Abstract
Hepatocellular Carcinoma (HCC) is one of the most common malignant tumor, which is causing the second leading cancer-related death worldwide. The tumor tissues and the adjacent noncancerous tissues obtained from HCC patients with single and multiple lesions were quantified using iTRAQ. A total of 5513 proteins (FDR of 1%) were identified which correspond to roughly 27% of the total liver proteome. And 107 and 330 proteins were dysregulated in HCC tissue with multiple lesions (MC group) and HCC tissue with a single lesion (SC group), compared with their noncancerous tissue (MN and SN group) respectively. Bioinformatics analysis (GO, KEGG and IPA) allowed these data to be organized into distinct categories. The data accompanying the manuscript on this approach (Xing et al., J. Proteomics (2015), http://dx.doi.org/10.1016/j.jprot.2015.08.007[1]) have been deposited to the iProX with identifier IPX00037601.
Specifications table
| Subject area | Biology |
| More specific subject area | Proteomics on the Hepatocellular Carcinoma |
| Type of data | List of identified proteins as tables (.xls), raw data in website |
| How data was acquired | The data was acquired by Liquid chromatography mass spectrometry in tandem (LC–MS/MS).The samples were separated by a Acquity UPLC system (Waters Corporation, Milford, MA) and detected by a Nano-Aquity UPLC system (Waters Corporation, Milford, MA) connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive) (Thermo Fisher Scientific, Bremen, Germany). |
| Data format | Filtered and analyzed |
| Experimental factors | Non-applied |
| Experimental features | Proteins were extracted from tumor tissues of HCC patients with single and multiple lesions, iTRAQ labeled and then prepared for liquid chromatography-mass spectrometry (LC–MS/MS) analysis. |
| Data source location | Fuzhou, China, Mengchao Hepatobiliary Hospital of Fujian Medical University |
| Data accessibility | Filtered and analyzed data are supplied here and raw data have also been deposited to the integrated Proteome resources (iProX) with identifier IPX00037601 (http://www.iprox.org/index). |
Value of the data
-
•
The proteome of hepatocellular carcinoma with single and multiple lesions analyzed using iTRAQ technology.
-
•
A total of 5513 proteins (FDR of 1%) were identified which correspond to roughly 27% of the total liver proteome.
-
•
The in-depth proteomics analysis of the HCC tumor tissues with a single and multiple lesions might be useful for further study of the mechanisms.
1. Data, experimental design, materials and methods
1.1. Data and experimental design
The data show the lists of proteins identified and quantified in the HCC tumor tissues with single and multiple lesions. The tissues were divided into 4 groups: cancerous tissues from HCC patients with multiple observed lesions (MC group, n=30); surrounding noncancerous tissues from HCC patients with multiple observed lesions (MN group, n=30); cancerous tissues from primary HCC patients with a single observed lesion (SC group, n=30); surrounding noncancerous tissues from primary HCC patients with a single observed lesion (SN group, n=30). The detailed characteristics of the selected HCC patients were listed in Table 1. For each group, every 5 individual samples with equal tissue weight were mixed, and then the proteins were extracted from the mixed samples. And then the samples were labeled with the iTRAQ 8-plex reagent as follows: four groups (MC group, MN group, SC group and SN group) were labeled with 113, 114, 115 and 116 isobaric tag, respectively; and the peptides from the biological repetitions of the above 4 groups were labeled with 117, 118, 119 and 121, respectively. The iTRAQ 8-plex labeling was independently repeated 3 times, defining as A, B and C. So we have 6 repeated protein extracts for each group to minimize the individual differences of the patients.
Table 1.
Basic information and characteristics of the HCC patients with a single or multiple observed lesions, who were enrolled in this dataset.
| HCC with multiple lesions | HCC with a single lesion | |
|---|---|---|
| Gender | ||
| Male | 30 | 30 |
| Female | 0 | 0 |
| Age (years) | ||
| ≤55 | 17 | 17 |
| >55 | 13 | 13 |
| AFP (ng/ml) | ||
| ≤400 | 12 | 20 |
| >400 | 18 | 10 |
| Tumor size (cm) | ||
| ≤5 | 9 | 6 |
| 5–10 | 21 | 24 |
| Progression of cirrhosis | ||
| None | 3 | 5 |
| Mild | 13 | 12 |
| Moderate | 13 | 12 |
| Severe | 1 | 1 |
| Tumor boundaries | ||
| Distinct | 18 | 22 |
| Indistinct | 12 | 8 |
| Differentiation degree | ||
| I–II | 8 | 4 |
| II–III | 17 | 22 |
| III–IV | 5 | 4 |
| Vascular tumor thrombosis | ||
| No | 26 | 25 |
| Yes | 4 | 5 |
| Tumor encapsulation | ||
| No | 2 | 6 |
| Incomplete | 14 | 10 |
| Complete | 14 | 14 |
1.2. Materials and methods
Tissue samples, including the cancerous and surrounding noncancerous tissues, were obtained from 30 primary HCC patients with multiple observed lesions and 30 primary HCC patients with a single observed lesion, respectively. All patients have undergone radical surgery at Mengchao Hepatobiliary Hospital of Fujian Medical University from August 2010 to January 2013. The protein from these two type HCC tissues was determined by BCA assay (TransGen Biotech, Beijing, China) following the manufacture’s protocol. Afterwards, 100 μg proteins per condition were treated with DTT (8 mM) and iodoacetamide (50 mM) for reduction and alkylation. Afterwards, the proteins were typically digested by sequence-grade modified trypsin (Promega, Madison, WI), and then the resultant peptides mixture was further labeled using chemicals from the iTRAQ reagent kit (AB SCIEX, USA).
The peptide mixture was fractionated by high pH separation using a Acquity UPLC system (Waters Corporation, Milford, MA) connected to a reverse phase column (BEH C18, 1.7 µm, 2.1×50 mm2, Waters Corporation, Milford, MA). High pH separation was performed using a linear gradient starting from 5% B to 35% B in 20 min (solution B: 20 mM ammonium formate in 90% ACN, the pH was adjusted to 10.0 with ammonium hydroxide). The column flow rate was maintained at 600 μl/min and column temperature was maintained at room temperature. Finally 40 fractions were collected, and two fractions with the same time interval were pooled together to reduce the fraction numbers, such as 1 and 21, 2 and 22, and so on [2]. Twenty fractions at the end were dried in a vacuum concentrator for further usage.
