Abstract
Yeast can use a wide variety of nitrogen compounds. However, the ability to synthesize enzymes and permeases for catabolism of poor nitrogen sources is limited in the presence of a rich one. This general mechanism of transcriptional control is called nitrogen catabolite repression. Poor nitrogen sources, such as leucine, γ-aminobutyric acid (GABA), and allantoin, enable growth after the synthesis of pathway-specific catabolic enzymes and permeases. This synthesis occurs only under conditions of nitrogen limitation and in the presence of a pathway-specific signal. In this work we studied the temporal order in the induction of AGP1, BAP2, UGA4, and DAL7, genes that are involved in the catabolism and use of leucine, GABA, and allantoin, three poor nitrogen sources. We found that when these amino acids are available, cells will express AGP1 and BAP2 in the first place, then DAL7, and at last UGA4. Dal81, a general positive regulator of genes involved in nitrogen utilization related to the metabolisms of GABA, leucine, and allantoin, plays a central role in this coordinated regulation.
1. Introduction
Yeast cells can use a variety of compounds as nitrogen source through different specific pathways. Synthesis of proteins involved in each of these pathways is tightly regulated. The availability of readily transported and metabolized nitrogen sources, which are known as good nitrogen sources, results in the strong repression of genes involved in transport and metabolism of poor ones [1, 2]. The utilization of poor sources such as leucine and other branched amino acids, γ-aminobutyric acid (GABA), and allantoin requires the synthesis of pathway-specific catabolic enzymes and permeases. This synthesis occurs only under conditions of nitrogen limitation and in the presence of a pathway-specific signal such as a substrate or an intermediate of a metabolic pathway [3].
Extracellular GABA is imported into Saccharomyces cerevisiae cells through three proteins: the general amino acid permease (Gap1), the proline specific permease (Put4), and the GABA permease (Uga4) [4]. Within the cells, GABA is catabolized by the GABA transaminase and succinate semialdehyde dehydrogenase enzymes encoded by UGA1 and UGA2 genes, respectively [5]. The induction of UGA genes by GABA requires at least two positive regulatory proteins: the specific Uga3 factor and the pleiotropic Dal81 factor (also called Uga35) that act through a 19 bp CG-rich Upstream Activating Sequence named UASGABA [6–8]. The UASGABA present in UGA4 and UGA1 promoters contains two independent Uga3 binding sites that consist of the asymmetric sequence 5′-SGCGGNWTT-3′ (S = G or C, W = A or T, and N = no nucleotide or G) whereas UGA2 promoter contains only one site [9, 10]. Both factors Uga3 and Dal81 interact in vivo with the UGA4 promoter in a GABA-dependent manner [11].
Leucine is a major amino acid in nutrients and proteins. Its uptake is mediated by multiple amino acid permeases, including the high-affinity leucine permeases Bap2 and Bap3 and the high capacity permease Agp1. When leucine is available in the environment, the SPS (Ssy1, Ptr3, and Ssy5) sensor is activated and hence the transcriptional induction mediated by the homologous transcription factors Stp1 and Stp2 takes place. These factors bind to the sequence known as UASaa first identified in the BAP3 promoter [13] and then in the BAP2 promoter [14] but later also found in the promoter sequences of several amino acid permease genes including AGP1 [15]. In the absence of amino acids, Stp1 and Stp2 are present mainly in the cytoplasm. When amino acids are detected in the environment, 10 kDa of the N terminus of these transcription factors is endoproteolytically cleaved off, resulting in their relocation to the nucleus [16]. Then, Stp1 and Stp2 can act on AGP1 and BAP2 promoters. Dal81 acts through the UAS sequence facilitating the binding of Stp1 and Stp2 to AGP1 promoter [17, 18]. It was proposed that Dal81 is also essential for the induction of BAP2 [19] and that Leu3 modifies the expression of BAP2 [14].
