACCGCAGCTAGGAATAATGGA) and m18s-R (GCCTCAGTTCCGAAAACCA), hCdc14A-F (CAAAACATGGAACGATTTGGA) and hCdc14A-R (GATGAACTTAATCTGAAAGGC), hCdc14B-F (CAAACGCTTTACGGATGCTGG) and hCdc14B-R (TGATGTAGCAGGCTATCAGAGT), and hGAPDH-F (GAAATCCCATCACCATCTTCCAGG) and hGAPDH-R (GAGCCCCAGCCTTCTCCATG) (18). The data were analyzed by the comparative threshold cycle (CT) method (20, 21) with the m18s or human glyceraldehyde-3-phosphate dehydrogenase (hGAPDH) gene as the internal control gene. The relative expression levels of the target genes were normalized to that in control cells, and statistical analyses were performed by analysis of variance (ANOVA) with data from three independent experiments.
Western blotting and IF staining.
For Western blotting, cells were lysed with radioimmunoprecipitation assay (RIPA) buffer supplemented with proteinase inhibitors and phosphatase inhibitors (Roche), and the protein concentrations were determined with the Bradford assay (Bio-Rad). The primary antibodies for Western blotting included anti-BRCA1 (sc-6954; Santa Cruz), anti-53BP1 (NB100-304; Novus), anti-Cdh1 (CC43; Calbiochem), anti-GAPDH (MAB374; Millipore), and anti-tubulin-α (625902; BioLegend).
For immunofluorescent (IF) staining, MEFs were seeded on coverslips and fixed in 4% paraformaldehyde (PFA) at 0.5, 2, 6 and 24 h after treatment with 10 Gy of ionizing radiation (IR). The cells were then permeabilized in 0.5% Triton X-100, blocked in 5% bovine serum albumin (BSA) in phosphate-buffered saline–Tween 20 (PBST), and incubated with primary antibodies overnight at 4°C. After three PBS washes, cells were incubated with Cy3- or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (115-165-146 or 111-095-045; Jackson ImmunoResearch) for 1 h at room temperature. Cells were counterstained with 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) and mounted on slides before imaging. The primary antibodies for IF staining included anti-γH2AX (05-636; Millipore) and anti-53BP1 (NB100-304; Novus).
Neutral comet assay.
MEFs were collected at 0 h (no IR) and at 0.5 h or 6 h after treatment with 10 Gy IR for neutral comet assay to analyze DSB damage in the cells. The neutral comet assay was performed according to the manual for the CometAssay kit (Trevigen). After SYBR gold (Invitrogen) staining, the images of the comet slides were taken by fluorescence microscopy. The comet parameter tail moment (TM) was gauged for at least 50 cells in each experiment by CometScore software (TriTik). The cells with TMs of >5 were considered damaged cells, and the percentage of damaged cells was counted for each slide. The counting results from three independent experiments were analyzed by ANOVA.
HR assay.
The HR assay was performed as previously described (11, 22). At 48 h after siRNA transfection, U2OS (DR-GFP) cells were transfected with pCBA-SceI or pCAGGS (mock transfection). pCAGGS-mCherry was cotransfected as a control for transfection efficiency. Another 48 h later, cells were collected and flow cytometry was performed to detect GFP+ and mCherry+ cells with LSR Fortessa at the Cytometry and Cell Sorting Center of Baylor College of Medicine. Data were analyzed with BD FACSDiva software.
NHEJ assay.
The NHEJ assay was performed as previously described (23). At 72 h after siRNA transfection, U2OS cells were electroporated with 5 μg of linearized pCSCMV:tdTomato (digested with BamHI between the promoter and tdTomato coding sequence) and 5 μg of pEGFP-N1 per 106 cells. The electroporation was performed at 250 V and 950 μF in 800-μl cuvettes (VWR) according to the instructions for the Gene Pulser II (Bio-Rad). The cells were returned to culture for 5 h and then collected for fluorescence-activated cell sorter (FACS) analyses of tdTomato+ and GFP+ cells.
Cell cycle analysis and BrdU staining.
To analyze the cell cycle distribution, MEFs or U2OS cells were fixed in 70% ethanol for at least 2 h at 4°C, and then the cells were washed twice in PBS and resuspended in DAPI staining solution (1 μg/ml DAPI in 0.1% Triton X-100–PBS) for 30 min in the dark before FACS. Cell singlets were gated by the pulse width-pulse area signal of DAPI, and the cell cycle distribution was analyzed with FlowJo software.
Bromodeoxyuridine (BrdU) pulse-labeling and staining were performed according to the manual of the FITC BrdU Flow kit (BD Biosciences). MEFs were pulse-labeled with 10 μM BrdU for 45 min and then collected for BrdU staining using the kit. FACS was performed to detect FITC+ (BrdU+) cells and measure total DNA stained by 7-aminoactinomycin D (7-AAD).
2D gel analysis.
MEFs with or without IR treatment (10 Gy, 0.5 h) were collected for two-dimensional (2D) PAGE according to the manual of the 2-D Starter kit (Bio-Rad). The first-dimension isoelectric focusing (IEF) was performed in a Protean IEF Cell (Bio-Rad), followed by the second-dimension SDS-PAGE. The proteins were transferred to polyvinylidene difluoride (PVDF) membranes for Western blotting with anti-Cdh1 (CC43; Calbiochem).
RESULTS
Cdc14B−/− MEFs show a DSB repair defect only at late passages.
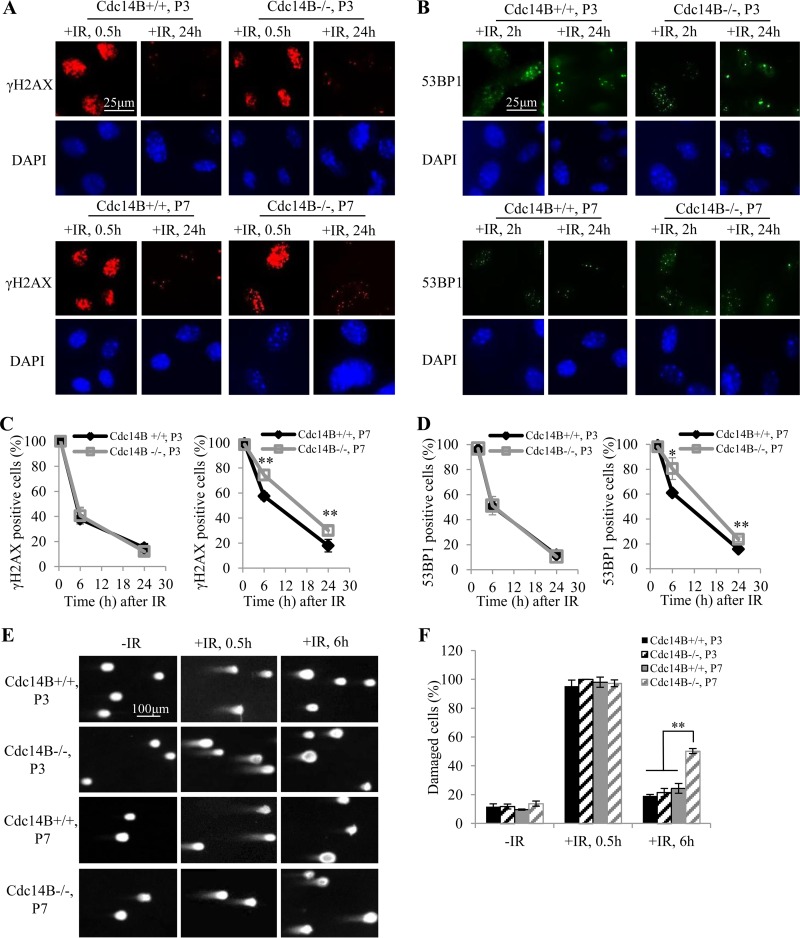
We previously reported that Cdc14B−/− MEFs underwent premature senescence and accumulated DNA damage at late passages (P7 or later) (10). We compared the DNA damage responses (DDR) in early- and late-passage Cdc14B−/− MEFs after 10 Gy of IR exposure to induce DSB. γH2AX was used as a marker for DNA damage, as it forms foci at DSBs. At 0.5 h after IR, nearly 100% of the cells were positive for γH2AX (>20 foci/cell). With repairing of the damage, the γH2AX focus number was reduced, and consequently, the percentage of γH2AX-positive cells decreased (Fig. 1A). There was no significant difference between Cdc14B+/+ and Cdc14B−/− MEFs in percentage of γH2AX-positive cells at early passages (Fig. 1C, left panel). At late passages, however, Cdc14B−/− MEFs showed significantly higher percentages of γH2AX-positive cells than Cdc14B+/+ MEFs at 6 h and 24 h after IR (Fig. 1C, right panel). The staining of 53BP1 foci, a late DDR marker, showed dynamics similar to those for γH2AX staining (Fig. 1B). At 2 h after IR, almost 100% of the cells were positive for 53BP1 (>10 foci/cell). More 53BP1 foci remained at 6 h and 24 h after IR in late-passage Cdc14B−/− MEFs than that in Cdc14B+/+ MEFs (Fig. 1D). These results indicate defective DSB repair in late-passage Cdc14B−/− MEFs.