The fractions were then separated by nano-LC and analyzed by on-line electrospray tandem mass spectrometry. The experiments were performed on a Nano-Aquity UPLC system (Waters Corporation, Milford, MA) connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive) (Thermo Fisher Scientific, Bremen, Germany) equipped with an online nano-electrospray ion source. 8 μl peptide sample was loaded onto the trap column (Thermo Scientific Acclaim PepMap C18, 100 μm×2 cm)with a flow of 10 μl/min, and subsequently separated on the analytical column (Acclaim PepMap C18, 75 μm×50 cm) with a linear gradient, from 2% D to 40% D in 135 min (solution D: 0.1% formic acid in ACN). The Q-Exactive mass spectrometer was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full-scan MS spectra (m/z 350–1200) was acquired with a mass resolution of 70 K, followed by 15 sequential high energy collisional dissociation (HCD) MS/MS scans with a resolution of 17.5 K. In all cases, one microscan was recorded using dynamic exclusion of 30 s.
1.3. Data analysis
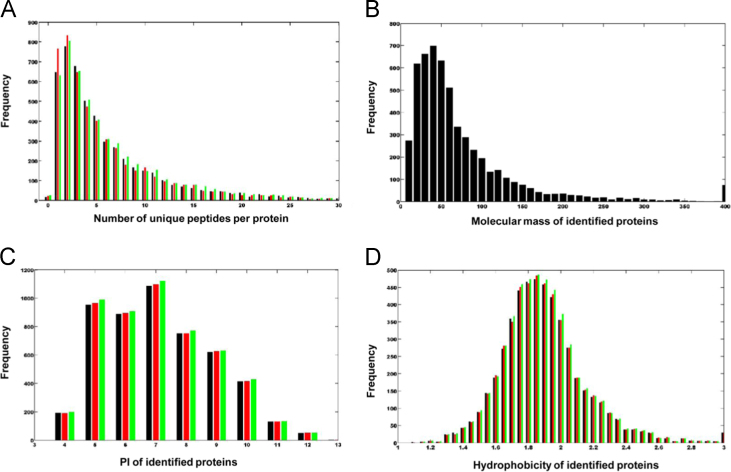
All the raw files generated by the Q-Exactive instrument were converted into mzXML and MGF files using the ms convert module in Trans-Proteomic Pipeline (TPP 4.6.2). All MGF files were searched using Mascot (Matrix Science, London, UK; version 2.3.0) against a human_database provided by The Universal Protein Resource (http://www.uniprot.org/uniprot, released at 2014-04-10, with 20,264 entries). Using the results from Scaffold_4.3.2, we quantified 5513 proteins in three iTRAQ 8-plex labeling replicates. The complete list of identified proteins in our dataset is shown in Table S1. The detailed characteristics of proteomes of the primary HCC with single and multiple lesions, including Molecular Weight (MW), Isoelectric Point (PI), Hydrophobicity, exponentially modified Protein Abundance Index (emPAI), Quantitative Clustering, Average Coefficient of Variance (CV), quantification results with percentage variability, were included in the list as well. The distribution of unique peptide numbers per protein, MW, PI and hydrophobicity also clearly showed that the overall proteome datasets of the primary HCC with single and multiple lesions had no strong bias (Fig. 1).
Fig. 1.
The qualities of the proteome dataset. (A) Frequency distribution of the identified proteins with ≥1 unique peptides. (B) Molecular weight distribution of identified proteins proved that there is no bias in the protein extraction process. (C) Isoelectric point distribution of the identified proteins to show the unbias of the protein extraction. (D) Protein hydrophobicity distribution of the identified proteins.
2. Analysis of the dataset
2.1. The analysis of the quantitative proteomics
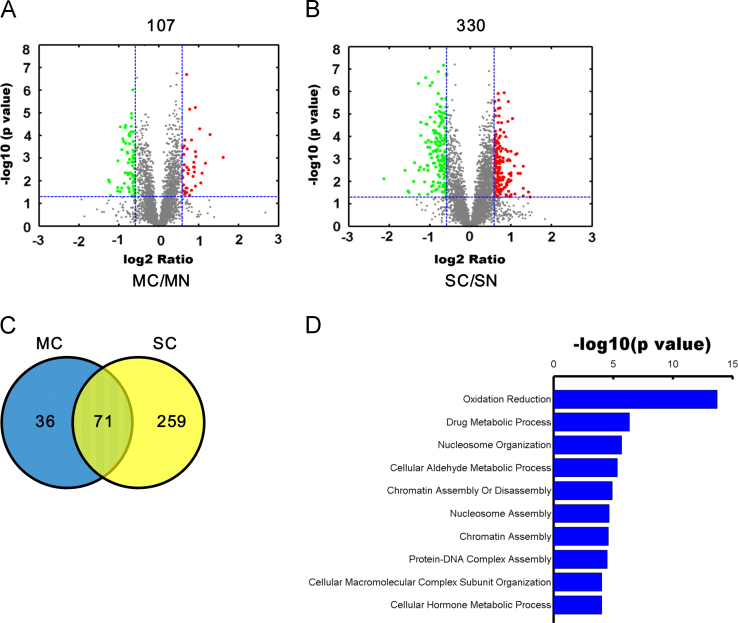
In this dataset, 107 and 330 proteins were classified as differentially expressed in HCC tumor tissues with single and multiple lesions compared to surrounding noncancerous tissues (Fig. 2A, B). All of the differentially expressed proteins presented a mean expression fold change of ±1.5 (log2 0.58) or even more with a p value less than 0.05 (paired T-test), meanwhile these proteins should have the same change trends in all six biological replicates. Among these differentially expressed proteins, 71 proteins altered their expression in both HCC types (Fig. 2C). GO annotation analysis showed that these proteins were the major participants in the oxidation reduction process and the cellular metabolic processes (Fig. 2D).