Allantoin, the heterocyclic compound produced during purine degradation, is rich in nitrogen, and S. cerevisiae among many other organisms is able to degrade it and recycle it in order to use it as nitrogen source. The allantoin degradation pathway converts allantoin to ammonia and carbon dioxide. Conversion of allantoin to ammonia is carried out by the DAL1, DAL2, and DAL3 gene products, which work sequentially to generate urea. Urea is then degraded to ammonia in a two-step process by the Dur1,2 protein, a multifunctional single enzyme originally thought to be encoded by two tightly linked genes. DAL7 encodes for an enzyme recycling the glyoxylate generated during allantoin degradation. All the enzymes associated with allantoin degradation are inducible and repressible. Their production is contingent on the presence of allophanate, the last intermediate in the allantoin-degradative pathway or the gratuitous inducer, oxaluric acid [20]. The cis-acting element mediating inducer responsiveness of the allantoin pathway genes is the dodecanucleotide element UIS ALL(Upstream Induction Sequence). The DAL81 and DAL82/DURM gene products are required for this inducer responsiveness. Dal82 has been shown to be the UIS ALL DNA-binding protein whose binding to DNA is independent of inducer. Whereas Dal82 appears to be a pathway-specific regulatory element, Dal81 functions more broadly, being required for induced expression of the UGA (GABA catabolic pathway) and AGP1 (amino acid uptake) genes as well as those of the allantoin pathway [21].
Dal81 is a general positive regulator of genes involved in nitrogen utilization related to metabolisms of GABA, urea, leucine, and allantoin [22, 23]; moreover, Dal81 is involved in the amino acid SPS sensor pathway [17, 18, 24]. In all these induction processes, Dal81 acts together with an inducer-specific protein; this specific factor is Uga3 in GABA-induction of UGA genes [6, 11, 25], Dal82 in allophanate-induction of DUR and DAL genes [26–28], and Stp1 in amino acid induction of amino acid permease genes such as BAP2, BAP3, and AGP1 [17, 18, 24].
A hierarchy for different inducible processes mediated by the Dal81 factor has been proposed [24]. Our results showing that the decrease caused by leucine in the recruitment of HA-Dal81 to the UGA4 promoter depended on Ssy1 supported this hypothesis [11]. The aim of this work was to study the mechanisms that lead to the temporal order in the expression of genes involved in the use of poor nitrogen sources.
2. Methods
2.1. Strains and Media
The S. cerevisiae strains used in this study are isogenic to the wild type Σ1278b and are listed in Table 1.
Table 1.
Strains used in this work.
| Strain | Genotype | Parent | Primer | Source or reference |
|---|---|---|---|---|
| Σ1278b | Matα | — | — | [12] |
| 23344c | Matα ura3 | — | — | Grenson et al. [4] |
| SBCY10 | Matα ura3 6HA-DAL81 | — | — | [11] |
| SBCY17 | Matα ura3 dal81Δ::natMX4 | — | — | [11] |
| SBCY20 | Matαura3 uga35::natMX4 leu3::KanMX4 | — | [11] | |
| MPY09 | Matα ura3 leu3::loxP | 23344c | This study | |
| DEBY01 | Matα ura3 STP1-3HA-KanMX6 | 23344c | This study | |
| DEBY02 | Matα ura3 dal81Δ::natMX4 STP1-3HA-KanMX6 | SBCY17 | This study |
Cells were grown in minimal medium containing 0.17% Difco yeast nitrogen base (YNB without amino acids and ammonium sulfate) with 2% glucose as carbon source and 10 mM proline as nitrogen source. The final concentrations of the inducers GABA, leucine, and oxalurate were 0.1 mM, 1.3 mM, and 1 mM, respectively.
MPY09, DEBY01, and DEBY02 mutant strains were generated using the PCR-based gene-deletion strategy [29, 30] or modified versions of it [31].
The MPY09 strain (leu3Δ deletion) was generated using the pUG6 plasmid [32] to amplify the loxP-KanMX-loxP cassette with primers F-leu3 and R-leu3 (Table 2). After generating the strain, KanMX cassette was excised by recombination mediated by Cre recombinase (pSH47 plasmid).
Table 2.
Primers used in this work.