FIG 1.
Cdc14B−/− MEFs are defective in DNA damage repair at late passages. (A and B) Immunofluorescence (IF) staining of the DNA damage markers γH2AX (A) and 53BP1 (B) in early-passage (P3) and late-passage (P7) MEFs after 10 Gy of IR. (C) Quantification of γH2AX-positive cells (>20 foci) in the P3 and P7 MEFs from panel A. (D) Quantification of 53BP1-positive cells (>10 foci) in the P3 and P7 MEFs from panel B. At least 100 cells were counted for each experiment. The error bars indicate standard deviations for three independent experiments. (E) Neutral comet assay in P3 and P7 MEFs after 10 Gy of IR. (F) Quantification of damaged cells with DNA double-strand breaks from panel E. Tail moments (TMs) were gauged for at least 50 cells in each experiment. The cells with TMs of >5 were counted as damaged cells. The error bars indicate standard deviations for three independent experiments. Statistical significance was assessed by ANOVA (*, P < 0.05; **, P < 0.01).
We performed a neutral comet assay to further demonstrate the DSB repair defect in late-passage Cdc14B−/− MEFs. Damaged DNA with DSBs migrates faster than undamaged DNA in gel electrophoresis, giving rise to a comet tail. The cells with tail moments of more than 5 were considered damaged. At 0.5 h after IR, most cells were damaged, showing large comet tails (Fig. 1E and F). At 6 h after IR, the damage was largely repaired in most cells, which showed smaller or no comet tails. There were still large comet tails in about half of late-passage Cdc14B−/− MEFs, and the percentage of damaged cells was significantly higher than that in early-passage MEFs or late-passage Cdc14B+/+ MEFs (Fig. 1E and F). Together, the above results demonstrate the DSB repair defect in late-passage Cdc14B−/− MEFs.
The genetic background of the MEFs might have an impact on the DSB repair. Previous work in MEFs isolated from mice that had been backcrossed to a C57BL/6 background for six generations showed defective repair of IR-induced DNA damage in early-passage (P3) Cdc14B−/− MEFs (10). However, after backcrossing for more than 10 generations, this defect of DSB repair in Cdc14B−/− MEFs diminished with purer C57BL/6 background, as shown in left panels of Fig. 1C and D. The difference between the early-passage and late-passage MEFs in DSB repair as shown in Fig. 1 is consistent with a previous report of lower growth rates and more cellular senescence in Cdc14−/− MEFs at late passages only (10). Therefore, the MEFs from Cdc14B knockout mice after backcrossing for more than 10 generations were used in the rest of experiments involving MEFs.
Cdc14A and Cdc14B are redundant in DSB repair.
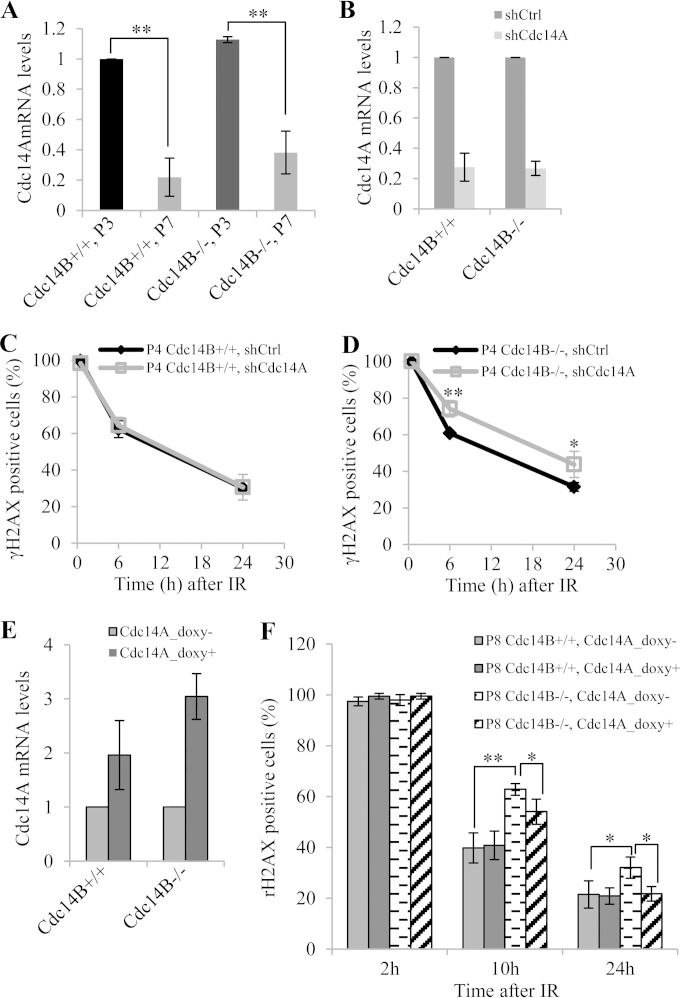
What is the factor(s) that causes the difference between the early-passage and late-passage MEFs in DSB repair? As Cdc14A is another mammalian homologue of Cdc14 and may have functional overlap with Cdc14B, we examined whether Cdc14A could be the factor. It is possible that at early passages Cdc14A is compensating for the loss of Cdc14B but at late passages the compensation is no longer present, perhaps due to reduced expression. Therefore, we measured the mRNA levels of Cdc14A by RT-PCR in early- and late-passage MEFs. As shown in Fig. 2A, Cdc14A levels were 3- to 5-fold lower in both Cdc14B+/+ and Cdc14B−/− MEFs at late passages than at early passages. To demonstrate that it is Cdc14A that is compensating for the loss of Cdc14B in early passages, we depleted its expression through shRNA-mediated knockdown (Fig. 2B). Indeed, while wild-type MEFs repaired DSBs with same kinetics regardless of the status of Cdc14A (Fig. 2C), Cdc14B−/− MEFs with Cdc14A knocked down showed significantly higher percentages of γH2AX-positive cells than those with control shRNA at 6 h and 24 h after IR (Fig. 2D). Moreover, with induced Cdc14A overexpression (Fig. 2E), the high percentage of remaining γH2AX-positive cells in late-passage Cdc14B−/− MEFs at 10 h and 24 h after IR was at least partially restored (Fig. 2F). These results indicate that Cdc14A and Cdc14B are redundant in DSB repair. It is unclear at present why the expression of Cdc14A would go down at late passages.
