Abstract
A thermo-tolerant NADP(H)-preferring xylose pathway was constructed in Kluyveromyces marxianus for ethanol production with xylose at elevated temperatures (Zhang et al., 2015 [25]). Ethanol production yield and efficiency was enhanced by pathway engineering in the engineered strains. The constructed strain, YZJ088, has the ability to co-ferment glucose and xylose for ethanol and xylitol production, which is a critical step toward enabling economic biofuel production from lignocellulosic biomass. This study contains the fermentation results of strains using the metabolic pathway engineering procedure. The ethanol-producing abilities of various yeast strains under various conditions were compared, and strain YZJ088 showed the highest production and fastest productivity at elevated temperatures. The YZJ088 xylose fermentation results indicate that it fermented well with xylose at either low or high inoculum size. When fermented with an initial cell concentration of OD600=15 at 37 °C, YZJ088 consumed 200 g/L xylose and produced 60.07 g/L ethanol; when the initial cell concentration was OD600=1 at 37 °C, YZJ088 consumed 98.96 g/L xylose and produced 33.55 g/L ethanol with a productivity of 0.47 g/L/h. When fermented with 100 g/L xylose at 42 °C, YZJ088 produced 30.99 g/L ethanol with a productivity of 0.65 g/L/h, which was higher than that produced at 37 °C.
Keywords: Kluyveromyces marxianus, Xylose, Ethanol, Co-assimilation, Elevated temperature
Specifications table
| Subject area | Biology |
| More specific subject area | Xylose metabolism |
| Type of data | Table; figure |
| How data was acquired | The metabolic products were acquired by HPLC using an Agilent 1100 series HPLC system. XR and XDH activity were determined using a spectrophotometer to monitor the change in A340 upon oxidation of NAD(P)H. |
| Data format | Raw and analyzed |
| Experimental factors | No pretreatment |
| Experimental features | Batch fermentation; HPLC; enzyme activity |
| Data source location | Not applicable |
| Data accessibility | The data are supplied with this article. |
The value of the data
-
●
Comparison of the fermentation results of the different engineered strains during pathway engineering revealed the specific role of genes related to xylose metabolism under oxygen-limited conditions.
-
●
Compared with other reported yeast strains, K. marxianus YZJ088 showed considerable ethanol production and the highest ethanol productivity.
-
●
Strain YZJ088 fermented xylose well with an initial OD=1 or 15 at 37 °C, which indicates this strain produced more ethanol with relative lower productivity [25].
-
●
K. marxianus YZJ088 fermented xylose well with an initial OD=1 at 42 °C, and the co-fermentation of glucose and xylose indicates that it has great potential for application in simultaneous saccharification and fermentation at elevated temperatures.
-
●
Though it produced relative less ethanol, the productivity of YZJ088 at 42 °C was faster.
1. Data, experimental design, materials and methods
1.1. Comparison of the xylose fermentation ability of constructed strains
To compare the effects of over-expression or disruption of downstream genes, K. marxianus strains YZJ020, YZJ051, YZJ061, YZJ069, YZJ071, YZJ077, YZJ084, YZJ086, YZJ088, YZJ089, and YZJ091 (Table 2 in Ref. [25]), which were constructed during pathway engineering, were fermented with YP medium that contained 100 g/L xylose at 42 °C with 250 rpm and initial OD600=15 under oxygen-limited conditions [25]. The over-expression of genes involved in xylose metabolic promoted ethanol production in the engineered strains (Table 1). KmFPS1 disruption reduced xylitol accumulation and utilization but blocked the production of glycerol (Table 1).
Table2.
