Abstract
Nuclear receptors play an important role in transcriptional regulation of diverse cellular processes and is also relevant in diseases such as cancer. In breast cancer, the nuclear receptors – estrogen receptor (ER) and progesterone receptor (PR) are classical markers of the disease and are used to classify breast cancer subtypes. Using a recently developed affinity purification MS technique (RIME) [1], we investigate the protein interactors of ER and PR in breast cancer cell lines upon stimulation by the ligands – estrogen and progesterone. The data is deposited at proteomeXchange (PXD002104) and is part of a publication [2] that explains the link between the two nuclear receptors and potential consequences of this in breast cancer. In this manuscript, we describe the methodology used and provide details on experimental procedures, analysis methods and analysis of raw data. The purpose of this article is to enable reproducibility of the data and provide technical recommendations on performing RIME in hormonal contexts.
Specifications table
| Subject area | Biology, Chemistry |
| More specific subject area | Chromatin biology and Mass spectrometry |
| Type of data | MS data and annotations |
| How data was acquired | Thermo Orbitrap Velos |
| Data format | Raw (.raw files), MGF files, analysed Excel lists |
| Experimental factors | Breast cancer cell lines were grown in SILAC labeled media, hormone treated, formaldehyde crosslinked and RIME performed of ER and PR |
| Experimental features | Interactomes of ER and PR with differential hormone treatments |
| Data source location | Cambridge, United Kingdom |
| Data accessibility | Data deposited at proteomeXchange (PXD002104) |
Value of the data
-
•
This data exhibits quantitative (SILAC) RIME proteomics experiments and details on the methodology.
-
•
Comprehensive analysis of the ER and PR interactome under estrogen alone or Estrogen+ Progesterone conditions.
-
•
Co-stimulation of estrogen and progesterone leads to an interaction between the two receptors.
1. Experimental design – materials and methods
The data presented here uses Rapid Immunoprecipitation Mass spectrometry of Endogenous protein (RIME) [1] to interrogate the interactomes of the estrogen Receptor (ER) and progesterone receptor (PR) under either estrogenic conditions (complete media) or with additional progesterone treatment. This proteomics study describes the effect of activating a single nuclear receptor or two receptors with their respective ligands in breast cancer cell lines and is part of a study that identifies the synergistic roles of ER and PR in breast cancer [2]. All experiments were performed in the breast cancer cell lines MCF-7 and T47-D, both of which express sufficient levels of ER and PR.
1.1. Growth conditions and hormone treatments
The ER+/PR+ cell lines, MCF-7 and T47-D cells were grown in SILAC DMEM (PAA; E15-086) supplemented with 10% dialysed serum (Sigma-Aldrich; F0392) and 800 mM L-lysine 13C615N2 hydrochloride and 482 mM L-arginine 13C6 15N4 hydrochloride (Sigma-Aldrich) for ‘heavy’-labelled media or 800 mM L-lysine 12C614N2 hydrochloride and 482 mM L-arginine 12C614N4 hydrochloride for ‘light’-labelled media. Alternative SILAC DMEM compatible with MCF-7 and T47-D cells also yield similar results (Thermo; 89985). Cells switching to SILAC media were also supplemented with additional glutamate. To avoid clonal effects, cells are expanded in SILAC media and harvested immediately after an appropriate level of label incorporation is achieved, thus heavy or light labeled cells were not maintained long term. Cells were obtained from ATCC. Culture cells were supplemented with 100 nM of progesterone or 10 nM R5020 for 4 h. The ligands were diluted in DMEM (no serum) and all dilutions and stock solutions were maintained in glass vials. Control cells were supplemented with an equivalent amount of vehicle (ethanol).
Cells were grown in 15 cm culture (Nunc) dishes and harvested at ~80% confluency. Cells were crosslinked by replacing media with fresh pre-warmed DMEM (10 ml, without FBS) containing 1% formaldehyde (TEBU Bioscience; 18814). After 8 min of crosslinking, 1 ml of 2.5 M glycine was added to each plate (final volume 11 ml). Plates of cells were then washed twice with cold PBS before the cells were harvested in 1 ml of PBS using a cell scraper. The cells were pelleted at 2000 G and PBS was removed. Cells were then frozen at −80 °C for storage and subsequent steps were performed at a later date.