FIG 2.
Cdc14A and Cdc14B redundantly regulate DNA DSB repair. (A) RT-PCR quantification of Cdc14A expression in MEFs. The error bars indicate standard deviations for three independent experiments. (B) RT-PCR analysis of Cdc14A knockdown efficiency in P4 MEFs. shCtrl, control shRNA. (C and D) Quantification of γH2AX-positive cells (>20 foci) among P4 Cdc14B+/+ (C) and Cdc14B−/− (D) MEFs with or without Cdc14A knockdown. IF staining of γH2AX was performed with MEFs fixed at 0.5 h, 6 h, and 24 h after 10 Gy of IR. (E) RT-PCR analysis of Cdc14A overexpression in P8 MEFs. Cdc14A overexpression with pInducer20-Cdc14A was induced by 0.05 μg/ml doxycycline. (F) Quantification of γH2AX-positive cells among P8 Cdc14B+/+ and Cdc14B−/− MEFs with or without Cdc14A overexpression. IF staining of γH2AX was performed with MEFs fixed at 2 h, 10 h, and 24 h after 10 Gy of IR. Statistical significance was assessed by ANOVA (*, P < 0.05; **, P < 0.01).
Cdc14B and Cdc14A redundantly regulate both HR- and NHEJ-mediated DSB repair.
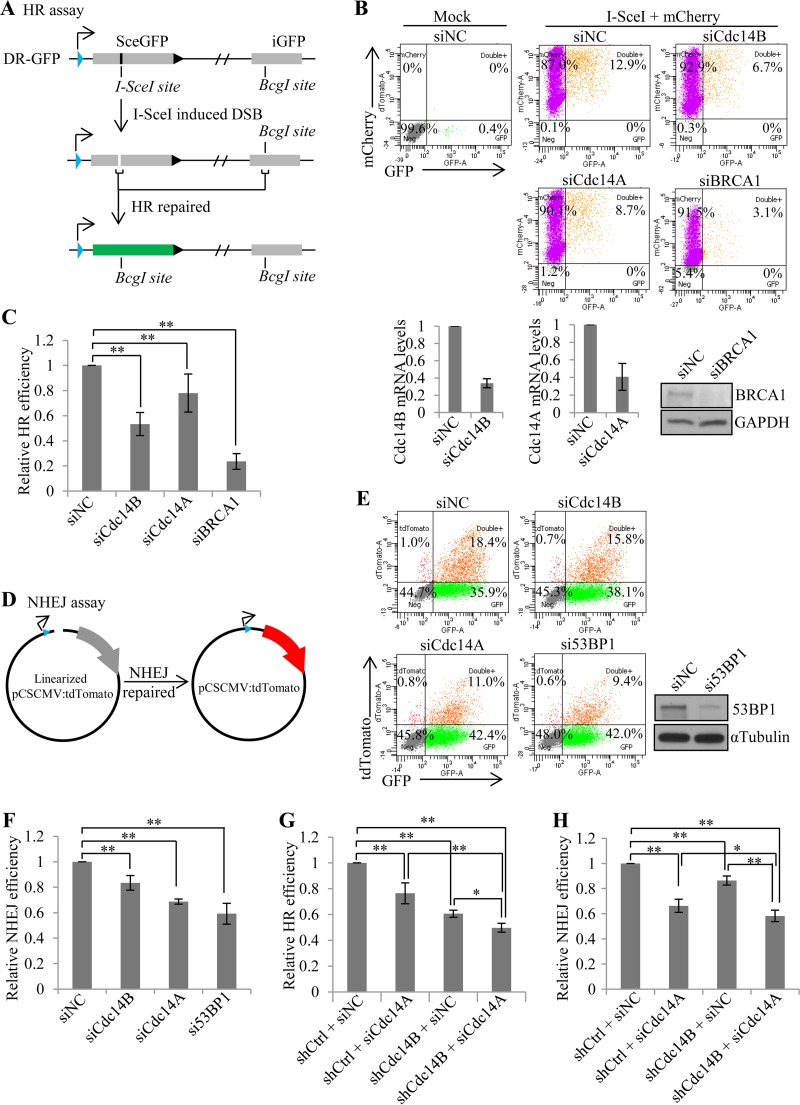
DSBs are repaired through two different pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ) (24). The choice between HR and NHEJ is determined by a number of factors, including the stage of the cell cycle (24–26). We wondered which repair pathway would require Cdc14B and Cdc14A. To answer that, we first looked at HR repair. We took advantage of the established HR reporter assay (22, 27). This assay was established in U2OS cells with the integration of a construct (DR-GFP) containing two direct repeats of GFP separated by some distance. Both GFPs are nonfunctional. The first is a mutant GFP that contains early-terminating stop codons within an I-SceI site in the middle, and the second is an internal truncate of wild-type GFP. When the first GFP is broken by I-SceI cutting, it can be repaired by either NHEJ or HR with the second GFP as a donor. Only if it is repaired by HR will the reading frame of the first GFP be restored and become functional (Fig. 3A). We depleted Cdc14B, Cdc14A, or BRCA1 (an essential HR protein) in the reporter cell line with siRNA (Fig. 3B, lower panels) and then transfected in pCBA-SceI along with an mCherry expression plasmid to control transfection efficiency. At 48 h after the transfection, the cells were collected for FACS analysis (Fig. 3B, upper panels). The HR efficiency, as indicated by the percentage of GFP+ cells among mCherry+ cells, in Cdc14B-depleted cells reached only about 50% of that in the control cells (Fig. 3C), which is comparable to that of BRCA1 knockdown (Fig. 3C). Cdc14A depletion reduced the HR efficiency to about 80% of that in the control cells. The HR assays with shRNA targeting different sequences of Cdc14B (Fig. 3G) or Cdc14A (data not shown) showed similar results. These results suggest that both Cdc14A and Cdc14B play a role in DSB repair through HR. The difference in the HR efficiency affected by their depletion might be simply due to the difference in their respective knockdown efficiencies.
FIG 3.