| Primer group and name | Sequence (5′ to 3′) |
|---|---|
| Oligonucleotides for strain construction | |
| F-leu3 | TGCAATTATGGAAGGAAGATCAGATTTTGTGGCGACTTCACACAGCTGAAGCTTCGTACGC |
| R-leu3 | GGACTTTAAACCTTGGGATTGAACGCAAATTCATTCATTAAACATAGGCCACTAAGTGGATCTG |
| F-STP1-HA | CAATATTTGAATTTTTACAATGACAACTTTGGGTCACAATTTCGGATCCCCGGGTTAATTAA |
| R-STP1-HA | TTCCAATATGATACCCTTATTTTTATCCCGTGTTATATTTAAGAATTCGAGCTCGTTTAAAC |
| F-LEU3 prom | AGGTGCCGCCTAATTTATCG |
| R-LEU3 int | ACTTCTGCTGACGACATTCC |
| F-KanMX6 int | CATCCTATGGAACTGCCTCG |
| R-KanMX6 int | GATAGATTGTCGCACCTGATTG |
| F-STP1 int | GCACAAGATAATCCTTCGTTCC |
| R-STP1 down | TCGGCTTTCCAATATGATACCC |
| R-Kan int | CTATACCTGAGAAAGCAACCTG |
|
| |
| Oligonucleotides for RT-qPCR | |
| F-qRT-UGA4 | CTGCTGCTGTCACATTAACC |
| R-qRT-UGA4 | AATACACATAACCACCACTGC |
| F-qRT-DAL7 | AACCGAACAAATCAGGAAC |
| R-qRT-DAL7 | CAAGTTGGAGATGAAGAGTC |
| F-qRT-BAP2 | TAGAGGATGGCGTTGAGTC |
| R-qRT-BAP2 | ACCAAGATGTAACCAATTATTAGC |
| F-qRT-AGP1 | ATCTTATTCCTATTCTTGGCTACC |
| R-qRT-AGP1 | CGGCGTTAATGAAGTGTGG |
| F-qRT-TBP1 | TATAACCCCAAGCGTTTTGC |
| R-qRT-TBP1 | GCCAGCTTTGAGTCATCCTC |
|
| |
| Oligonucleotides for ChIP | |
| F-UGA4 | GGAACTGATTACTGTGCCAAG |
| R-UGA4 | AATCGCTTATCGCTTATCGTG |
| F-AGP1 | TTATACCTCGGCGGCTTC |
| R-AGP1 | GCAAGATTTCTCCAAAGTCC |
| F-BAP2 | AGGAGGCTACTGACACTGC |
| R-BAP2 | GCTGACATATTTACCGTTGAAGG |
| F-DAL7 | AATCTCCGCTGAAGTTGC |
| R-DAL7 | TTTCACGATGTACCTTATCCAAGA |
| F-UC | AGTCCAATACCTCTGTCCTC |
| R-UC | AGCCGCAACTTCATTCTG |
DEBY01 and DEBY02 strains were generated using pFA6a-3HA-KanMX6 plasmid [31] and the F-STP1-HA and R-STP1-HA primers (Table 2). The correct insertion of the tag was corroborated by PCR using the F-STP1 int, R-STP1 down, R-Kan int primers.
All yeast transformations were carried out using the lithium method [33]. Transformants were selected on rich medium containing 200 µg/ml G418.
To complement the deficiency in DAL81 gene, cells were transformed with the pSBC-HA-DAL81 plasmid that contains the complete DAL81 gene [34].
2.2. Quantitative RT-PCR
RT-qPCR experiments were performed according to Cardillo et al. [34]. cDNAs were quantified by RT-PCR using an Opticon Monitor 3 (Bio-Rad) with the primers listed in Table 2. Expression values correspond to the ratio of concentrations of UGA4, AGP1, BAP2, and DAL7 over TBP1 specific mRNAs determined in each sample and represent the mean ± SEM of three independent experiments.
2.3. Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) experiments were performed according to Cardillo et al. [25]. Normal mouse IgG (Santa Cruz) or monoclonal anti-HA antibody (HA probe (F-7), Santa Cruz) was used. Real time quantitative PCR was carried out in an Opticon Monitor 3 (Bio-Rad) with primers that amplified promoter regions of UGA4, AGP1, DAL7, and BAP2 genes (Table 2). A pair of primers that amplified a region located 2.5 Kb downstream of UGA4 promoter (F-UC/R-UC) was used as an unbound control. ChIP DNA was normalized to input DNA and calculated as a signal-to-noise ratio over IgG control ChIP. The ΔΔCt method was used to calculate fold change of binding to the promoter of interest [35]. Results are expressed as the mean ± SEM of three independent experiments.
3. Results and Discussion
It has been proposed that the expression of genes that encode permeases of different poor nitrogen sources follows a certain order rather than occurring simultaneously.