Fig. 2.
The iTRAQ ratio distribution and involved biological processes of the differentially expressed proteins in the HCC with single and multiple lesions. (A) Volcano plot represented the protein abundance changes in the HCC cancerous tissue with multiple lesions comparing to their adjacent noncancerous tissues. A total of 107 dysregulated proteins with fold change ≥1.5 and p values <0.05 were identified. (B) Volcano plot represented the protein abundance changes in the HCC cancerous tissues with a single lesion comparing to their adjacent noncancerous tissue. A total of 330 dysregulated proteins with fold change ≥1.5 and p values <0.05 were identified. (C) Venn diagrams showed the overlaps and number of differentially expressed proteins in the HCC with single and multiple lesions. (D) GO analysis of the involved biological processes of the common dysregulated proteins in both types of the HCC. All of the biological processes were ranked in term of enrichment of the differentially expressed proteins, and the top 10 are presented here.
2.2. Bioinformatics analysis
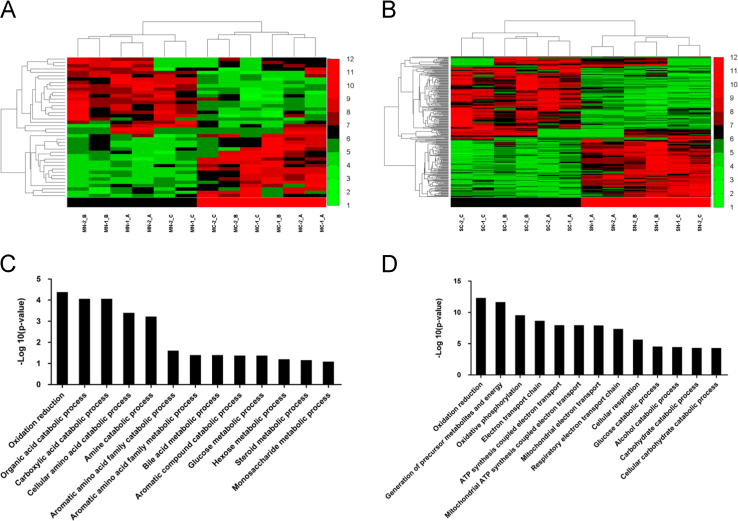
The Gene Ontology (GO) annotation and pathway enrichment analysis of all the identified proteins and differentially expressed proteins were implemented using the online tool DAVID (http://david.abcc.ncifcrf.gov/). The quantitative iTRAQ ratios of 36 proteins, which dysregulated in MC group comparing to MN group, but these proteins were not dysregulated in primary HCC with a single lesion, were plotted on a heatmap (Fig. 3A). The names of the dysregulated proteins are listed in Table 2. We further analyzed these protein involved biological process by GO analysis (Fig. 3C). Meanwhile, 142 up-regulated proteins and 117 down-regulated proteins were specifically appeared in HCC with a single lesion group, but not in HCC with multiple lesions group; and the up and down regulated proteins also form clearly distinct clusters in the heatmap (Fig. 3B). The list of protein names is also displayed in Table 3. We further analyzed these protein involved biological process by GO analysis (Fig. 3D). Gene ontology (GO) analysis of the molecular function and cell component of differentially expressed proteins which is only dysregulated in HCC with a single lesion or HCC with multiple lesions are also displayed in Fig. 4.
Fig. 3.
The hierarchical clustering and involved biological processes analysis of differentially expressed proteins in the primary HCC with single and multiple lesions. (A) Hierarchical clustering of the 107 dysregulated proteins in the HCC with multiple lesions (MC vs. MN). (B) Hierarchical clustering of the 330 dysregulated proteins in the HCC with a single lesion (SC vs. SN). (C, D) GO analysis of the dysregulated proteins involved biological processes in the HCC with multiple lesions (C) and in the HCC with a single lesion (D).
Table 2.
List of the differentially expressed proteins which is only dysregulated in HCC with multiple lesions.
| Differentially expressed proteins | Gene | Fold change | Fold change |
|---|---|---|---|
| MC/MN | SC/SN | ||
| UTP-glucose-1-phosphate uridylyltransferase | UGP2 | 0.62 | 0.68 |
| Bile acyl-CoA synthetase | SLC27A5 | 0.58 | 0.67 |
| Cytochrome P450 2A6 | CYP2A6 | 0.6 | 0.73 |
| Glycine dehydrogenase (decarboxylating), mitochondrial | GLDC | 0.65 | 0.72 |
| 17-Beta-hydroxysteroid dehydrogenase 13 | HSD17B13 | 0.56 | 0.71 |
| Glycogen [starch] synthase, liver | GYS2 | 0.65 | 0.7 |
| Sequestome-1 | SQSTM1 | 1.77 | 1.59 |
| 4-Hydroxyphenylpyruvate dioxygenase | HPD | 0.63 | 0.76 |
| Kynurenine 3-monooxygenase | KMO | 0.56 | 0.76 |
| Beta-enolase | ENO3 | 0.64 | 0.71 |
| Urocanate hydratase | UROC1 | 0.66 | 0.76 |
| Keratin, type I cytkeletal 20 | KRT20 | 1.53 | 1.97 |
| Synembryn-A | RIC8A | 1.56 | 1.4 |
| Cadherin-related family member 2 | CDHR2 | 0.64 | 0.78 |
| Cytochrome P450 2B6 | CYP2B6 | 0.64 | 0.69 |
| NAD(P)H dehydrogenase [quinone] 1 | NQO1 | 1.89 | 1.52 |
| Anterior gradient protein 2 homolog | AGR2 | 1.71 | 1.14 |
| Peripherin | PRPH | 0.64 | 0.7 |
| Fuce mutarotase | FUOM | 0.6 | 0.61 |
| Coiled-coil domain-containing protein 57 | CCDC57 | 1.54 | 1.4 |
| Gangliide-induced differentiation-associated protein 1 | GDAP1 | 1.51 | 1.44 |
| Histone H1.1 | HIST1H1A | 1.62 | 1.12 |
| Choline transporter-like protein 2 | SLC44A2 | 0.65 | 0.63 |
| RAS protein activator like-3 | RASAL3 | 0.63 | 0.79 |
| Non-histone chromomal protein HMG-17 | HMGN2 | 3.06 | 1.65 |
| FH1/FH2 domain-containing protein 1 | FHOD1 | 1.66 | 1.44 |
| Copine-6 | CPNE6 | 0.58 | 0.78 |
| Myeloblastin | PRTN3 | 1.51 | 1.94 |
| 24-Hydroxycholesterol 7-alpha-hydroxylase | CYP39A1 | 0.61 | 0.71 |
| Sodium/hydrogen exchanger 10 | SLC9C1 | 0.63 | 0.68 |
| Steroid 17-alpha-hydroxylase/17,20 lyase | CYP17A1 | 1.57 | 2.15 |
| HLA class I histocompatibility antigen, alpha chain G | HLA-G | 0.66 | 1.08 |
| MICAL C-terminal-like protein | MICALCL | 0.49 | 0.4 |
| Nucleolysin TIA-1 isoform p40 | TIA1 | 0.66 | 0.91 |
| Immunoglobulin-binding protein 1 | IGBP1 | 1.57 | 1.63 |
| Protein FAM171A1 | FAM171A1 | 0.59 | 0.63 |
Table 3.