Cdc14B and Cdc14A redundantly regulate DNA DSB repair by both homologous recombination (HR) and nonhomologous end joining (NHEJ). (A) Diagram of the HR reporter assay. U2OS cells were integrated with the DR-GFP reporter construct, which carries a nonfunctional mutant GFP (SceGFP) and an internal truncate of GFP (iGFP). The reporter U2OS cells were transfected with I-SceI along with an mCherry-expressing plasmid to control transfection efficiencies. I-SceI digested SceGFP and induced DSB. Only by HR repair with iGFP as the homologous template could damaged SceGFP be restored to a functional GFP. GFP+ cells were HR-repaired cells. (B) FACS analyses of HR assay in the reporter cells with siRNA-mediated gene silencing of Cdc14B, Cdc14A, or BRCA1. The bottom panels show knockdown efficiency by RT-PCR (Cdc14B and Cdc14A) or Western blotting (BRCA1). (C) Quantification of the HR assay (B). The HR efficiencies (the percentage of GFP+ cells among mCherry+ cells) were normalized to that in control (siNC) cells. (D) Diagram of the NHEJ reporter assay. A tdTomato expression plasmid was linearized by enzyme digestion between the promoter and the tdTomato sequence, making it nonfunctional. U2OS cells were transfected with the linearized tdTomato plasmid and a GFP-expressing plasmid to control for transfection efficiencies. The DSB ends between the promoter and the tdTomato sequence could be joined by NHEJ, and tdTomato+ cells were NHEJ-repaired cells. (E) FACS analyses of NHEJ assay in U2OS cells with siRNA-mediated gene silencing of Cdc14B, Cdc14A, or 53BP1. The right panel shows 53BP1 knockdown by Western blotting. (F) Quantification of the NHEJ assay (E). The NHEJ repair efficiencies (the percentage of tdTomato+ cells among GFP+ cells) were normalized to that in control (siNC) cells. (G and H) Quantification of the HR assay (G) or NHEJ assay (H) performed in the reporter U2OS (DR-GFP) cells with shRNA- and siRNA-mediated single or double knockdown of Cdc14A and Cdc14B. shCtrl, control shRNA. The error bars indicate standard deviations for at least three independent experiments. Statistical significance was assessed by one-way ANOVA (*, P < 0.05; **, P < 0.01).
Next, we asked if Cdc14B and Cdc14A also play a role in NHEJ. The NHEJ assay (23) is based on the rejoining of a plasmid cut between the promoter and the coding sequence of a fluorescent protein tdTomato (Fig. 3D). The rejoining makes the expression of tdTomato possible, and the cells become tdTomato positive, which is quantifiable through FACS analysis (Fig. 3E). The depletion of Cdc14B with either siRNA (Fig. 3F) or shRNA (Fig. 3H) resulted in a slight (∼20%) but significant reduction in NHEJ efficiency, as indicated by the percentage of tdTomato+ cells among GFP+ cells. Cdc14A depletion resulted in a greater reduction (∼30%) in NHEJ efficiency, similar to the case for the depletion of 53BP1, an essential NHEJ protein.
Since the γH2AX staining results indicate the redundancy between Cdc14B and Cdc14A in DSB repair, we further tested their redundancy in HR and NHEJ assays. Indeed, double depletion of Cdc14B and Cdc14A reduced the HR efficiency (Fig. 3G) and NHEJ efficiency (Fig. 3H) more than single depletion, indicating a redundant role of Cdc14B and Cdc14A in both HR- and NHEJ-mediated DSB repair.
Cdc14B and Cdc14A depletion does not alter cell cycle distribution.
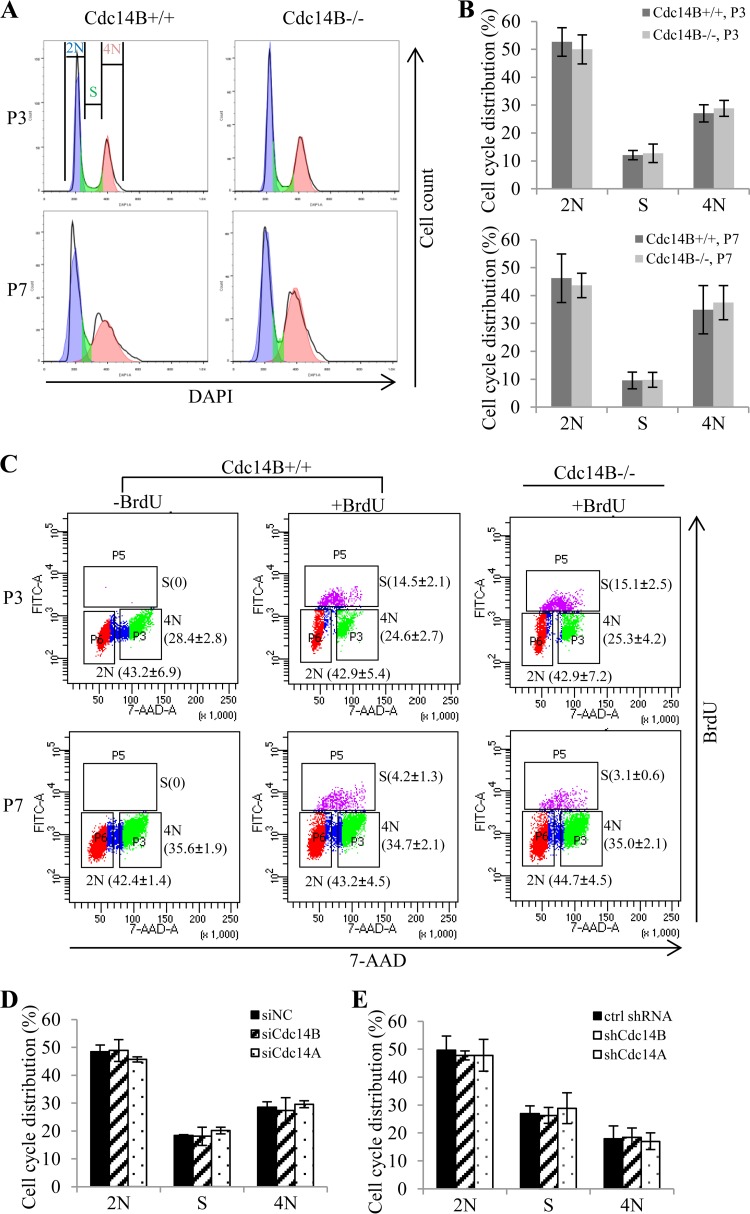
The cell cycle stage is a major determinant for the choice and efficiency of HR and NHEJ, with HR dominating S/G2 phase and NHEJ functioning throughout the cell cycle but most significantly in G1 (24–26). We therefore analyzed the cell cycle distribution after Cdc14B depletion and Cdc14A depletion. By FACS analyses of the DNA content after DAPI staining, we did not observe a significant change in cell cycle distribution between Cdc14B−/− and Cdc14B+/+ MEFs either at early or late passages (Fig. 4A and B). BrdU pulse-labeling and staining showed a lower percentage of S phase cells in the less proliferative late-passage MEFs, but there was no significant difference between Cdc14B−/− and Cdc14B+/+ MEFs at either early or late passages when the DNA damage repair assays were performed (Fig. 4C). Similarly, the cell cycle distribution was not altered in U2OS cells after Cdc14B depletion or Cdc14A depletion (Fig. 4D and E).
FIG 4.
Cdc14 deficiency does not alter cell cycle distribution in the DSB repair assays. (A) Cell cycle analyses of P3 and P7 MEFs by detection of DNA content (DAPI staining) with flow cytometry. (B) Quantification of the cell cycle distribution of the MEFs from panel A. (C) BrdU staining of P3 and P7 MEFs. The MEFs were labeled with 10 μM BrdU for 45 min and stained with FITC-conjugated anti-BrdU. Unlabeled cells were included as negative controls for BrdU staining. Total DNA was stained with 7-AAD for cell cycle analysis by flow cytometry. The numbers (mean ± standard deviation for three independent experiments) indicate the percentage of cells with a DNA content of 2N or 4N and cells in S phase (BrdU positive). (D and E) Cell cycle analyses of the U2OS cells with siRNA (D)- or shRNA (E)-mediated Cdc14 knockdown in the HR assay and NHEJ assay. The error bars indicate standard deviations for at least three independent experiments. ctrl, control. Statistical analyses were performed by unpaired t test (B and C) or one-way ANOVA (D and E). *, P < 0.05; **, P < 0.01.