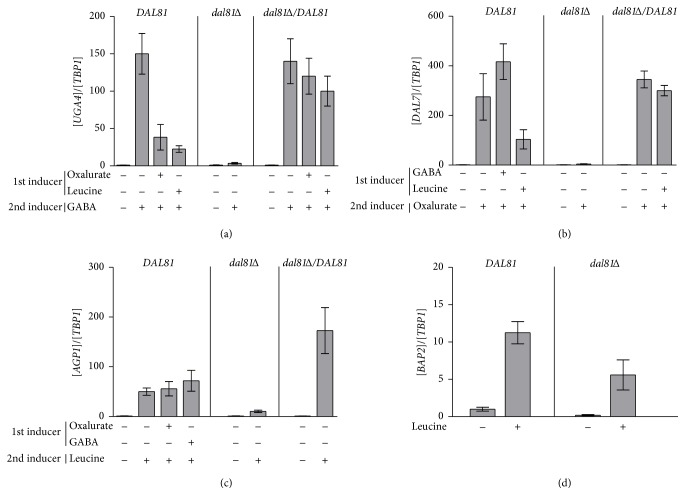
In order to determine this hierarchy, we measured the induction of each gene in the presence of the inducer of another gene. For instance, to analyze the effect of the inducers of DAL7, AGP1, and BAP2 genes on GABA-induction of UGA4, exponentially grown cells were incubated for 30 minutes with oxalurate or leucine before the 30-minute incubation with GABA. The induction of UGA4 produced by the presence of GABA was strongly inhibited by leucine, the inducer of AGP1 and BAP2 genes, and also by oxalurate, the inducer of DAL7 (Figure 1(a)). This is indicating that GABA is incorporated into the cells for its catabolism only after the leucine and allantoin added to the culture medium are used. The induction of DAL7 by oxalurate was significantly lower when cells were previously incubated by leucine while preincubation with GABA did not produce any effect (Figure 1(b)), suggesting that the use of allantoin might occur in the presence of GABA but only after the consumption of leucine. The expression of AGP1 and BAP2 genes was induced by leucine and it was not affected by GABA or oxalurate (Figures 1(c) and 1(d)). These results suggest that the order of the use of the analyzed poor nitrogen sources could be leucine, allantoin, and then GABA.
Figure 1.
Effect of Dal81 on gene induction. mRNA levels of UGA4 (a), DAL7 (b), AGP1 (c), and BAP2 (d) were determined in wild type cells (23344c strain), dal81Δ cells (SBCY17 strain), and dal81Δ cells transformed with the pSBC-HA-DAL81 plasmid. The second inducer, the specific one, was added 30 minutes after the addition of the first one and cells were incubated for another 30 minutes. mRNA levels were quantified by RT-qPCR. UGA4, DAL7, AGP1, and BAP2 values were normalized with TBP1 and results are the mean ± SEM of three independent experiments. Within each strain the values measured were normalized to the value obtained for the uninduced condition.
As expected, the induction of UGA4, DAL7, and AGP1 by GABA, oxalurate, and leucine, respectively, strictly depended on the activity of the transcription factor Dal81 since no induction was detected in dal81Δ cells (Figures 1(a), 1(b), and 1(c)). On the other hand, the induction of BAP2 measured in dal81Δ cells was significantly lower than that observed in wild type cells although some induction was still detected (Figure 1(d)). Our results suggest that although Dal81 is involved in BAP2 transcription as it was reported earlier [19], it is not essential for this process. Both BAP2 and AGP1 are regulated by the pair of transcription factors Dal81 and Stp1; however the role of Dal81 on the regulation of each gene seems to be different.
The fact that Dal81 is involved in these induction processes makes this factor a good candidate as the protein that establishes the hierarchy. So, this hierarchy might be due to the recruitment of Dal81 in the firstly induced genes with the consequently lower availability of this transcription factor to act on the other promoters. To test this hypothesis, we measured the induction of each gene in the presence of the inducers of other genes in cells that overexpressed Dal81. For this purpose, dal81Δ cells were transformed with the pSBC-HA-DAL81 plasmid [34] that contains the complete DAL81 open reading frame under the control of the GPD1 promoter. When the expression of DAL81 was under the regulation of the strong and constitutive promoter GPD1, the induction of UGA4 by GABA was not inhibited by the presence of leucine nor oxalurate and the induction of DAL7 by oxalurate was not inhibited by the presence of leucine (Figures 1(a) and 1(b)). So, this finding confirms our hypothesis since when Dal81 is not a limiting factor, the hierarchy is not observed.