List of the differentially expressed proteins which is only dysregulated in HCC with a single lesion.
| Differentially expressed proteins | Gene | Fold change | Fold change |
|---|---|---|---|
| MC/MN | SC/SN | ||
| Keratin, type II cytkeletal 8 | KRT8 | 0.73 | 0.62 |
| Keratin, type I cytkeletal 18 | KRT18 | 0.72 | 0.6 |
| Tenascin-X | TNXB | 0.72 | 0.65 |
| C-1-tetrahydrofolate synthase, cytoplasmic | MTHFD1 | 0.72 | 0.56 |
| Trifunctional enzyme subunit beta, mitochondrial | HADHB | 0.91 | 0.6 |
| Acetyl-CoA acetyltransferase, mitochondrial | ACAT1 | 0.78 | 0.61 |
| Long-chain-fatty-acid–CoA ligase 1 | ACSL1 | 0.72 | 0.65 |
| Haptoglobin | HP | 0.68 | 0.5 |
| 3-Ketoacyl-CoA thiolase, mitochondrial | ACAA2 | 0.81 | 0.5 |
| Non-specific lipid-transfer protein | SCP2 | 0.91 | 0.65 |
| Lumican | LUM | 0.86 | 0.63 |
| Fatty acid-binding protein, liver | FABP1 | 0.68 | 0.6 |
| d-Beta-hydroxybutyrate dehydrogenase, mitochondrial | BDH1 | 0.69 | 0.65 |
| Betaine–homocysteine S-methyltransferase 1 | BHMT | 0.67 | 0.58 |
| Putative hexokinase HKDC1 | HKDC1 | 1.28 | 1.54 |
| l-Lactate dehydrogenase A chain | LDHA | 1.24 | 1.6 |
| Pyruvate kinase PKM | PKM | 1.25 | 1.59 |
| X-ray repair crs-complementing protein 6 | XRCC6 | 1.36 | 1.63 |
| Enoyl-CoA hydratase, mitochondrial | ECHS1 | 0.86 | 0.55 |
| Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1 | 0.87 | 0.65 |
| ATP synthase subunit d, mitochondrial | ATP5H | 0.78 | 0.56 |
| Laminin subunit beta-1 | LAMB1 | 1.42 | 1.53 |
| S-adenylmethionine synthase isoform type-1 | MAT1A | 0.74 | 0.66 |
| X-ray repair crs-complementing protein 5 | XRCC5 | 1.49 | 1.77 |
| Electron transfer flavoprotein subunit alpha, mitochondrial | ETFA | 0.83 | 0.65 |
| ATP-citrate synthase | ACLY | 1.16 | 1.7 |
| Myeloperoxidase | MPO | 1.37 | 1.58 |
| Glucose-6-phosphate isomerase | GPI | 1.14 | 1.83 |
| Villin-1 | VIL1 | 1.36 | 1.75 |
| Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial | ACADSB | 0.86 | 0.61 |
| Endoplasmic reticulum resident protein 29 | ERP29 | 0.77 | 0.59 |
| Superoxide dismutase [Cu–Zn] | SOD1 | 0.83 | 0.64 |
| C4b-binding protein alpha chain | C4BPA | 1.24 | 1.62 |
| DnaJ homolog subfamily B member 9 | DNAJB9 | 0.81 | 0.45 |
| 3-Ketoacyl-CoA thiolase, peroxisomal | ACAA1 | 0.8 | 0.63 |
| Decorin | DCN | 0.8 | 0.64 |
| Transketolase | TKT | 1.42 | 1.66 |
| Ferritin light chain | FTL | 0.73 | 0.62 |
| Elongation factor 1-gamma | EEF1G | 1.18 | 1.55 |
| Cytochrome b-c1 complex subunit 7 | UQCRB | 0.79 | 0.53 |
| Transferrin receptor protein 1 | TFRC | 1.49 | 1.89 |
| Glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | GPD1 | 0.77 | 0.64 |
| Peroxiredoxin-4 | PRDX4 | 0.7 | 0.64 |
| Mimecan | OGN | 0.77 | 0.62 |
| Cytochrome c oxidase subunit 6B1 | COX6B1 | 0.79 | 0.58 |
| NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | NDUFV2 | 0.82 | 0.55 |
| Phenylalanine-4-hydroxylase | PAH | 0.68 | 0.66 |
| 6-Phosphogluconate dehydrogenase, decarboxylating | PGD | 1.22 | 1.73 |
| ATP synthase subunit O, mitochondrial | ATP5O | 0.93 | 0.66 |
| Cytochrome b-c1 complex subunit Rieske, mitochondrial | UQCRFS1 | 0.85 | 0.65 |
| Cytochrome c oxidase subunit 5B, mitochondrial | COX5B | 0.79 | 0.55 |
| Dehydrogenase/reductase SDR family member 4 | DHRS4 | 0.94 | 0.6 |
| Gamma-glutamyltransferase 5 | GGT5 | 0.67 | 0.6 |
| Sulfotransferase 1A1 | SULT1A1 | 0.81 | 0.58 |
| Carboxypeptidase D | CPD | 1.44 | 1.61 |
| Spliceome RNA helicase DDX39B | DDX39B | 1.18 | 1.53 |
| Core histone macro-H2A.1 | H2AFY | 1.48 | 1.51 |
| Polymerase I and transcript release factor | PTRF | 0.83 | 0.62 |
| Apolipoprotein D | APOD | 0.84 | 0.66 |
| ATP synthase-coupling factor 6, mitochondrial | ATP5J | 0.78 | 0.56 |
| Glucose-6-phosphate 1-dehydrogenase | G6PD | 1.43 | 1.61 |