Cdh1/Fzr1 is a downstream mediator of Cdc14B in DSB repair.
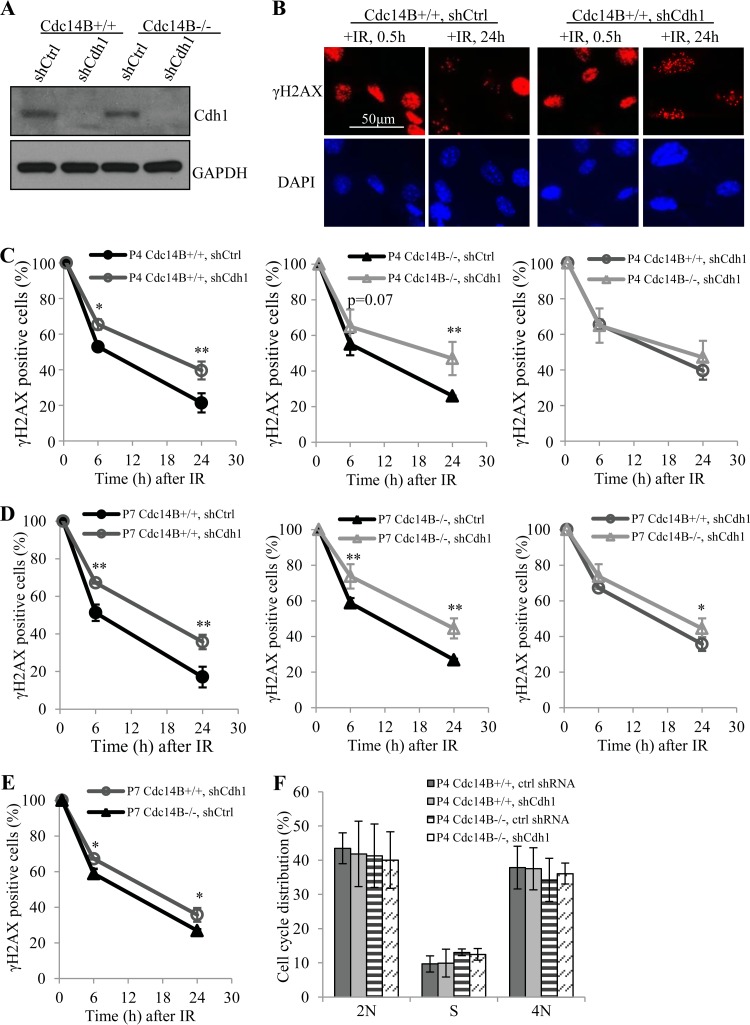
In budding yeast, Cdc14 is known to reverse the phosphorylation by CDC28 on a number of proteins involved in mitotic exit, among which is Cdh1/Fzr1, one of the two adaptor proteins of the anaphase-promoting complex (APC) (28). APC/Cdh1 functions as an E3 ubiquitin ligase, and Cdh1 works better in its dephosphorylated form (29). Mammalian Cdh1 is also very likely a substrate of Cdc14s (30–32). Further, Cdh1 has been reported to regulate the repair of DNA damage induced by UV (33, 34). Thus, it is reasonable to believe that the DNA damage repair defects seen in the absence of Cdc14B are due to the impaired function of Cdh1. To test that, we first examined whether the depletion of Cdh1 would exaggerate the repair defects in Cdc14B-deficient cells. If Cdc14B and Cdh1 work independently in DNA repair, we would see an exaggeration. Otherwise, if Cdc14B works through Cdh1, we would not see such an exaggeration. To that end, we depleted Cdh1 expression in wild-type and Cdc14B−/− MEFs via shRNA-mediated gene silencing (Fig. 5A). These cells were then subjected to IR, and the DSB repair process was assessed by γH2AX focus staining and quantification (Fig. 5B). At early passages, the depletion of Cdh1 caused significant delays in repair regardless of the status of Cdc14B, and the absence of Cdc14B did not further delay the repair, as expected (Fig. 5C). At late passages, again, the depletion of Cdh1 caused repair delays in both wild-type and Cdc14B−/− MEFs. The cells lacking both Cdc14B and Cdh1 experienced slightly worse repair delays than those with either single deficiency (Fig. 5D), suggesting that Cdc14B function in DSB repair goes primarily through Cdh1 but might have a small portion independent of Cdh1. More importantly, the depletion of Cdh1 caused much more significant repair delays than that of Cdc14B deficiency (Fig. 5E), indicating that Cdh1 plays a much more important role than Cdc14B in DNA damage repair and that its dephosphorylation could be accomplished by other phosphatases, including Cdc14A.
FIG 5.
Cdh1 depletion leads to severe DNA DSB repair defects in MEFs. (A) Western blotting of Cdh1 knockdown in MEFs. shCtrl, control shRNA. (B) IR-induced focus staining of γH2AX in early-passage (P4) MEFs with Cdh1 knockdown after 10 Gy of IR. shCtrl, control shRNA. (C) Quantification of γH2AX-positive cells (>20 foci) in early-passage MEFs from panel B. (D) Quantification of γH2AX-positive cells (>20 foci) in late-passage MEFs from panel B. (E) Comparison of Cdh1 depletion and Cdc14B deficiency (P7) for γH2AX-positive cells after 10 Gy of IR. (F) Cell cycle analyses of P4 MEFs with Cdh1 knockdown. ctrl, control. Statistical significance was assessed by ANOVA (*, P < 0.05; **, P < 0.01).