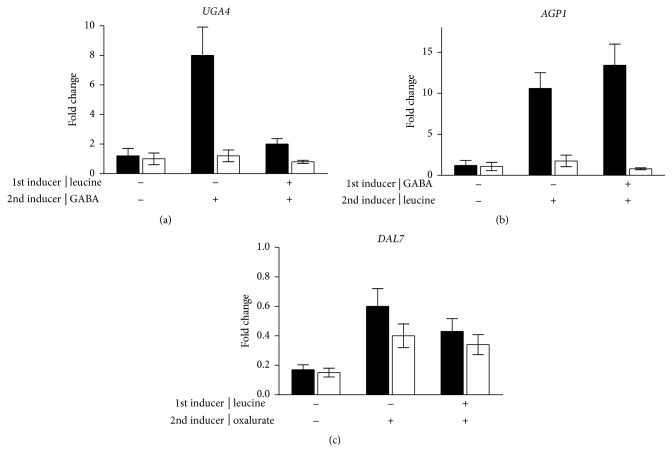
Then we analyzed the binding that occurs in vivo between Dal81 and UGA4, AGP1, and DAL7 promoters. We found that the strong binding of Dal81 with the regulatory region of UGA4 detected after incubating the cells with GABA significantly weakened when the cells were preincubated with leucine (Figure 2(a)). This inhibition by leucine was earlier shown [11]. In the contrary, the amount of Dal81 bound to the promoter of AGP1 did not change after the preincubation with GABA (Figure 2(b)). These results reinforce our idea of Dal81 being the limiting factor in these induction processes. On the other hand, we did not find any effect of leucine on the oxalurate-dependent binding of Dal81 to DAL7 promoter (Figure 2(c)). However, it must be noticed that the binding measured after the incubation with oxalurate was already very weak. So, we think that this result is probably due to technical difficulties in these ChIP assays.
Figure 2.
Interaction of Dal81 with UGA4, AGP1, and DAL7 promoters. Wild type cells, expressing the HA-Dal81 (SBCY10 strain) fusion protein, were treated or not with the indicated inducers. ChIP assays were carried out using antibodies against the HA epitope. qPCR was performed with specific primers (black bars) that amplify a region of UGA4 promoter (a), a region of AGP1 promoter (b), a region of DAL7 promoter (c), and a region 2.5 kb downstream of UGA4 promoter used as a negative control (white bars). Results are expressed as the fold change of binding to the promoter of interest and are the mean ± SEM of three independent experiments.
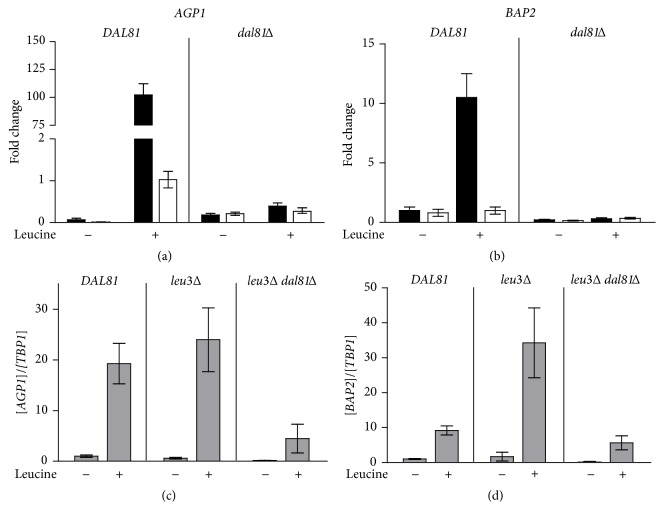
We were not able to detect Dal81 bound to the regulatory region of BAP2 gene (data not shown). This suggests that the mild effect of Dal81 detected on BAP2 (Figure 1(d)) is not due to a direct interaction. Again, Dal81 acts in a different way on BAP2 compared to on AGP1 in contrast with previous reports [18]. To assess this, we measured the binding of Stp1 to AGP1 and BAP2 genes. We found that Stp1 strongly bound to AGP1 promoter after the incubation with the inducer leucine and this interaction was avoided in a dal81Δ strain (Figure 3(a)). This result is in agreement with those obtained by Boban and Ljungdahl [17]. Stp1 also bound to BAP2 promoter in a leucine dependent manner (Figure 3(b)). However, it was rather surprising to find that this interaction also depended on the presence of Dal81. As Dal81 did not seem to bind to BAP2, the mechanism by which it facilitates Stp1 binding to this gene might be indirect, probably through Leu3. The expression of BAP2 gene was significantly higher in leu3Δ cells, indicating that in our growth conditions Leu3 factor has a negative effect on BAP2 (Figure 3(d)). It was earlier reported that the Leu3 binding site in the BAP2 promoter is required for full promoter activity [14]. This apparent contradiction is probably due to different growth conditions since it is known that Leu3 is a transcription factor that can act as an activator or as a repressor depending on the content of α-isopropylmalate within the cells [36]. Different growth conditions lead to changes in this content. Our results also reveal a substantial difference in the regulation of AGP1 and BAP2 since AGP1 is not regulated by Leu3. In the leu3Δ dal81Δ double mutant the expression of BAP2 was significantly lower than in the single mutant leu3Δ, confirming that Dal81 is somehow participating in the expression of BAP2. It must be noted that we have already found a negative effect of Leu3 on UGA4 gene, although we were not able to detect Leu3 bound to this gene [11]. Moreover, De Boer et al. demonstrated that Leu3 acts as a repressor on BAP3 expression but they also failed in detecting any interaction [13].
