Abstract
Exposure to the common environmental contaminant arsenic impacts the epigenetic landscape, including DNA methylation and histone modifications, of several cell types. Developmental arsenic exposure (DAE) increases acetylation and methylation of histone proteins and the protein expression of several chromatin-modifying enzymes in the dentate gyrus (DG) subregion of the adult male mouse brain [26]. To complement and support these data, ChIP-Seq analysis of DNA associated with trimethylation of histone 3 lysine 4 (H3K4me3) derived from the adult male DG after DAE was performed. DAE induced differential H3K4me3 enrichment on genes in pathways associated with cellular development and growth, cell death and survival, and neurological disorders, particularly as they relate to cancer, in the adult male brain. Comparison of H3K4me3 enrichment in controls revealed mechanisms that are potentially lacking in arsenic-exposed animals, including neurotransmission, neuronal growth and development, hormonal regulation, protein synthesis, and cellular homeostasis. New pathways impacted by arsenic include cytoskeleton organization, cell signaling, and potential disruption of immune function and warrant further investigation using this DAE paradigm in the mouse brain.
Specifications Table
| Subject area | Biology |
|---|---|
| More specific subject area | Neurotoxicology and epigenetics |
| Type of data | Tables, graph |
| How data was acquired | Genomic sequencing (HiSeq2000) and bioinformatics |
| Data format | Filtered for peak annotation, and analyzed for differential enrichment and gene ontology |
| Experimental factors | Developmental exposure to 50 parts-per-billion sodium arsenate in drinking water (DAE) |
| Experimental features | Exposure to sodium arsenate (50 parts-per-billion, ppb) in drinking water occurred during all three trimesters of fetal/neonatal development in C57BL/6 mice. Arsenic exposure ceased after the third postnatal trimester of development at approximately postnatal day 23. Animals were then placed on tap water (consisting of 5 ppb arsenic) until adulthood. Brain tissue, specifically the dentate gyrus sub-region of the hippocampus, was harvested from control and arsenic-exposed adult male mice (postnatal day 70). Chromatin immunoprecipitation was performed for the histone 3 lysine 4 trimethylation modification; immunoprecipitated DNA was isolated and sent to the University of Southern California for sequencing analysis on a HiSeq2000. Data was analyzed for specific H3K4me3 enrichment after arsenic exposure compared to controls. |
| Data source location | N/A |
| Data accessibility | Filtered raw data files from sequencing provided in table format Analyzed data files provided in table format Analyzed data files provided in graphical format |
Background
The data provided in this article are intended to support research demonstrating that developmental arsenic exposure (DAE) increases the levels of H3K4me3 and H3K9ac histone modifications along with associated histone methyltransferase and acetyltransferase proteins in the dentate gyrus (DG) of the adult mouse brain [26]. The DG contains neural progenitor cells that actively undergo proliferation, differentiation and integration into the hippocampal neural circuitry in adulthood [19]. These processes are collectively referred to as adult neurogenesis and are important for cognitive function and disease susceptibility including depression [15]. Epidemiological studies have shown arsenic exposure, a common contaminant found in drinking water, correlates with cognitive dysfunction, particularly in children, and psychiatric disorders like depression in adults [3,30,4,5]. Our DAE paradigm reduces differentiation of neural progenitor cells, induces deficits in memory, and increased depressive behaviors in adult male mice [18,24,27]. Epigenetic mechanisms within neural progenitor cells, particularly histone modifications, are paramount for proper specification of gene expression for all the processes of neurogenesis [11,14]. Arsenic exposure has been shown to alter histone modifications in the blood of humans exposed to high levels of this toxin [22,29,7]. Thus, to determine potential mechanisms of arsenic-induced toxicity in the DG, chromatin immunoprecipitation followed by sequencing (ChIP-Seq) for histone 3 lysine 4 trimethylation (H3K4me3) was performed. While research demonstrating arsenic’s impact on the brain’s epigenome brain is limited to a handful of studies [10,17,28], several reports have established that arsenic adversely alters histone posttranslational modifications and DNA methylation in the mammalian body [13,17,22,29,7–9]. While arsenic speciation likely plays a role in damage to methylation capacity in the body [16], generally, excessive exposure to arsenic inhibits one-carbon metabolism, effectively depleting S-adenosyl methionine (SAM) [21]. However, both hypo- and hyper-methylation of DNA has been observed in response to arsenic toxicity [20]; as such, simple depletion of SAM and altered methylation status is likely not the mechanism of arsenic toxicity in context of the brain [23]. Using ChIP-seq analysis we sought to identify new pathways for mechanisms of action, particularly in this region of the brain that contains stem cells. As arsenic exposure has been shown to adversely impact males more than females, this analysis was performed on the male brain [25].