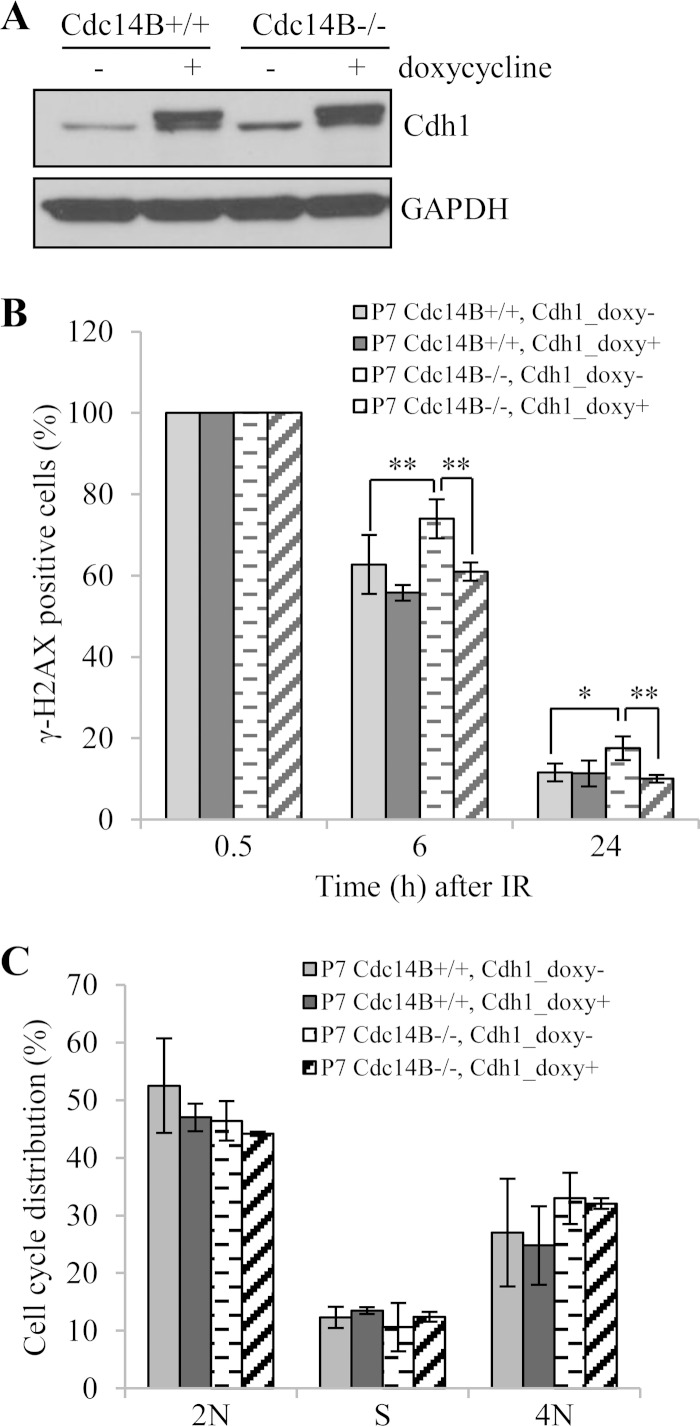
The above results place Cdc14B upstream of Cdh1 in DNA damage repair. To further that conclusion, we overexpressed Cdh1 to determine if the overexpression could rescue the repair defects in Cdc14B-deficient cells. We therefore established conditional expression of Cdh1 in both wild-type and Cdc14B−/− MEFs (Fig. 6A). Upon the induction of Cdh1 expression, we observed that the percentage of γH2AX-positive cells at 6 h and 24 h after IR in late-passage Cdc14B−/− MEFs was reduced to a level similar to that in Cdc14B+/+ MEFs (Fig. 6B), strongly supporting the conclusion that Cdc14B works primarily through Cdh1 in DNA damage repair.
FIG 6.
Cdh1 overexpression rescues DSB repair defect in late-passage Cdc14B−/− MEFs. (A) Western blotting of induced Cdh1 overexpression in MEFs. (B) Quantification of γH2AX-positive cells in late-passage Cdc14B+/+ and Cdc14B−/− MEFs with or without Cdh1 overexpression after 10 Gy of IR. (C) Cell cycle analyses of P7 MEFs with induced Cdh1 overexpression. Statistical significance was assessed by ANOVA (*, P < 0.05; **, P < 0.01).
Cdh1 is known to target a variety of cell cycle regulators for degradation in late mitosis and G1 phase, and it is also generally regarded as a cell cycle regulator (35, 36). However, there is no consistent conclusion about the effect of Cdh1 deficiency on cell cycle distribution regarding the different observations on cell growth and the cell cycle in different cells with either Cdh1 knockout or knockdown (37–41). As the cell cycle plays a critical role in DNA repair, we examined the cell cycle distribution of the above-described MEFs with Cdh1 knockdown or overexpression to test whether Cdh1 regulates DSB repair via altering cell cycle progression. FACS analysis of the DNA content showed that the cell cycle distribution was not significantly changed in the MEFs with either Cdh1 knockdown (Fig. 5F) or induced Cdh1 overexpression (Fig. 6C), indicating that it is unlikely that the effects of Cdh1 depletion or overexpression on DSB repair as observed are caused by changes in cell cycle distribution.
The Cdc14B-regulated Cdh1 phosphorylation level plays a critical role in DSB repair.
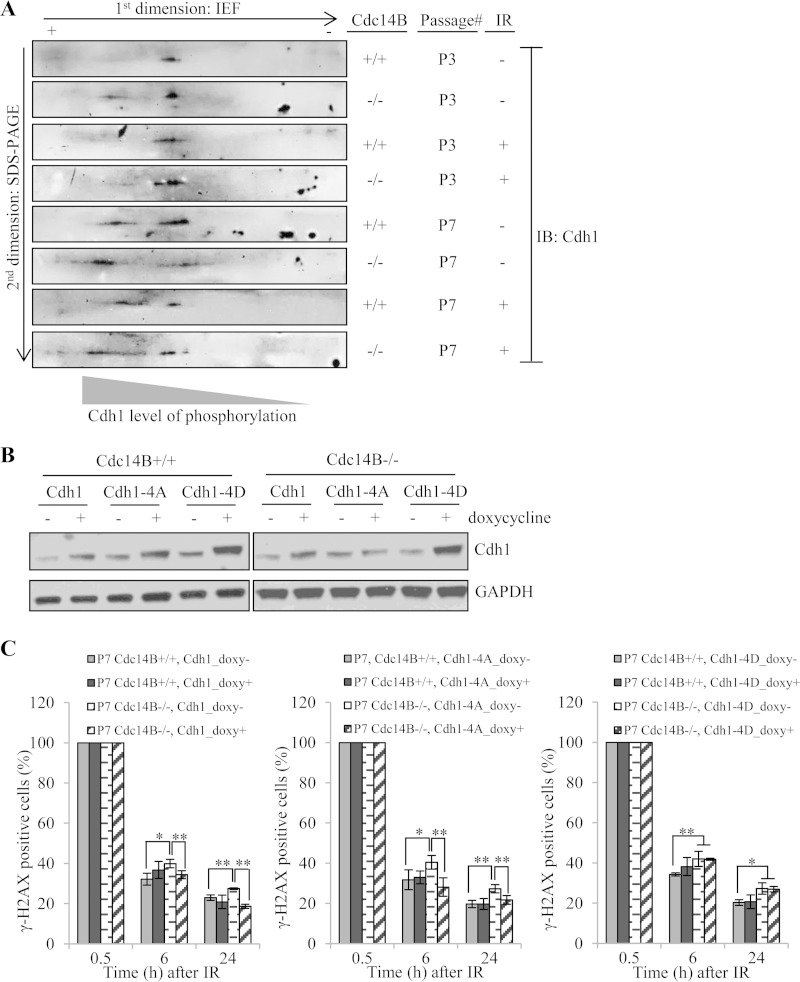
To further address the link between Cdc14B and Cdh1 in DSB repair, we tested whether Cdc14B deficiency affected the phosphorylation levels of endogenous Cdh1. There have been reports indicating direct biochemical interaction between these two proteins (30–32). However, direct evidence demonstrating Cdc14B's role in regulating endogenous Cdh1 phosphorylation levels in DNA repair or in general is lacking. Due to the lack of appropriate antibodies capable of detecting Cdh1 phosphorylation, we took the 2D gel approach as previously described (42) to assess the phosphorylation levels of Cdh1 in wild-type and Cdc14B-deficient MEFs. Protein extracts from these cells were separated first by charge through isoelectric focusing (IEF) and then by size through SDS-PAGE. As shown in Fig. 7A, at early passage (P3), the absence of Cdc14B already caused a shift of Cdh1 toward the more phosphorylated state. This shift was largely erased upon IR treatment, suggesting that Cdh1 is dephosphorylated by a different phosphatase(s) (perhaps Cdc14A) after DNA damage. At late passage, the level of more phosphorylated Cdh1 increased in both wild-type and Cdc14B-deficient MEFs compared to early-passage cells, with the increase more pronounced in Cdc14B-deficient cells than in wild-type cells. The increase in Cdh1 phosphorylation at late passage, regardless of Cdc14B status, probably reflects the now less proliferative state of these cells. IR treatment caused a shift of Cdh1 toward the less phosphorylated state in wild-type MEFs. However, the shift is negligible in Cdc14B mutant MEFs. These results indicate a role of Cdc14B in maintaining the steady state of Cdh1 phosphorylation (either directly or indirectly) and in activating Cdh1 upon DNA damage in late-passage MEFs.