Figure 3.
Role of Dal81 in AGP1 and BAP2 regulation. Wild type cells, expressing the Stp1-HA (DEBY01 strain) fusion protein, were treated or not with leucine. ChIP assays were carried out using antibodies against the HA epitope. qPCR was performed with specific primers (black bars) that amplify a region of AGP1 promoter (a), a region of BAP2 promoter (b), and a region 2.5 kb downstream of UGA4 promoter (white bars) used as a negative control. Results are expressed as the fold change of binding to the promoter of interest and are the mean ± SEM of three independent experiments. mRNA levels of AGP1 (c) and BAP2 (d) were determined in wild type cells (23344c strain), leu3Δ cells (MPY09 strain), and leu3Δ dal81Δ cells (SBCY20 strain). The cells were incubated with or without leucine for 30 minutes. mRNA levels were quantified by RT-qPCR. AGP1 and BAP2 values were normalized with TBP1 and results are the mean ± SEM of three independent experiments.
4. Conclusions
In this work we showed that the expression of the proteins responsible for the uptake and catabolism of different poor nitrogen sources occurs sequentially following a certain order determined by a tight regulation. This order could promote the utilization of a given nitrogen source whereas the utilization of others, which may be less useful, could be downregulated in some growth media. We demonstrated here that the transcription factor Dal81 is central in the regulation that leads to the hierarchical expression of the genes studied here and, consequently, in the utilization of leucine, allantoin, and GABA. BAP2, the gene of one of the permeases of leucine, seems not to be included in this hierarchical regulation since the regulation of BAP2 by Dal81 occurs through an indirect mechanism. This may have a physiological significance since Iraqui and collaborators [18] failed to show any contribution of Bap2 to the utilization of leucine as the sole nitrogen source and Cohen and Engelberg [37] suggested that Bap2 is the only functional leucine transporter on rich media.
The doubling time of wild type cells in minimal medium-cultures containing leucine as the sole nitrogen source was twice that of cultures in minimal medium containing GABA (http://www.doubling-time.com/compute.php). Other authors obtained similar values of doubling time of cells grown on allantoin or GABA as the sole nitrogen sources [38]. It is commonly assumed that preferred nitrogen sources allow higher growth rates. This is true when comparing rich and poor sources. However, if the order of the expression of AGP1, DAL7, and UGA4 was determining a hierarchy in the use of their substrates, that assumption would not be fulfilled for poor nitrogen sources. Probably, the importance of leucine as a ketogenic amino acid that supplies energy to the cells and its high frequency in protein composition led to a regulation that ends in the use of this poor nitrogen source before others that provide a higher growth rate.