Value of the data
-
•
First H3K4me3 ChIP-seq analysis in the dentate gyrus of a mouse model.
-
•
First genomic data to demonstrate that developmental arsenic exposure induces long-lasting transcriptional activation via altered epigenetic status in the adult male mouse brain.
-
•
Analyses indicate that arsenic alters epigenetic regulation of genes involved in cell death and survival, cell development and growth, abnormal cell morphology and organization, gene expression, some immune function, and a host of neurological diseases, including cancer and neuropathy, in the brain.
Data
Using a mouse model of developmental arsenic exposure (DAE), we have previously shown deficits in learning and memory, depressive-like symptoms, and reduced adult neurogenesis in adult male mice [18,24,27]. A region important in these processes is the dentate gyrus (DG) of the hippocampus, for which we have shown that arsenic increases H3K4 trimethylation and alters protein expression of MLL and KDM5B, two H3K4me3 chromatin modifiers [26]. To complement this data, next generation sequencing of H3K4me3 enriched DNA from the DG of control and arsenic-exposed male mice was performed on an Illumina HiSeq 2000 with 50 bp single end reads with a 98% alignment of approximately 30 million reads to the mouse genome. Peak calling comparisons between the arsenic and control sequences were performed using the HOMER (Hypergeometric Optimization of Motif EnRichment) package; annotation of peaks differentially enriched for H3K4me3 in the arsenic sequences and in the control sequences, along with the most significant gene ontology (GO) categories and functional annotations, are provided.
The data is as follows:
-
1.Pdf files of the original sequencing reads that have been filtered, aligned, and annotated to the mouse genome (mm10), indicating all genes with H3K4me3 enrichment in both the control and arsenic-exposed animals relative to input, labeled as the following
-
a.Supplementary Table S1: H3K4me3 ChIP-Seq for arsenic-exposed adult male dentate gyrus (PD70), Arsenic sample 1
-
b.Supplementary Table S2: H3K4me3 ChIP-Seq for arsenic-exposed adult male dentate gyrus (PD70), Arsenic sample 2
-
c.Supplementary Table S3: H3K4me3 ChIP-Seq for control adult male dentate gyrus (PD70), Control sample 1
-
d.Supplementary Table S4: H3K4me3 ChIP-Seq for control adult male dentate gyrus (PD70), Control sample 2
-
a.
-
2.Pdf files comparing the H3K4me3 enrichment of arsenic versus control animals and the significant gene ontologies, labeled as the following:
-
a.Supplementary Table S5: HOMER Analysis of H3K4me3 differential peak binding after developmental arsenic exposure in adult male dentate gyrus samples
- b.
-
a.
-
3.Pdf files comparing the H3K4me3 enrichment of control versus arsenic animals and the significant gene ontologies, labeled as the following:
-
a.Supplementary Table S7: HOMER Analysis of H3K4me3 differential peak binding in control male dentate gyrus samples
- b.
-
a.
-
4.Graphs of gene expression validation for Gas6, Rora, Sulf2 genes labeled as:
-
a.Fig. 1. mRNA gene expression validation of ChIP-Seq analysis in the adult male dentate gyrus after developmental arsenic exposure
-
i.ChIP-Seq analysis indicated these genes have increased H3K4me3 enrichment in arsenic-exposed animals; mRNA expression of these three genes is significantly increased in the adult male dentate gyrus after developmental arsenic exposure validating results provided by sequencing analysis
-
a.