1.2. Antibody conjugation to beads (RIME)
20 μg of ER (Santa Cruz; sc-543, lot-A2213) or PR (Santa Cruz; sc-7208, lot H2312) antibodies conjugated to 100 μl of magnetic Protein A beads were used for each RIME experiment. To prepare the conjugate, 100 μl of beads were washed three times with 1 ml of 0.5% BSA in PBS. The beads were then resuspended in 100 μl of this buffer and 20 μgs of antibody was added. The mixture was topped up with additional BSA/PBS buffer (250 μl) and then incubated overnight at 4 °C (rotating). The beads were then washed again 3 times in BSA/PBS to remove any unconjugated antibody and then resuspended in 100 μl of BSA/PBS buffer. This final antibody-bead conjugate is then subsequently added to the cell lysate for immunoprecipitation.
1.3. Cell lysis, sonication and immunoprecipitation (RIME)
For each RIME experiment, approximately 20 million cells each (equivalent to 2×15 cm2 dish) of treatment and control grown in heavy or light SILAC conditions were mixed. The cells are then resuspended in 10 ml of LB1 buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 or Igepal CA-630, and 0.25% Triton X-100) and incubated at 4 °C for 10 min (rotating). The cells were then pelleted at 2000 g for 5 mins (4 °C) and the supernatant removed. The resulting pellet was then resuspended in 10 ml of LB2 buffer (10 mM Tris–HCL [pH 8.0], 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) and incubated again at 4 °C for 5 min (rotating). This was then centrifuged at 2000 g for 5 mins and the supernatant was removed. This pellet was then resuspended in LB3 buffer (10 mM Tris–HCl [pH 8], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, and 0.5% N-lauroylsarcosine). Each plate worth of cell pellets (approx. 10–20 million cells) were resuspended in 300 μl of LB3 and transferred to a 1.5 ml eppendorf tube and sonicated in a bioruptor sonicator for 15 min (30 s on/30 s off, high setting). The sonicated lysates for each plate were then pooled and 10% volume of 10%Triton-X solution was added. This mixture was then centrifuged at 12,000 g for 10 min to pellet cell debris. The supernatant was carefully removed and 100 μl of beads pre-incubated with antibody was added. This IP mixture was then incubated overnight at 4 °C (rotating). Subsequently, the lysate-bead mixture was then placed on a magnetic rack and the supernatant removed. The resulting beads were then washed 10 times in RIPA buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCL) and then twice in 1 ml of 100 mM ammonium hydrogen carbonate (freshly prepared). After the last wash, the beads were briefly spun down and any residual liquid was removed. Beads were then either frozen (−20 °C) or directly processed for peptide digestion.
1.4. Tryptic digestion and MS
Bead bound protein complexes were then digested by adding 100 ng of trypsin in 10 μL of 100 mM NH4HCO3 onto the beads. The bead mixture was briefly vortexed and then incubated overnight at 37 °C. The next day, an additional 10 μl of trypsin was added, vortexed and incubated for an additional 4 h. The supernatant was then removed and transferred to a glass vial containing 1 μl of formic acid (5% final concentration). The acidified samples were then bound to a C18 Ultra-Micro Spin Column (Harvard Apparatus) and washed using a 0.1% solution of Formic Acid. Elution was then carried out using a 60% solution of acetonitrile.
The samples were then re-suspended in 0.1% formic acid and loaded on-to an LTQ Orbitrap Velos (Thermo) mass spectrometer and then run using settings previously described [1].
1.5. Data analysis
The acquired MS spectra were then analysed using Proteome Discoverer v1.3 (Thermo Scientific) in combination with the Mascot search engine (Matrix Science) against the SwissProt human database. The search parameters allowed for one missed cleavage and a precursor ion search tolerance of 10 ppm with 0.6 Da for MS/MS ions. Additionally, the variable modifications oxidized methionine and de-amidated asparagine and glutamate residues were included as search parameters. The appropriate heavy amino acid SILAC settings were set as fixed modifications. A false discovery rate (FDR) was calculated using a reversed decoy database with a target of less than 1%. For each RIME experiment, only proteins identified in both cell lines were considered (Fig. 1).