Acknowledgments
This research was supported by grants from the University of Buenos Aires (UBA) and the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Argentina. The authors thank Darío E. Balcazar for technical assistance and Terrance Cooper for providing the oxalurate.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Cooper T. G. The regulation of yeast gene expression by multiple control elements. Basic Life Sciences. 1982;19:143–161. doi: 10.1007/978-1-4684-4142-0_14. [DOI] [PubMed] [Google Scholar]
- 2.Wiame J.-M., Grenson M., Ars H. N., Jr. Nitrogen catabolite repression in yeasts and filamentous fungi. Advances in Microbial Physiology. 1985;26:1–88. doi: 10.1016/s0065-2911(08)60394-x. [DOI] [PubMed] [Google Scholar]
- 3.Marzluf G. A. Genetic regulation of nitrogen metabolism in the fungi. Microbiology and Molecular Biology Reviews. 1997;61(1):17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grenson M., Muyldermans F., Broman K., Vissers S. 4-Aminobutyric acid (GABA) uptake in Baker's yeast Saccharomyces cerevisiae is mediated by the general amino acid permease, the proline permease and a GABA specific permease integrated into the GABA-catabolic pathway. Life Sciences: Advances in Biochemistry. 1987;6:35–39. [Google Scholar]
- 5.Ramos F., El Guezzar M., Grenson M., Wiame J.-M. Mutations affecting the enzymes involved in the utilization of 4-aminobutyric acid as nitrogen source by the yeast Saccharomyces cerevisiae . European Journal of Biochemistry. 1985;149(2):401–404. doi: 10.1111/j.1432-1033.1985.tb08939.x. [DOI] [PubMed] [Google Scholar]
- 6.André B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Molecular and General Genetics. 1990;220(2):269–276. doi: 10.1007/bf00260493. [DOI] [PubMed] [Google Scholar]
- 7.André B., Hein C., Grenson M., Jauniaux J.-C. Cloning and expression of the UGA4 gene coding for the inducible GABA-specific transport protein of Saccharomyces cerevisiae . MGG Molecular & General Genetics. 1993;237(1-2):17–25. doi: 10.1007/bf00282779. [DOI] [PubMed] [Google Scholar]
- 8.Talibi D., Grenson M., Andre B. Cis- and trans-acting elements determining induction of the genes of the γ-aminobutyrate (GABA) utilization pathway in Saccharomyces cerevisiae . Nucleic Acids Research. 1995;23(4):550–557. doi: 10.1093/nar/23.4.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Idicula A. M. Binding and Transcriptional Activation by Uga3p, a Zinc Binuclear Cluster Protein of Saccharomyces cerevisiae: Redefining the UASGABA and the Uga3p Binding Site. Grahamstown, South Africa: Department of Biochemestry, Microbiology and Biotechnology, Rhodes University; 2002. [Google Scholar]
- 10.Idicula A. M., Blatch G. L., Cooper T. G., Dorrington R. A. Binding and activation by the zinc cluster transcription factors of Saccharomyces cerevisiae: redefining the UASGABA and its interaction with Uga3p. The Journal of Biological Chemistry. 2002;277(48):45977–45983. doi: 10.1074/jbc.m201789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardillo S. B., Moretti M. B., García S. C. Uga3 and Uga35/Dal81 transcription factors regulate UGA4 transcription in response to γ-aminobutyric acid and leucine. Eukaryotic Cell. 2010;9(8):1262–1271. doi: 10.1128/ec.00117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae . European Journal of Biochemistry. 1970;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 13.De Boer M., Bebelman J.-P., Gonçalves P. M., Maat J., Van Heerikhuizen H., Planta R. J. Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae . Molecular Microbiology. 1998;30(3):603–613. doi: 10.1046/j.1365-2958.1998.01094.x. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen P., van den Hazel B., Didion T., et al. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Molecular and General Genetics. 2001;264(5):613–622. doi: 10.1007/s004380000347. [DOI] [PubMed] [Google Scholar]
- 15.Crépin L., Nidelet T., Sanchez I., Dequin S., Camarasa C. Sequential use of nitrogen compounds by Saccharomyces cerevisiae during wine fermentation: a model based on kinetic and regulation characteristics of nitrogen permeases. Applied and Environmental Microbiology. 2012;78(22):8102–8111. doi: 10.1128/aem.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ljungdahl P. O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochemical Society Transactions. 2009;37(1):242–247. doi: 10.1042/BST0370242. [DOI] [PubMed] [Google Scholar]