| 2-Oxoisovalerate dehydrogenase subunit alpha, mitochondrial | BCKDHA | 0.83 | 0.63 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | NDUFB10 | 0.83 | 0.58 |
| Glycine N-acyltransferase | GLYAT | 0.73 | 0.6 |
| Cytochrome c oxidase subunit 5A, mitochondrial | COX5A | 0.82 | 0.59 |
| DNA replication licensing factor MCM3 | MCM3 | 1.5 | 1.62 |
| Ribonuclease UK114 | HRSP12 | 0.82 | 0.65 |
| Phenazine biosynthesis-like domain-containing protein | PBLD | 0.72 | 0.58 |
| Asparagine–tRNA ligase, cytoplasmic | NARS | 1.43 | 1.67 |
| Lamin-B receptor | LBR | 1.36 | 1.68 |
| Polypeptide N-acetylgalactaminyltransferase 2 | GALNT2 | 1.22 | 1.58 |
| Paralemmin-3 | PALM3 | 0.71 | 0.63 |
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | 1.37 | 1.51 |
| Heme-binding protein 1 | HEBP1 | 0.83 | 0.54 |
| Apolipoprotein C-III | APOC3 | 0.92 | 0.66 |
| Phosphoglucomutase-2 | PGM2 | 1.16 | 1.85 |
| Complement factor H-related protein 5 | CFHR5 | 0.89 | 0.63 |
| l-Lactate dehydrogenase B chain | LDHB | 1.2 | 1.62 |
| Serine–tRNA ligase, cytoplasmic | SARS | 1.09 | 1.55 |
| ATP synthase subunit e, mitochondrial | ATP5I | 0.79 | 0.56 |
| Creatine kinase B-type | CKB | 0.95 | 1.92 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | NDUFA5 | 0.74 | 0.53 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 8, mitochondrial | NDUFS8 | 0.89 | 0.66 |
| Desmin | DES | 0.71 | 0.51 |
| DNA replication licensing factor MCM6 | MCM6 | 1.34 | 1.54 |
| Serum deprivation-response protein | SDPR | 0.79 | 0.55 |
| Acyl-coenzyme A synthetase ACSM3, mitochondrial | ACSM3 | 0.75 | 0.6 |
| Clathrin light chain B | CLTB | 0.88 | 0.65 |
| Probable d-lactate dehydrogenase, mitochondrial | LDHD | 0.75 | 0.63 |
| Beta-2-microglobulin | B2M | 0.87 | 0.63 |
| Four and a half LIM domains protein 1 | FHL1 | 0.86 | 0.66 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 | NDUFA2 | 0.85 | 0.58 |
| Perilipin-2 | PLIN2 | 1.23 | 1.65 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | NDUFA8 | 0.72 | 0.47 |
| Tubulointerstitial nephritis antigen-like | TINAGL1 | 0.81 | 0.61 |
| Farnesyl pyrophosphate synthase | FDPS | 1.37 | 1.6 |
| Minor histocompatibility antigen H13 | HM13 | 1.3 | 1.68 |
| Glutathione peroxidase 1 | GPX1 | 0.81 | 0.64 |
| DNA-(apurinic or apyrimidinic site) lyase | AX1 | 1.34 | 1.67 |
| Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 | PLOD3 | 1.15 | 1.58 |
| Angiotensinogen | AGT | 1.14 | 1.55 |
| Transmembrane protein 2 | TMEM2 | 1.1 | 1.51 |
| Alpha/beta hydrolase domain-containing protein 14B | ABHD14B | 0.86 | 0.66 |
| EF-hand domain-containing protein D1 | EFHD1 | 0.81 | 0.65 |
| Protein mago nashi homolog 2 | MAGOHB | 1.3 | 1.73 |
| 3-Hydroxyanthranilate 3,4-dioxygenase | HAAO | 0.79 | 0.66 |
| Cofilin-2 | CFL2 | 0.78 | 0.64 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 6, mitochondrial | NDUFS6 | 0.82 | 0.47 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | NDUFA12 | 0.8 | 0.62 |
| Alpha-fetoprotein | AFP | 1.02 | 2.13 |
| Proliferating cell nuclear antigen | PCNA | 1.43 | 1.62 |
| Transmembrane 9 superfamily member 4 | TM9SF4 | 1.35 | 1.54 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 5 | NDUFS5 | 0.82 | 0.56 |
| Nuclear cap-binding protein subunit 1 | NCBP1 | 1.34 | 1.55 |
| Dermatopontin | DPT | 0.74 | 0.62 |
| Glycine N-methyltransferase | GNMT | 0.8 | 0.65 |
| Ataxin-10 | ATXN10 | 1.38 | 1.57 |
| UPF0553 protein C9orf64 | C9orf64 | 1.41 | 1.64 |
| BRO1 domain-containing protein BROX | BROX | 1.38 | 1.51 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 4, mitochondrial | NDUFS4 | 0.78 | 0.5 |
| l-Serine dehydratase/l-threonine deaminase | SDS | 1.01 | 0.65 |
| Protein transport protein Sec23B | SEC23B | 1.39 | 1.65 |
| Mitochondrial import inner membrane translocase subunit Tim8 A | TIMM8A | 0.82 | 0.56 |
| Nicastrin | NCSTN | 1.29 | 1.53 |
| Cytochrome P450 3A7 | CYP3A7 | 1.38 | 1.94 |