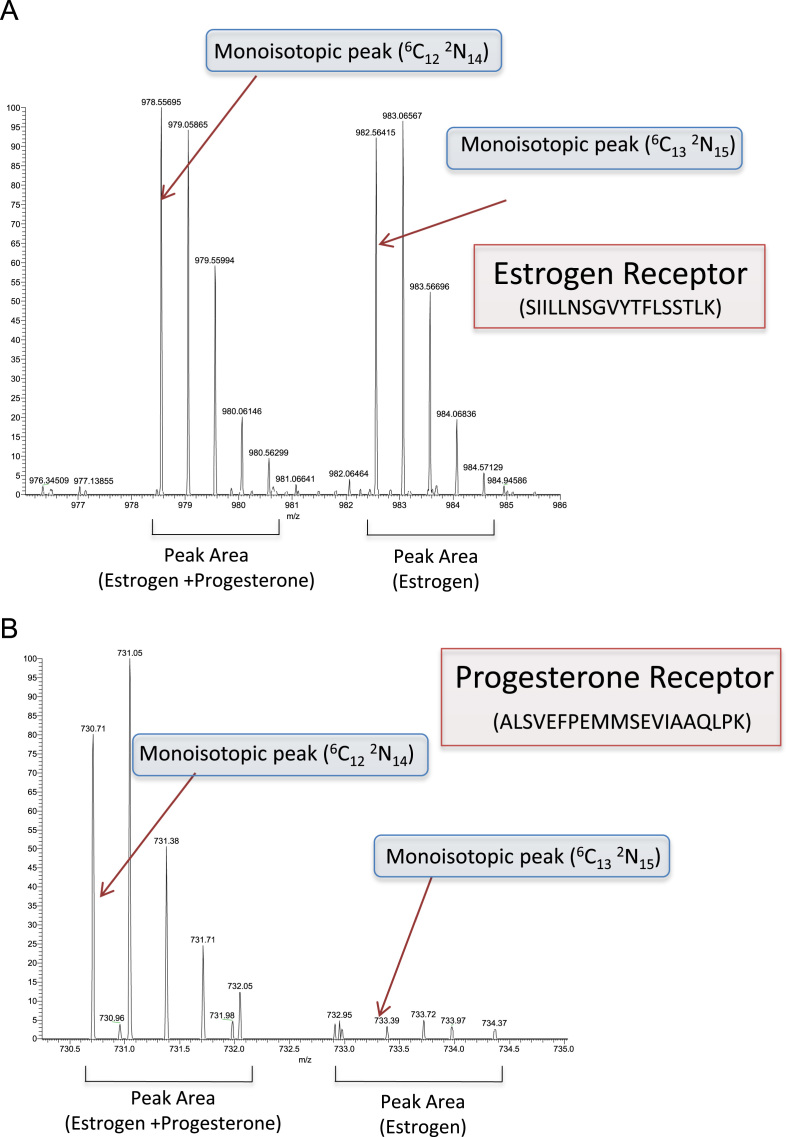
Fig. 1.
Example mass spectra (MS1) of peptide ions used for quantification in an ER RIME experiment with or without progesterone treatment where ER and PR association is seen only after progesterone treatment (PRIDE Accession PDX002104>52234/JP517). (A) MS1 spectrum of the 2+ peptide ions corresponding to the peptide sequence SIILLNSGVYTFLSSTLK from the human estrogen receptor (AC# P03372). Both the light (SIILLNSGVYTFLSSTLK [6C122N14], monoisotopic peak at m/z 978.55695) and heavy (SIILLNSGVYTFLSSTLK [6C132N15], monisotopic peak at m/z 982. 56415) versions of the peptide are indicated. These originate from the estrogen+progesterone treated cells and the estrogen alone cells respectively. Quantification of the peak areas resulted in a 1:1 (H:L) ratio. (B) MS1 spectrum of the 3+ peptide ions corresponding to the peptide sequence ALSVEFPEMMSEVIAAQLPK from the human progesterone receptor (AC# P06401). Both the light (ALSVEFPEMMSEVIAAQLPK [6C122N14], monoisotopic peak at m/z 730.71130) and heavy (ALSVEFPEMMSEVIAAQLPK [6C132N15], monoisotopic peak at m/z 732.95355) versions of the peptide are indicated. These originate from the estrogen+progesterone treated cells and the estrogen alone cells respectively. Quantification of the peak areas resulted in a 1:15 (H:L)ratio.
Acknowledgments
We would like to acknowledge the University of Cambridge and Cancer Research UK for funding and support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.08.019.
Appendix A. Supplementary materials
Supplementary material
References
- 1.Mohammed H. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohammed H. Progesterone receptor modulates oestrogen receptor-a action in breast cancer. Nature. 2015 doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material