FIG 7.
The Cdc14B-regulated Cdh1 phosphorylation level is critical for DSB repair. (A) Cell lysates of P3 and P7 MEFs with or without IR treatment (0.5 h after 10 Gy) were used for 2D gel analyses of Cdh1 phosphorylation levels. + and −, direction of the first-dimension isoelectric focusing. Cdh1 was detected by Western blotting following the second-dimension SDS-PAGE. The spot closer to the “+” end indicates highly phosphorylated Cdh1 with more phosphorylated residues, and the spot closer to the “−” end indicates less phosphorylated Cdh1. (B) Western blotting of induced overexpression of wild-type, phosphodeficient (Cdh1-4A), or phosphomimetic (Cdh1-4D) Cdh1 in MEFs. (C) Quantification of γH2AX-positive cells in late-passage Cdc14B+/+ and Cdc14B−/− MEFs with or without overexpression of wild-type (left panel), phosphodeficient (Cdh1-4A, middle panel), or phosphomimetic (Cdh1-4D, right panel) Cdh1 after 10 Gy of IR. Statistical significance was assessed by ANOVA (*, P < 0.05; **, P < 0.01).
To further demonstrate the relevance of Cdh1 dephosphorylation by Cdc14B in DSB repair, we tested the performance of phosphodeficient and phosphomimetic Cdh1 mutants in rescuing the repair defects of late-passage Cdc14B−/− MEFs. Given that Cdc14B reverses Cdk phosphorylation, we mutated the four Cdk consensus sites (S40, T121, S151, and S163) that have been previously described to regulate Cdh1 activity in S and G2 phases (15, 30, 43). Figure 7B shows conditional expression of the wild-type, phosphodeficient (Cdh1-4A), or phosphomimetic (Cdh1-4D) Cdh1 in both Cdc14B+/+ and Cdc14B−/− MEFs. Both wild-type and constitutively active mutant (Cdh1-4A) Cdh1 rescued the DSB repair defects in late-passage Cdc14B−/− MEFs (Fig. 7C, left and middle panels), while the phosphomimetic mutant (Cdh1-4D) failed to do so (Fig. 7C, right panel). These results indicate a critical role of Cdc14B in activating Cdh1 through dephosphorylation during DSB repair.
DISCUSSION
Phosphorylation/dephosphorylation is a major tool used by cells to control a large number of processes, including cell division. In the cell cycle, a huge number of phosphorylation events catalyzed by cyclin-dependent kinases culminate at the S phase entry as well as at mitotic entry (44–46). It is necessary to undo all the phosphorylation before the cell can exit either S or M phase. Cdc14 was a phosphatase identified in budding yeast to undo mitotic phosphorylation for mitotic exit. However, that function of Cdc14 has not been strictly conserved. In fission yeast, the Cdc14 homologue, Clp1, is not essential as in budding yeast (47). In mammals, a completely different phosphatase, PP2A-B55α, is required for mitotic exit (48). Instead, Cdc14 has been given new functions over evolution, along with its duplication into two homologues, Cdc14A and Cdc14B. In zebrafish, Cdc14B was implicated in ciliogenesis, although the exact role or its substrates in that process remain to be defined. Even this function in ciliogenesis seems not to be conserved in mammals, as Cdc14B-deficient mice do not show any signs of ciliopathy (10).
Cdc14A in human cells was proposed to maintain centrosome integrity in a knockdown study (49). However, knockout of Cdc14A in HCT116 cells did not cause any noticeable deleterious effects on the cells except a mild defect in DNA damage repair (9). Similarly, Cdc14B knockout HCT116 cells did not show any defects in the cell cycle except mild DNA damage repair defects (9), which was confirmed by our knockout studies in mice (10). It is not clear how and why mammalian Cdc14s became involved in DNA damage repair, nor is their exact role in the process understood. Cdc14B was initially reported to function in the G2 DNA damage checkpoint response through activating Cdh1 (30). However, such a checkpoint function was not observed in Cdc14B-deficient MEFs (10) or HCT116 cells (9). Interestingly, early-passage Cdc14B-deficient MEFs did not show defects in DNA damage repair; only the late-passage cells did. We provide evidence here that the relatively high levels of Cdc14A in early-passage MEFs probably compensate for the loss of Cdc14B. At late passages, the level of Cdc14A expression goes down for unknown reasons and is no longer sufficient to compensate for Cdc14B loss, making DNA damage repair defects apparent.
With the established requirement of Cdc14B and Cdc14A in DSB repair, it was unclear how they regulate DSB repair. Here we report that the two Cdc14s are redundantly required in both HR- and NHEJ-mediated repairs, although the two repair processes were affected to different degrees when the two phosphatases were depleted, suggesting that one process is more sensitive than another in terms of the requirement of the two phosphatases. If the phosphatases are completely absent (i.e., knocked out), both HR and NHEJ pathways may be affected to the same degree.
We also show that Cdh1 is a downstream mediator of Cdc14B in DSB repair. In that regard, we provide evidence that the phosphorylation status of Cdh1 is regulated by Cdc14B (directly or indirectly), and regulated Cdh1 phosphorylation is critical in DSB repair (Fig. 7). Cdh1 is best known for its roles in promoting protein degradation during mitotic exit and G1/S regulation (38, 39, 50, 51), while recent reports also revealed its emerging roles in DDR. Cdh1-mediated Plk1 degradation was proposed to regulate the G2/M DNA damage checkpoint (30). Cdh1 was also reported to regulate the degradation of USP1 to allow nucleotide excision repair (NER) of UV-induced DNA damage (34) and the degradation of Rad17 for checkpoint termination at the end of the UV-induced DNA damage response (33). More recently, Cdh1 was proposed to regulate the clearance of CtIP, an essential HR protein, at a late time of HR repair to prevent excessive end resection for optimal HR efficiency (52). The Cdc14B-Cdh1-CtIP axis might be part of the mechanism underlying the requirement of Cdc14B in HR repair. Our results showing slightly more severe DSB repair defects in cells lacking both Cdc14B and Cdh1 than in singly deficient cells also indicate that there might be some other downstream targets of Cdc14B in DSB repair and that Cdh1 activation might be accomplished by some other phosphatases, including Cdc14A. In the future, it is necessary to identify these unknown factors. More importantly, it will be of great interest to determine if Cdh1 (and the APC itself) is recruited to the damage site (which would imply that this E3 ligase participates in DDR more directly) and what are the (other) substrates of APC/Cdh1 in DNA damage repair.
ACKNOWLEDGMENTS
We thank S.-Y. Lin (M. D. Anderson Cancer Center) for cell lines, J. Rosen (Baylor College of Medicine) for reagents, C.-H. Chang (Baylor College of Medicine) for discussion, and the Cytometry and Cell Sorting Center (CCSC) at Baylor College of Medicine for assistance with FACS. We also thank other members of the Zhang lab for helpful discussion and support.