Fig. 1.
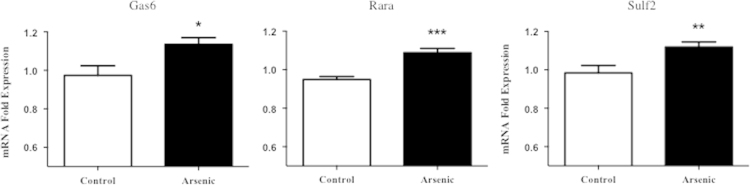
mRNA gene expression validation of ChIP-Seq analysis in the adult male dentate gyrus after developmental arsenic exposure.
Experimental design
A mouse model of developmental arsenic exposure during all three-trimester equivalents was used, as previously described [18,24], to determine the impact of arsenic on the epigenome. DNA from H3K4me3 chromatin immunoprecipitation of the dentate gyrus region of the male brain was sequenced using via HiSeq2000 and analyzed using HOMER software suite and Ingenuity Pathway Analysis. A selection of genes with significant H3K4me3 enrichment in either control or arsenic-exposed animals was analyzed for mRNA expression in dentate gyrus tissue to confirm ChIP-Seq findings.
Developmental arsenic exposure paradigm
The Institutional Animal Care and Use Committee at the University of New Mexico (UNM) approved the animal protocols, including the arsenic exposure paradigm, used in this study. C57BL/6 mice obtained from Jackson Labs were maintained on a reverse light/dark cycle (lights off at 0800) with ad libitum access to food and water in the Animal Resource Facility at UNM. Arsenic exposure was performed as previously described [27]; briefly, singly-housed female mice aged 55 days were acclimated to drinking 50 parts-per-billion arsenic water (sodium arsenate, Sigma Aldrich) for 10 days prior to mating. Arsenic water was prepared weekly using standard tap and MilliQ water. Control mice were administered tap water from UNM, which contains approximately 2–5 ppb arsenic. Mating occurred for five days; dams continued to drink arsenic-laced water throughout pregnancy until offspring were weaned at postnatal day (PD) 23. Offspring were group housed separately by sex, four per cage, with ad libitum access to food and tap water. At PD70, animals were euthanized via rapid decapitation, and the dentate gyrus from male animals was microdissected and snap frozen and stored at −80 °C until further analysis. For each experiment, n represents the number of different litters used with one animal per litter to avoid litter effects.
Chromatin immunoprecipitation
Dentate gyrus tissue obtained from adult male mice was homogenized in a Biomasher II disposable microhomogenizer (Kimble Chase) using a 1% formaldehyde crosslinking solution (1% formaldehyde, 1 mM EDTA, 0.5 mM EGTA, 50 mM HEPES, pH 8.0) for 15 min. Reagents from the ChIP-IT® Express Chromatin Immunoprecipitation Kits from Active Motif (53008) were used for some steps in the protocol. A 1× Glycine solution (Active Motif) was added to each reaction for 5 min. Homogenates were centrifuged at 1000×g for 6 min at 4 °C and washed with 1× PBS containing 1 μg/μl protease inhibitor cocktail (Sigma, P8340). Cell pellets were resuspended in cell lysis buffer containing 0.5% Triton-X 100 (v/v), 85 mM KCl, 5 mM HEPES, and 2 μg/μl protease inhibitor cocktail and allowed to sit on ice for 30 min. Cells were centrifuged as before, resuspended in cell lysis buffer, and repelleted as before. The pellet was resuspended in nuclear lysis buffer containing 50 mM Tris, 10 mM EDTA, 1% SDS, and 2 μg/μl protease inhibitor cocktail. Crosslinked chromatin was sheared via sonication. Samples were sonicated for 10 s with 2 min on ice between intervals (10×) to yield approximately 300–500 base pair chromatin, determined by 1.5% (w/v) agarose gel separation. Samples were centrifuged at 14,000×g for 10 min at 4 °C. The supernatant was snap frozen and stored at –80 °C. The antibody of interest was incubated with magnetic beads for 4 hours at 4 °C prior to the IP. ChIP-ready chromatin was incubated overnight with magnetic beads (Active Motif) and the histone 3 lysine 4 trimethyl (H3K4me3) antibody (Millipore, 04-745). Antibody validation was performed using RNA Polymerase II (Qiagen, GAM-111), IgG (Qiagen, GAM-8208), and the H3K4me3 antibody for the genes Gapdh and Myod1 as positive and negative controls, respectively. A small fraction of the chromatin was saved to derive the input DNA. The following day, samples were washed at 4 °C and the DNA eluted using a ChIP DNA Purification Kit (Active Motif; 58002).