Comparison of the xylose consumption and the ethanol production among the various yeast strainsa.
| Strains | Temperature (°C) | Xylose (g/L) | Initial OD | Xylose consumption (g/L) | Xylitol production (g/L) | Xylitol yield (g/g) | Ethanol production (g/L) | Ethanol yield (g/g) | Ethanol productivity (g/L/h) | Time of fermentation (h) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| K. marxianus SUB-80- S | 35 | 20 | 1/20 volume | 20 | NR | NR | 5.6 | 0.28 | 0.12 | 48 | [12] |
| K. marxianus IMB4 | 40 | 20 | 0.22 g/L | 13.61 | 7.36 | 0.54 | 2.08 | 0.15 | 0.022 | 96 | [19] |
| K. marxianus DMKU3-1042 | 40 | 20 | OD600=1 | 20 | ~6.5 | ~0.33 | 2.2±0.2 | 0.11±0.01 | 0.046±0.001 | 48 | [15] |
| Kluyveromyces sp. IIPE453 | 50 | 20 | OD600=1 | ~17.5 | 11.5±0.4 | 0.66±0.02 | 1.75±0.05 | 0.10±0.01 | 0.025±0.001 | 80 | [9] |
| K. marxianus YZB014 | 42 | 20 | OD600=10 | 19.00±1.00 | 11.32±0.36 | 0.60±0.02 | 3.55±0.19 | 0.19±0.01 | 0.110±0.006 | 32 | [21] |
| K. marxianus YRL002 | 42 | 50 | OD600=10 | 30.15 | – | – | 11.52 | 0.38 | 0.069 | 168 | [17] |
| H. polymorpha CBS4732 | 48 | 120 | OD600~5 | ~16 | 0.02 | 0.00125 | 1.31 | 0.08 | 0.054 | 24 | [2] |
| H. polymorpha 2EthOH− /XYL1m/XYL2/XYL3/BrPA | 45 | 92 | 2 g/L | 32.67 | 0 | 0 | 9.8 | 0.3 | 0.18 | 55 | [10] |
| S. cerevisiae SXA-R2P-E | 30 | 40 | OD600=20 | 36.67 | – | – | 16.5 | 0.45 | 0.28 | 60 | [11] |
| S. cerevisiae PUA6-9 | 30 | 20 | 1/10 volume | 19.65 | 9.88 | 0.50 | 3.08 | 0.16 | 0.04 | 76 | [8] |
| S. cerevisiae TMB 3057 | 30 | 50 | OD600=10 | 39.6±3.4 | 8.71±1.19 | 0.22±0.03 | 13.30±1.70 | 0.33±0.02 | 0.133±0.017 | 100 | [4] |
| S. cerevisiae F106KR | 30 | 165 | OD600=10 | 161.2 | 20.6 | 0.13 | 58.5 | 0.36 | 1.22 | 48 | [20] |
| S. cerevisiae F106KR | 30 | 221.1 | OD600=10 | 212.0 | 21.7 | 0.10 | 77.6 | 0.37 | 1.08 | 72 | [20] |
| S. cerevisiae DA24-16 | 30 | 80 | 4 g/L | 79.7 | 3.2 | 0.04 | 27.9 | 0.35 | 0.47 | 60 | [3] |
| S. cerevisiae CIBTS0735 | 30 | 40 | OD600=10 | 39.7 | – | – | 17.47 | 0.44 | 1.09 | 16 | [1] |
| S. cerevisiae DGX23 | 30 | 40 | OD600=1.3 | 32.28 | 2.00 | 0.06 | 9.36 | 0.29 | 0.13 | 72 | [7] |
| S. cerevisiae Y-ARSdRb | 30 | 15c | NR | 13.6 | 4.00 | 0.29 | 7.02 | 0.46 | 0.10 | 72 | [18] |
| S. cerevisiae MA-N5b | 30 | 45 | OD600=15 | 40.56 | 2.64 | 0.07 | 14.6 | 0.36 | 0.20 | 72 | [13] |
| S. cerevisiae D-XR/XDH/XKb | 30 | 15c | OD600=10 | 12.75 | 2.74 | 0.21 | 8.00 | 0.43 | 0.11 | 72 | [14] |
| S. cerevisiae SK-N2b | 30 | 55d | NR | ~55 | ~3.8 | 0.07 | 30.1 | 0.41 | 0.18 | 168 | [6] |
| S. cerevisiae SK-NNb | 30 | 20 | NR | 15 | ~4.4 | 0.29 | 4.02 | 0.27 | 0.03 | 144 | [5] |
| S. passalidarum NN245 | 25 | 150 | OD600=15 | 150 | – | – | 53.3 | 0.36 | 0.44 | 120 | [16] |
| K. marxianus YZJ088 | 42 | 128.46±3.91 | OD600=15 | 118.39±2.91 | 11.09±1.47 | 0.09±0.01 | 44.95±3.21 | 0.38±0.02 | 2.49±0.18 | 18 | This study |
NR: not reported.