| 40S ribomal protein S15 | RPS15 | 1.08 | 0.55 |
| Integrin alpha-IIb | ITGA2B | 1.1 | 0.65 |
| Acyl-CoA:lysophosphatidylglycerol acyltransferase 1 | LPGAT1 | 1.14 | 1.51 |
| Apolipoprotein L1 | APOL1 | 1.36 | 1.61 |
| Peptidyl-prolyl cis–trans isomerase FKBP2 | FKBP2 | 0.9 | 0.66 |
| Complement factor H-related protein 1 | CFHR1 | 0.7 | 0.63 |
| Plasma serine protease inhibitor | SERPINA5 | 1.08 | 1.52 |
| Mitochondrial import inner membrane translocase subunit Tim13 | TIMM13 | 0.81 | 0.48 |
| Tropomodulin-1 | TMOD1 | 0.83 | 0.64 |
| Myin regulatory light polypeptide 9 | MYL9 | 0.82 | 0.62 |
| ER lumen protein retaining receptor 1 | KDELR1 | 1.1 | 1.58 |
| NAD-dependent malic enzyme, mitochondrial | ME2 | 1.33 | 1.6 |
| Ceramide synthase 2 | CERS2 | 1.47 | 1.51 |
| Monocarboxylate transporter 4 | SLC16A3 | 1.44 | 1.66 |
| Glutaredoxin-1 | GLRX | 0.82 | 0.66 |
| Collagen alpha-6(VI) chain | COL6A6 | 0.75 | 0.63 |
| Group XIIB secretory phospholipase A2-like protein | PLA2G12B | 0.79 | 0.59 |
| Latent-transforming growth factor beta-binding protein 2 | LTBP2 | 1.33 | 1.53 |
| Myin-7 | MYH7 | 1.14 | 2.46 |
| 15 kDa selenoprotein | 15-Sep | 0.69 | 0.58 |
| Metalloproteinase inhibitor 1 | TIMP1 | 1.33 | 1.52 |
| Protein RCC2 | RCC2 | 1.37 | 1.65 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 | NDUFA7 | 0.86 | 0.57 |
| Calmegin | CLGN | 1.41 | 1.75 |
| Apolipoprotein(a) | LPA | 0.74 | 0.64 |
| Elongation factor 1-alpha 2 | EEF1A2 | 1.13 | 1.74 |
| Cytochrome c oxidase protein 20 homolog | COX20 | 1.38 | 1.63 |
| Translin | TSN | 1.2 | 1.57 |
| Folate receptor beta | FOLR2 | 0.72 | 0.65 |
| Secretory carrier-associated membrane protein 3 | SCAMP3 | 1.38 | 1.53 |
| Chitinase-3-like protein 1 | CHI3L1 | 1.11 | 1.86 |
| Mitochondrial intermembrane space import and assembly protein 40 | CHCHD4 | 0.9 | 0.57 |
| Fibrocystin-L | PKHD1L1 | 0.71 | 0.55 |
| Girdin | CCDC88A | 0.82 | 0.38 |
| Flap endonuclease 1 | FEN1 | 1.3 | 1.74 |
| Solute carrier family 43 member 3 | SLC43A3 | 1.29 | 1.65 |
| Complement factor H-related protein 3 | CFHR3 | 0.84 | 0.59 |
| Cleavage stimulation factor subunit 3 | CSTF3 | 1.39 | 1.88 |
| Protein kinase C delta-binding protein | PRKCDBP | 0.76 | 0.54 |
| Transmembrane protein 176B | TMEM176B | 1.32 | 1.62 |
| 60 kDa lysophospholipase | ASPG | 0.7 | 0.62 |
| Spermidine synthase | SRM | 1.2 | 1.51 |
| B-cell receptor-associated protein 29 | BCAP29 | 1.11 | 1.56 |
| Retinol dehydrogenase 10 | RDH10 | 1.36 | 1.58 |
| PDZ and LIM domain protein 2 | PDLIM2 | 0.8 | 0.6 |
| Sodium/potassium-transporting ATPase subunit beta-3 | ATP1B3 | 1.46 | 1.56 |
| Uncharacterized protein C19orf52 | C19orf52 | 1.36 | 1.53 |
| Transmembrane protein 70, mitochondrial | TMEM70 | 1.24 | 1.51 |
| Insulin-like growth factor 2 mRNA-binding protein 2 | IGF2BP2 | 1.41 | 1.77 |
| C-reactive protein | CRP | 1.2 | 1.83 |
| Importin subunit alpha-1 | KPNA2 | 1.33 | 2.77 |
| COX assembly mitochondrial protein 2 homolog | CMC2 | 0.82 | 0.61 |
| Retinoic acid receptor responder protein 2 | RARRES2 | 1.38 | 1.54 |
| Oncoprotein-induced transcript 3 protein | OIT3 | 0.71 | 0.59 |
| Ficolin-1 | FCN1 | 0.9 | 0.65 |
| StAR-related lipid transfer protein 5 | STARD5 | 0.76 | 0.56 |
| Transmembrane protein 14C | TMEM14C | 1.33 | 1.64 |
| P2X purinoceptor 4 | P2RX4 | 1.39 | 1.58 |
| Bifunctional lysine-specific demethylase and histidyl-hydroxylase MINA | MINA | 1.28 | 1.63 |
| Myeloid leukemia factor 2 | MLF2 | 0.81 | 0.58 |
| C4b-binding protein beta chain | C4BPB | 1.16 | 1.65 |
| Astrocytic phosphoprotein PEA-15 | A15 | 1.07 | 1.53 |
| Pituitary tumor-transforming gene 1 protein-interacting protein | PTTG1IP | 1.36 | 1.68 |
| Unconventional myin-XIX | MYO19 | 1.4 | 1.55 |
| Ras-related protein Rab-3D | RAB3D | 1.49 | 1.61 |