ChIP-Seq library preparation and sequencing
The University of Southern California Norris Cancer Center Next Gen Sequencing Core Facility (http://epigenome.usc.edu) performed library preparation and sequencing on H3K4me3 immunoprecipitated DNA derived from control and arsenic-exposed adult male dentate gyrus tissue. Library preparation was performed using a NEB Ultra DNA library kit for biological replicates of control and arsenic samples (50 ng), and a separate library was generated from input DNA (no IP) from pooled tissue. Libraries were quantitated and size distribution and purity determined using a Qubit fluorometer and an Agilent 2100 Bioanalyzer, respectively. Sequencing was performed on an Illumina HiSeq 2000 sequencer using v3 chemistry with 50 bp single-end reads. Approximately 30 million reads per sample (two arsenic biological replicates, two control biological replicates, one input control) were obtained.
ChIP-Seq analyses and gene ontology
The USC Sequencing Core Facility performed quality control, using the CASAVA pipeline (version 1.8.4) and mapping to the USCS mouse genome (mm10), using BWA (version 0.6.2). ChIP-seq peaks were identified and analyzed using the HOMER software suite (http://biowhat.ucsd.edu/homer/). Tag directories were made for each of the five samples, and the findPeaks function in histone mode, with the FDR threshold 0.001, was used to filter approximately 16,000−18,000 peaks in each of the four ChIP-Seq samples against the input sample. The getDifferentialPeaks function was then used to identify differentially bound peaks with greater than 4-fold H3K4me3 enrichment, with a cumulative Poisson p-value less than 0.0001, in the arsenic samples compared to the control samples and vice versa. Annotation of the enriched peaks was performed using the HOMER annotatePeaks with the mm10 mouse genome as a reference and promoters defined as −1 kb to +1 kb relative to the position of the TSS. Functional clustering using Ingenuity Pathway Analysis (IPA) was performed to determine common processes affected by developmental arsenic exposure.
Gene expression validation
Trimethylation of lysine 4 on histone 3 (H3K4me3), particularly in the promoters and coding regions of genes, is associated with transcriptional activation [1,2]. To confirm results provided by ChIP-Seq data analysis via HOMER, we assessed mRNA gene expression for candidate genes with significant H3K4me3 enrichment in either arsenic-exposed or control dentate gyrus tissue. Tissue was derived from adult male mice (aged 70 days) from different litters than those used for sequencing to avoid litter confounds. Total RNA was purified from whole mouse dentate gyrus using the Ambion mirVana™ miRNA Isolation Kit (Life Technologies; AM1560) following the manufacturer’s instructions. Total RNA was quantified with the Qubit® RNA BR Assay Kit (Life Technologies; Q10211). gDNA was digested with DNase I recombinant (Roche Life Science; 04716728001) and cDNA reverse transcription was conducted with the Transcriptor First Strand cDNA Synthesis Kit (Roche Life Science; 04897030001). cDNA was quantified by NanoDrop 1000 spectrophotometer (Life Technologies). Quantitative PCR was performed on a Roche LightCycler 96 with Roche FastStart Essential DNA Green Master (06924204001) under standard cycle conditions. Results were analyzed by the relative quantification method using Hprt as an endogenous control. mRNA target primer efficiencies were validated and matched to control primer efficiencies. Primer sequences were as follows:
| Gene | Gene ID | Forward primer | Reverse primer | Source |
|---|---|---|---|---|
| Hprt (set a) | 15452 | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT | [12,6] |
| Hprt (set b) | 15452 | AAGACTTGCTCGAGATGTCATGAA | ATCCAGCAGGTCAGCAAAGAA | [6] |
| Gas6 | 14456 | TGCTGGCTTCCGAGTCTTC | CGGGGTCGTTCTCGAACAC | Primer Bank ID: 9506715a1 |
| Rara | 19401 | TTCTTTCCCCCTATGCTGGGT | GGGAGGGCTGGGTACTATCTC | Primer Bank ID: 22800363a1 |
| Sulf2 |
72043 |
CTGCCACTATGGCTGCTGTC |
GTTGGGCCGGATGTTCCTG |
Primer Bank ID: 26330131a1 |
Acknowledgments
We thank Dr. Daniel Purcell, Dr. Mary Ann Osley, and Dr. Charles Nicolet for assistance and helpful discussions. This work was supported by grants from the National Institute of Environmental Health Sciences [RO1ES019583 to AMA], the National Institute of Mental Health [F31 101984 to CRT], and the Pilot Project Award from the UNM HSC Environmental Health Signature Program [AMA & CRT].