If the literature described several strains, only the best one is shown.
Strains with NADPH-NADP+ xylose metabolic pathway.
Fermentation with 5 g/L glucose as co-substrate.
Fermentation with 20 g/L glucose as co-substrate.
Table 1.
Summary of the fermentation by engineered strains with YP medium containing 100 g/L xylose at 42 °C.
| Strains | Time (h) | Residual xylose (g/L) | Xylulose (g/L) | Xylitol (g/L) | Glycerol (g/L) | Acetate (g/L) | Ethanol (g/L) | Ethanol productivity (g/L/h) |
| YZJ020 | 18 | 21.14±1.25 | 2.8±0.69 | 10.78±1.02 | 5.56±1.54 | 1.06±0.52 | 25.48±0.57 | 1.42±0.24 |
| YZJ051 | 18 | 16.49±0.96 | 3.72±0.58 | 9.21±2.30 | 5.53±1.44 | 1.22±0.34 | 29.73±1.24 | 1.65±0.52 |
| YZJ061 | 18 | 12.2±1.56 | 3.9±1.34 | 10.29±2.11 | 6.84±1.63 | 1.34±0.40 | 31.99±2.31 | 1.78±0.30 |
| YZJ077 | 18 | 10.4±1.50 | 3.97±0.32 | 9.46±2.13 | 6.48±1.52 | 1.24±0.33 | 31.38±1.47 | 1.74±0.41 |
| YZJ084 | 18 | 11.6±2.41 | 1.88±0.64 | 4.80±1.02 | 6.70±2.01 | 0.98±0.29 | 33.90±1.38 | 1.88±0.74 |
| YZJ086 | 18 | 6.57±1.63 | 9.13±1.61 | 12.25±2.52 | 0.13±0.03 | 0.48±0.31 | 33.78±1.29 | 1.88±0.50 |
| YZJ088 | 18 | 3.9±0.96 | 9.00±1.85 | 11.86±3.44 | 0.15±0.04 | 0.70±0.41 | 35.94±1.24 | 2.00±0.34 |
| YZJ089 | 18 | 3.82±1.32 | 9.27±2.41 | 11.94±2.12 | 0.91±0.32 | 0.67±0.28 | 34.36±0.98 | 1.91±0.69 |
| YZJ091 | 18 | 5.39±1.21 | 5.11±2.31 | 8.32±2.84 | 0.19±0.08 | 0.62±0.19 | 33.21±2.07 | 1.85±0.34 |
1.2. Comparison of ethanol producing abilities from xylose with various previously reported yeast strains
The ethanol-producing ability with xylose at 42 °C of K. marxianus YZJ088 was compared with other ethanol fermentation yeast strains. K. marxianus YZJ088 exhibited considerable ethanol production and the highest ethanol productivity at elevated temperatures (Table 2).