| F-box only protein 22 | FBXO22 | 1.15 | 1.61 |
| UPF0364 protein C6orf211 | C6orf211 | 1.33 | 1.59 |
| PRA1 family protein 2 | PRAF2 | 1.28 | 1.53 |
| Serine incorporator 1 | SERINC1 | 1.41 | 1.53 |
| Spermatogenesis-defective protein 39 homolog | VIPAS39 | 1.33 | 1.53 |
| Ryanodine receptor 1 | RYR1 | 0.91 | 1.52 |
| AP-1 complex subunit gamma-like 2 | AP1G2 | 1.4 | 1.65 |
| Hexokinase-2 | HK2 | 1.37 | 1.97 |
| Uncharacterized protein C2orf42 | C2orf42 | 1.19 | 1.53 |
| Phospholipid transfer protein | PLTP | 1.05 | 2.05 |
| PC4 and SFRS1-interacting protein | PSIP1 | 1.3 | 1.53 |
| Rho guanine nucleotide exchange factor 18 | ARHGEF18 | 1.48 | 1.56 |
| Acylphosphatase-2 | ACYP2 | 0.82 | 0.6 |
| Claudin-1 | CLDN1 | 1.05 | 1.68 |
| Neutral amino acid transporter B(0) | SLC1A5 | 1.2 | 2 |
| CB1 cannabinoid receptor-interacting protein 1 | CNRIP1 | 0.91 | 0.66 |
| Cytolic Fe–S cluster assembly factor NUBP2 | NUBP2 | 0.84 | 0.6 |
| Sortilin | SORT1 | 1.12 | 1.51 |
| UPF0729 protein C18orf32 | C18orf32 | 1.15 | 1.77 |
| Protein YIPF4 | YIPF4 | 1.33 | 1.6 |
| Cell growth regulator with EF hand domain protein 1 | CGREF1 | 1.11 | 1.54 |
| Presenilins-associated rhomboid-like protein, mitochondrial | PARL | 1.3 | 1.59 |
| Bactericidal permeability-increasing protein | BPI | 1.23 | 1.66 |
| Protein S100-A1 | S100A1 | 1.27 | 1.63 |
| Ammonium transporter Rh type A | RHAG | 2.34 | 1.83 |
| Pleckstrin homology domain-containing family G member 3 | PLEKHG3 | 1.37 | 1.52 |
| Putative methyltransferase NSUN5 | NSUN5 | 0.71 | 0.59 |
| Secretory carrier-associated membrane protein 4 | SCAMP4 | 1.32 | 1.61 |
| Cochlin | COCH | 1.31 | 2.12 |
| tRNA (guanine(10)-N2)-methyltransferase homolog | TRMT11 | 1.33 | 2 |
| Protein YIPF3 | YIPF3 | 1.27 | 1.54 |
| Synaptogyrin-1 | SYNGR1 | 1.34 | 1.73 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | 1.26 | 1.57 |
| MAP kinase-activated protein kinase 2 | MAPKAPK2 | 1.41 | 1.52 |
| Proteoglycan 3 | PRG3 | 0.69 | 0.65 |
| Bcl-2 homologous antagonist/killer | BAK1 | 1.3 | 1.7 |
| TNF receptor-associated factor 6 | TRAF6 | 1.03 | 0.66 |
| Ethanolamine-phosphate phospho-lyase | ETNPPL | 0.66 | 0.46 |
| Proto-oncogene tyrine-protein kinase Src | SRC | 1.29 | 1.62 |
| Folate transporter 1 | SLC19A1 | 1.3 | 1.81 |
| Platelet factor 4 | PF4 | 0.76 | 0.54 |
| Chloride intracellular channel protein 5 | CLIC5 | 1 | 1.52 |
| Negative elongation factor E | NELFE | 1.32 | 1.64 |
| RNA polymerase II-associated protein 1 | RPAP1 | 1.26 | 1.65 |
| Zinc transporter SLC39A7 | SLC39A7 | 1.67 | 2 |
| Ankyrin repeat domain-containing protein 24 | ANKRD24 | 0.8 | 1.65 |
| Centromal protein of 85 kDa-like | CEP85L | 0.26 | 0.65 |
| Caspase-3 | CASP3 | 1.22 | 1.68 |
| Peroxisomal leader peptide-processing protease | TYSND1 | 1.3 | 1.6 |
| tRNA (guanine(37)-N1)-methyltransferase | TRMT5 | 1.26 | 1.86 |
| Mitochondrial inner membrane organizing system protein 1 | MINOS1 | 1.31 | 1.52 |
| Coiled-coil domain-containing protein 153 | CCDC153 | 1.36 | 2.47 |
| Conserved oligomeric Golgi complex subunit 8 | COG8 | 1.23 | 1.56 |
| Vesicle transport protein SFT2B | SFT2D2 | 1.13 | 1.51 |
| F-box only protein 10 | FBXO10 | 0.71 | 0.63 |
| Muskelin | MKLN1 | 1.09 | 1.57 |
| Tuftelin-interacting protein 11 | TFIP11 | 1.27 | 1.52 |
| Sulfotransferase 1C2 | SULT1C2 | 1.15 | 1.62 |
| Zinc transporter ZIP1 | SLC39A1 | 1.45 | 1.91 |
| Proton-coupled folate transporter | SLC46A1 | 1.19 | 1.52 |
| Putative heat shock protein HSP 90-beta 2 | HSP90AB2P | 1.29 | 2.25 |
| Asparagine synthetase [glutamine-hydrolyzing] | ASNS | 1.21 | 2.7 |
| Ubiquitin carboxyl-terminal hydrolase 38 | USP38 | 0.64 | 0.52 |
| Glycogen synthase kinase-3 alpha | GSK3A | 1.34 | 1.71 |
| Coiled-coil domain-containing protein 69 | CCDC69 | 0.8 | 0.66 |
| Retinoid-binding protein 7 | RBP7 | 0.95 | 1.51 |