This work was supported in part by NIH grants CA116097 and CA122623 to P.Z. and in part by an international collaboration grant from the Chinese Ministry of Science and Technology (grant 2013DFB30210). The CCSC is supported by grants from NIAID (P30AI036211), NCI (P30CA125123), and NCRR (S10RR024574).
REFERENCES
- 1.Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norbury CJ, Hickson ID. 2001. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol 41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- 3.Mocciaro A, Schiebel E. 2010. Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci 123:2867–2876. doi: 10.1242/jcs.074815. [DOI] [PubMed] [Google Scholar]
- 4.Eissler CL, Mazon G, Powers BL, Savinov SN, Symington LS, Hall MC. 2014. The Cdk/Cdc14 module controls activation of the Yen1 Holliday junction resolvase to promote genome stability. Molecular Cell 54:80–93. doi: 10.1016/j.molcel.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray CH, Good VM, Tonks NK, Barford D. 2003. The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J 22:3524–3535. doi: 10.1093/emboj/cdg348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk dependent phosphorylation. Mol Cell 2:709–718. doi: 10.1016/S1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 7.Clement A, Solnica-Krezel L, Gould KL. 2012. Functional redundancy between Cdc14 phosphatases in zebrafish ciliogenesis. Dev Dyn 241:1911–1921. doi: 10.1002/dvdy.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdougo E, Nachury MV, Jackson PK, Jallepalli PV. 2008. The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle 7:1184–1190. doi: 10.4161/cc.7.9.5792. [DOI] [PubMed] [Google Scholar]
- 9.Mocciaro A, Berdougo E, Zeng K, Black E, Vagnarelli P, Earnshaw W, Gillespie D, Jallepalli P, Schiebel E. 2010. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J Cell Biol 189:631–639. doi: 10.1083/jcb.200910057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei Z, Peddibhotla S, Lin H, Fang X, Li M, Rosen JM, Zhang P. 2011. Early-onset aging and defective DNA damage response in Cdc14b-deficient mice. Mol Cell Biol 31:1470–1477. doi: 10.1128/MCB.01330-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng G, Yim EK, Dai H, Jackson AP, Burgt I, Pan MR, Hu R, Li K, Lin SY. 2009. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 11:865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 13.Yuan B, Latek R, Hossbach M, Tuschl T, Lewitter F. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res 32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, Bernardi RJ, Fu X, Bland CS, Cooper TA, Schiff R, Rosen JM, Westbrook TF, Elledge SJ. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci U S A 108:3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukas C, Sørensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. 1999. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature 401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 16.Tumurbaatar I, Cizmecioglu O, Hoffmann I, Grummt I, Voit R. 2011. Human Cdc14B promotes progression through mitosis by dephosphorylating Cdc25 and regulating Cdk1/cyclin B activity. PLoS One 6:e14711. doi: 10.1371/journal.pone.0014711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munoz MC, Laulier C, Gunn A, Cheng A, Robbiani DF, Nussenzweig A, Stark JM. 2012. RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J Biol Chem 287:40618–40628. doi: 10.1074/jbc.M112.410951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Yuan JS, Reed A, Chen F, Stewart CN Jr. 2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sotiropoulou PA, Candi A, Mascre G, De Clercq S, Youssef KK, Lapouge G, Dahl E, Semeraro C, Denecker G, Marine JC, Blanpain C. 2010. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol 12:572–582. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]
- 24.Helleday T, Lo J, van Gent DC, Engelward BP. 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA repair 6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 25.Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- 26.Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi K, Cavallo F, Brunet E, Jasin M. 2011. Homologous recombination assay for interstrand cross-link repair. Methods Mol Biol 745:283–291. doi: 10.1007/978-1-61779-129-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaspersen SL, Charles JF, Morgan DO. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol 9:227–236. doi: 10.1016/S0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 29.Zachariae W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 30.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bembenek J, Yu H. 2001. Regulation of the anaphase-promoting complex by the dual specificity phosphatase human Cdc14a. J Biol Chem 276:48237–48242. [DOI] [PubMed] [Google Scholar]
- 32.Schindler K, Schultz RM. 2009. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod 80:795–803. doi: 10.1095/biolreprod.108.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Park CH, Wu J, Kim H, Liu W, Fujita T, Balasubramani M, Schreiber EM, Wang XF, Wan Y. 2010. Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J 29:1726–1737. doi: 10.1038/emboj.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. 2011. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 194:177–186. doi: 10.1083/jcb.201101062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Zhang P. 2009. The function of APC/CCdh1 in cell cycle and beyond. Cell Div 4:2. doi: 10.1186/1747-1028-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiao X, Zhang L, Gamper AM, Fujita T, Wang Y. 2010. APC/C-Cdh1: from cell cycle to cellular differentiation and genomic integrity. Cell Cycle 9:3904–3912. doi: 10.4161/cc.9.19.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudo T, Ota Y, Kotani S, Nakao M, Takami Y, Takeda S, Saya H. 2001. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J 20:6499–6508. doi: 10.1093/emboj/20.22.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 39.Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. 2008. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol 10:1083–1089. doi: 10.1038/ncb1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigl R, Wandke C, Rauch V, Kirk J, Hunt T, Geley S. 2009. Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J Cell Sci 122:4208–4217. doi: 10.1242/jcs.054197. [DOI] [PubMed] [Google Scholar]
- 41.Delgado-Esteban M, Garcia-Higuera I, Maestre C, Moreno S, Almeida A. 2013. APC/C-Cdh1 coordinates neurogenesis and cortical size during development. Nat Commun 4:2879. doi: 10.1038/ncomms3879. [DOI] [PubMed] [Google Scholar]
- 42.Huang X, Tran T, Zhang L, Hatcher R, Zhang P. 2005. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. Proc Natl Acad Sci U S A 102:1065–1070. doi: 10.1073/pnas.0409130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. 2001. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol Cell Biol 21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher DL, Nurse P. 1996. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J 15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 45.Morgan DO. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 46.Fisher D, Krasinska L, Coudreuse D, Novak B. 2012. Phosphorylation network dynamics in the control of cell cycle transitions. J Cell Sci 125:4703–4711. doi: 10.1242/jcs.106351. [DOI] [PubMed] [Google Scholar]
- 47.Trautmann S, Wolfe BA, Jorgensen P, Tyers M, Gould KL, McCollum D. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr Biol 11:931–940. doi: 10.1016/S0960-9822(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz MH, Held M, Janssens V, Hutchins JR, Hudecz O, Ivanova E, Goris J, Trinkle-Mulcahy L, Lamond AI, Poser I, Hyman AA, Mechtler K, Peters JM, Gerlich DW. 2010. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol 12:886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mailand N, Lukas C, Kaiser BK, Jackson PK, Bartek J, Lukas J. 2002. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat Cell Biol 4:317–322. [DOI] [PubMed] [Google Scholar]
- 50.Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M. 2004. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature 428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 51.Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WGJ. 2004. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature 428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 52.Lafranchi L, de Boer HR, de Vries EG, Ong SE, Sartori AA, van Vugt MA. 2014. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J 33:2860–2879. doi: 10.15252/embj.201489017. [DOI] [PMC free article] [PubMed] [Google Scholar]