- 17.Boban M., Ljungdahl P. O. Dal81 enhances Stp1- and Stp2-dependent transcription necessitating negative modulation by inner nuclear membrane protein Asi1 in Saccharomyces cerevisiae . Genetics. 2007;176(4):2087–2097. doi: 10.1534/genetics.107.075077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iraqui I., Vissers S., Bernard F., et al. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Molecular and Cellular Biology. 1999;19(2):989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernard F., André B. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae . Molecular Microbiology. 2001;41(2):489–502. doi: 10.1046/j.1365-2958.2001.02538.x. [DOI] [PubMed] [Google Scholar]
- 20.Sumrada R., Zacharski C. A., Turoscy V., Cooper T. G. Induction and inhibition of the allantoin permease in Saccharomyces cerevisiae . Journal of Bacteriology. 1978;135(2):498–510. doi: 10.1128/jb.135.2.498-510.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai R., Daugherty J. R., Cunningham T. S., Cooper T. G. Overlapping positive and negative GATA factor binding sites mediate inducible DAL7 gene expression in Saccharomyces cerevisiae . The Journal of Biological Chemistry. 1999;274(39):28026–28034. doi: 10.1074/jbc.274.39.28026. [DOI] [PubMed] [Google Scholar]
- 22.Vissers S., Andre B., Muyldermans F., Grenson M. Induction of the 4-aminobutyrate and urea-catabolic pathways in Saccharomyces cerevisiae. Specific and common transcriptional regulators. European Journal of Biochemistry. 1990;187(3):611–616. doi: 10.1111/j.1432-1033.1990.tb15344.x. [DOI] [PubMed] [Google Scholar]
- 23.Coornaert D., Vissers S., André B. The pleiotropic UGA35(DURL) regulatory gene of Saccharomyces cerevisiae: cloning, sequence and identity with the DAL81 gene. Gene. 1991;97(2):163–171. doi: 10.1016/0378-1119(91)90048-g. [DOI] [PubMed] [Google Scholar]
- 24.Abdel-Sater F., Iraqui I., Urrestarazu A., André B. The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae . Genetics. 2004;166(4):1727–1739. doi: 10.1534/genetics.166.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cardillo S. B., Levi C. E., Moretti M. B., García S. C. Interplay between the transcription factors acting on the GATA- and GABA-responsive elements of Saccharomyces cerevisiae UGA promoters. Microbiology. 2012;158(4):925–935. doi: 10.1099/mic.0.051235-0. [DOI] [PubMed] [Google Scholar]
- 26.Olive M. G., Daugherty J. R., Cooper T. G. DAL82, a second gene required for induction of allantoin system gene transcription in Saccharomyces cerevisiae . Journal of Bacteriology. 1991;173(1):255–261. doi: 10.1128/jb.173.1.255-261.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs P., Jauniaux J.-C., Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae . Journal of Molecular Biology. 1980;139(4):691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- 28.Andre B., Jauniaux J.-C. Nucleotide sequence of the DURM gene coding for a positive regulator of allophanate-inducible genes in Saccharomyces cerevisiae. Nucleic Acids Research. 1990;18(23):p. 7136. doi: 10.1093/nar/18.23.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae . Yeast. 1996;12(3):259–265. doi: 10.1002/(sici)1097-0061(19960315)12:3lt;259::aid-yea901>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 30.Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae . Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 31.Longtine M. S., McKenzie A., III, Demarini D. J., et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14(10):953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Güldener U., Heck S., Fiedler T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Research. 1996;24(13):2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods in Enzymology. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 34.Cardillo S. B., Correa García S., Bermúdez Moretti M. Common features and differences in the expression of the three genes forming the UGA regulon in Saccharomyces cerevisiae . Biochemical and Biophysical Research Communications. 2011;410(4):885–889. doi: 10.1016/j.bbrc.2011.06.086. [DOI] [PubMed] [Google Scholar]
- 35.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Wang D., Zheng F., Holmberg S., Kohlhaw G. B. Yeast transcriptional regulator Leu3p. Self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. The Journal of Biological Chemistry. 1999;274(27):19017–19024. doi: 10.1074/jbc.274.27.19017. [DOI] [PubMed] [Google Scholar]
- 37.Cohen R., Engelberg D. Commonly used Saccharomyces cerevisiae strains (e.g. BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiology Letters. 2007;273(2):239–243. doi: 10.1111/j.1574-6968.2007.00798.x. [DOI] [PubMed] [Google Scholar]
- 38.Gutiérrez A., Beltran G., Warringer J., Guillamón J. M. Genetic basis of variations in nitrogen source utilization in four wine commercial yeast strains. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067166.e67166 [DOI] [PMC free article] [PubMed] [Google Scholar]