| Signal-transducing adaptor protein 2 | STAP2 | 1.21 | 1.59 |
| Soluble calcium-activated nucleotidase 1 | CANT1 | 1.5 | 2 |
| Threonine synthase-like 2 | THNSL2 | 0.65 | 0.48 |
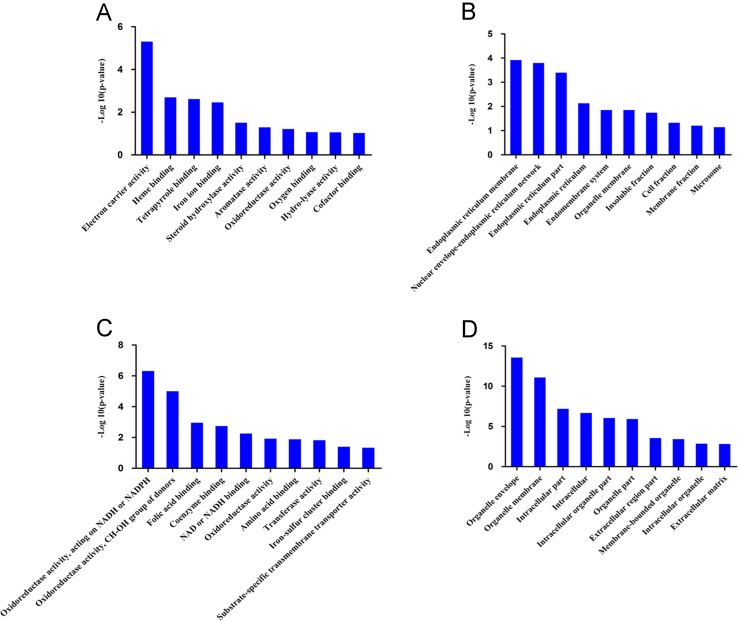
Fig. 4.
Gene ontology (GO) function analysis of differentially expressed proteins which is only dysregulated in HCC with a single lesion or HCC with multiple lesions. (A) GO analysis of the molecular function of the proteins which is only differentially expressed in HCC with multiple lesions. (B) GO analysis of the molecular function of the proteins which is only differentially expressed in HCC with a single lesion. (C) GO analysis of the cell component of the proteins which is only differentially expressed in HCC with multiple lesions. (D) GO analysis of the cell component of the proteins which is only differentially expressed in HCC with a single lesion. All of biological processes were ranked in term of the enrichment of the differentially expressed proteins, and the top 10 are presented.
The biological functions and signaling pathway annotations of the differentially expressed proteins were analyzed by Ingenuity Pathways Analysis (IPA) software (version 7.5), which is based on the Ingenuity Pathways database. The key functions of the differentially expressed proteins involved in the HCC with single and multiple lesions according to IPA analysis are also displayed in Fig. 5. The GO annotations, involved signaling pathways and networks were ranked in term of the enrichment of the differentially expressed proteins.
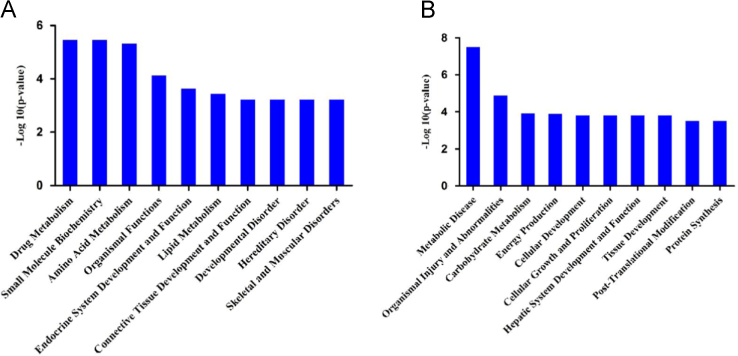
Fig. 5.
The key functions of the differentially expressed proteins involved in the HCC with single and multiple lesions according to IPA analysis. (A) Enriched Functions of the differentially expressed proteins that is only dysregulated in HCC with multiple lesions. (B) Enriched Functions of the differentially expressed proteins that is only dysregulated in HCC with a single lesion. All of pathways were ranked in term of the enrichment of the differentially expressed proteins, and the top 10 were presented.
Acknowledgments
This work is supported by the key clinical specialty discipline construction program of Fujian, P.R. China; the key project of National Science and Technology of China (Grant nos. 2012ZX10002010-001-006 and 2012ZX10002016-013); the National Natural Science Foundation of China (Grant nos. 31201008 and 31400634); the specialized Science and Technology Key Project of Fujian Province (Grant no. 2013YZ0002-3); the Science and Technology Infrastructure Construction Program of Fujian Province (Grant no. 2014Y2005); the Scientific Foundation of Fuzhou Health Department (Grant nos. 2014-S-w24, 2013-S-w12, 2013-S-125-4, and 2013-S-wp1); the Young Teacher Education Project of Fujian Province (Grant no. 2013 JB13125).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.08.036.
Conflict of interest associated with this article can be found in the online version at doi:10.1016/j.dib.2015.08.036.
Appendix A. Supplementary material
Supplementary material
Appendix B. Conflict of interest
Conflict of interest
References
- 1.Xiaohua Xing, Yao Huang, Sen Wang, Minhui Chi, Yongyi Zeng, Lihong Chen, Jinhua Zeng, Minjie Lin, Xiaolong Liu, Jingfeng Liu. Comparative analysis of the primary multiple and single hepatocellular carcinoma by iTRAQ based quantitative proteomics. J. Proteomics 128 (2015) 262–271. 〈http://dx.doi.org/10.1016/j.jprot.2015.08.007〉 (in press). [DOI] [PubMed]
- 2.Gilar M., Olivova P., Daly A.E., Gebler J.C. Two-dimensional separation of peptides using RP–RP–HPLC system with different pH in first and second separation dimensions. J. Sep. Sci. 2005;28:1694–1703. doi: 10.1002/jssc.200500116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Conflict of interest