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.08.037.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E., Hitz B.C., Gupta R., Rando T.A., Baker J.C., Snyder M.P., Cherry J.M., Brunet A. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein B.E., Humphrey E.L., Erlich R.L., Schneider R., Bouman P., Liu J.S., Kouzarides T., Schreiber S.L. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkel J., Khan M.H., Kraemer A. A systematic review of arsenic exposure and its social and mental health effects with special reference to Bangladesh. Int. J. Environ. Res. Public Health. 2009;6:1609–1619. doi: 10.3390/ijerph6051609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderon J., Navarro M.E., Jimenez-Capdeville M.E., Santos-Diaz M.A., Golden A., Rodriguez-Leyva I., Borja-Aburto V., Diaz-Barriga F. Exposure to arsenic and lead and neuropsychological development in Mexican children. Environ. Res. 2001;85:69–76. doi: 10.1006/enrs.2000.4106. [DOI] [PubMed] [Google Scholar]
- 5.Calderon R.L., Hudgens E.E., Carty C., He B., Le X.C., Rogers J., Thomas D.J. Biological and behavioral factors modify biomarkers of arsenic exposure in a U.S. population. Environ. Res. 2013;126:134–144. doi: 10.1016/j.envres.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell K.K., Sheema S., Paz R.D., Samudio-Ruiz S.L., Laughlin M.H., Spence N.E., Roehlk M.J., Alcon S.N., Allan A.M. Fetal alcohol spectrum disorder-associated depression: evidence for reductions in the levels of brain-derived neurotrophic factor in a mouse model. Pharmacol. Biochem. Behav. 2008;90:614–624. doi: 10.1016/j.pbb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chervona Y., Arita A., Costa M. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics: Integr. Biomet. Sci. 2012;4:619–627. doi: 10.1039/c2mt20033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chervona Y., Hall M.N., Arita A., Wu F., Sun H., Tseng H.C., Ali E., Uddin M.N., Liu X., Zoroddu M.A., Gamble M.V., Costa M. Association between arsenic exposure and global post-translational histone modifications among adults in Bangladesh. Cancer Epidemiol. Biomark. Prev. 2012 doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu F., Ren X., Chasse A., Hickman T., Zhang L., Yuh J., Smith M.T., Burlingame A.L. Quantitative mass spectrometry reveals the epigenome as a target of arsenic. Chem.-Biol. Interact. 2011;192:113–117. doi: 10.1016/j.cbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cronican A.A., Fitz N.F., Carter A., Saleem M., Shiva S., Barchowsky A., Koldamova R., Schug J., Lefterov I. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PloS One. 2013;8:e53478. doi: 10.1371/journal.pone.0053478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day J.J., Sweatt J.D. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilsbach R., Kouta M., Bonisch H., Bruss M. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. BioTechniques. 2006;40:173–177. doi: 10.2144/000112052. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Rubio P., Roberge J., Arendell L., Harris R.B., O'Rourke M.K., Chen Z., Cantu-Soto E., Meza-Montenegro M.M., Billheimer D., Lu Z., Klimecki W.T. Association between body mass index and arsenic methylation efficiency in adult women from southwest U.S. and northwest Mexico. Toxicol. Appl. Pharmacol. 2011;252:176–182. doi: 10.1016/j.taap.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh J., Eisch A.J. Epigenetics, hippocampal neurogenesis, and neuropsychiatric disorders: unraveling the genome to understand the mind. Neurobiol. Dis. 2010;39:73–84. doi: 10.1016/j.nbd.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnan V., Nestler E.J. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C., Zhao F., Sun D., Zhong Y., Yu X., Li G., Lv X., Sun G., Jin Y. Comparison of speciated arsenic levels in the liver and brain of mice between arsenate and arsenite exposure at the early life. Environ. Toxicol. 2014;29:797–803. doi: 10.1002/tox.21808. [DOI] [PubMed] [Google Scholar]
- 17.Martinez L., Jimenez V., Garcia-Sepulveda C., Ceballos F., Delgado J.M., Nino-Moreno P., Doniz L., Saavedra-Alanis V., Castillo C.G., Santoyo M.E., Gonzalez-Amaro R., Jimenez-Capdeville M.E. Impact of early developmental arsenic exposure on promotor CpG-island methylation of genes involved in neuronal plasticity. Neurochem. Int. 2011;58:574–581. doi: 10.1016/j.neuint.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Finley E.J., Ali A.M., Allan A.M. Learning deficits in C57BL/6J mice following perinatal arsenic exposure: consequence of lower corticosterone receptor levels? Pharmacol. Biochem. Behav. 2009;94:271–277. doi: 10.1016/j.pbb.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ming G.L., Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichard J.F., Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2:87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichard J.F., Schnekenburger M., Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem. Biophys. Res. Commun. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren X., McHale C.M., Skibola C.F., Smith A.H., Smith M.T., Zhang L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ. Health Perspect. 2011;119:11–19. doi: 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rios R., Santoyo M.E., Cruz D., Delgado J.M., Zarazua S., Jimenez-Capdeville M.E. Methyl group balance in brain and liver: role of choline on increased S-adenosyl methionine (SAM) demand by chronic arsenic exposure. Toxicol. Lett. 2012;215:110–118. doi: 10.1016/j.toxlet.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Tyler C.R., Allan A.M. Adult hippocampal neurogenesis and mRNA expression are altered by perinatal arsenic exposure in mice and restored by brief exposure to enrichment. PloS One. 2013;8:e73720. doi: 10.1371/journal.pone.0073720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyler C.R., Allan A.M. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: a review. Curr. Environ. Health Rep. 2014;1:132–147. doi: 10.1007/s40572-014-0012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyler C.R., Hafez A.K., Solomon E.R., Allan A.M. Developmental exposure to 50 parts-per-billion arsenic influences histone modifications and associated epigenetic machinery in a region- and sex-specific manner in the adult mouse brain. Toxicol. Appl. Pharmacol. 2015;288:40–51. doi: 10.1016/j.taap.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyler C.R., Solomon B.R., Ulibarri A.L., Allan A.M. Fluoxetine treatment ameliorates depression induced by perinatal arsenic exposure via a neurogenic mechanism. Neurotoxicology. 2014;44:98–109. doi: 10.1016/j.neuro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarazua S., Rios R., Delgado J.M., Santoyo M.E., Ortiz-Perez D., Jimenez-Capdeville M.E. Decreased arginine methylation and myelin alterations in arsenic exposed rats. Neurotoxicology. 2010;31:94–100. doi: 10.1016/j.neuro.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Sun H., Ellen T.P., Chen H., Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zierold K.M., Knobeloch L., Anderson H. Prevalence of chronic diseases in adults exposed to arsenic-contaminated drinking water. Am. J. Public Health. 2004;94:1936–1937. doi: 10.2105/ajph.94.11.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material