1.3. K. marxianus YZJ088 fermented well with a high concentration xylose at 37 °C
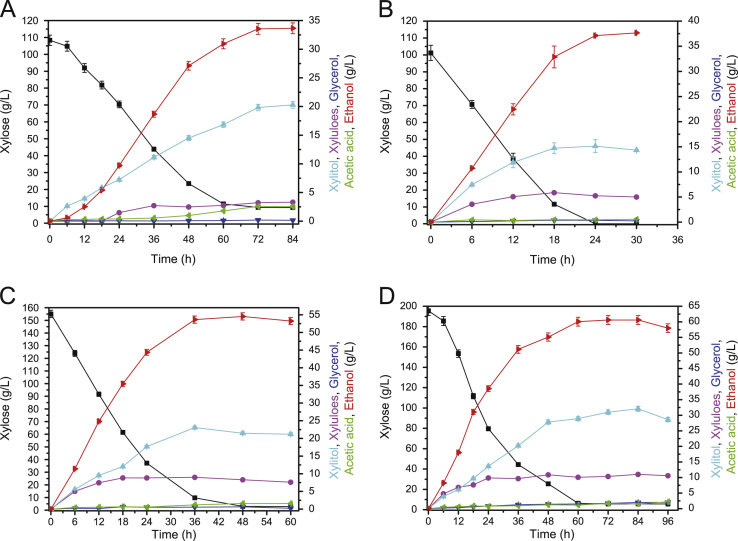
The fermentation ability of K. marxianus YZJ088 at 37 °C was explored. K. marxianus YZJ088 fermented 100 g/L xylose and produced 33.55 g/L ethanol in 72 h with an initial OD600=1. When increased to an initial OD600=15, YZJ088 could ferment 100, 150, and 200 g/L xylose and produced 37.13, 53.62, and 60.07 g/L ethanol with productivities of 1.55, 1.49, and 1.00 g/L/h, respectively (Fig. 1). Although YZJ088 used more xylose and produced more ethanol at 37 °C, faster productivity (2.49 g/l/h) was achieved at 42 °C (Table 1 and Fig. 1) [25].
Fig. 1.
Fermentation of YZJ088 in YP medium at 37 °C with 100 g/L xylose and initial OD600=1 (A), 100 g/L xylose and initial OD600=15 (B), 150 g/L xylose and initial OD600=15 (C), 200 g/L xylose and initial OD600=15 (D). The values are the means of three biological replicates±standard deviation (n=3).
1.4. K. marxianus YZJ088 fermented xylose at 42 °C with low inoculum size
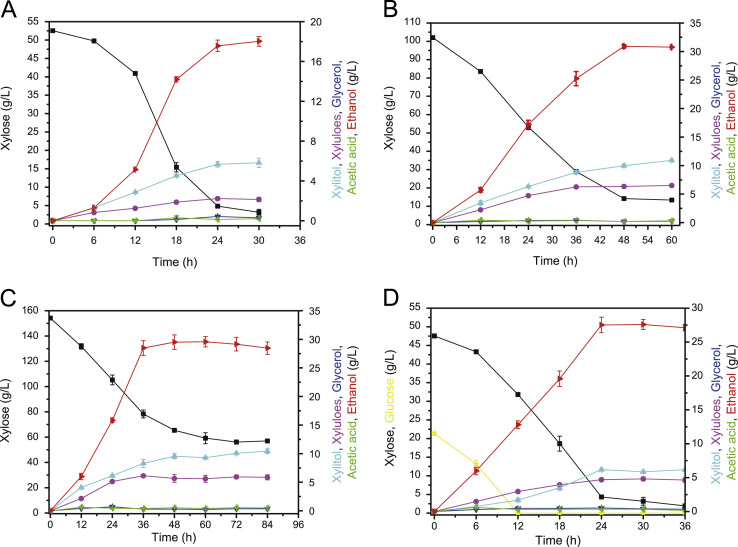
K. marxianus YZJ088 fermented 50, 100, 150 g/L xylose, and a 20 g/L glucose-50 g/L xylose mixture with an initial cell concentration of OD600=1 at 42 °C under oxygen-limited conditions and produced 18.03, 30.99, 28.48, and 27.52 g/L ethanol, respectively (Fig. 2). Although most xylose fermentation was conducted at high inoculum size, YZJ088 produced ethanol fairly well at 42 °C with low inoculum size. However, when xylose concentration reached 150 g/L, the ethanol production was limited. These results may have occurred because xylose tolerance decreased at higher temperatures [23,24].
Fig. 2.
Fermentation of YZJ088 in YP medium with 50 g/L xylose (A), 100 g/L xylose (B), 150 g/L xylose (C) and 50 g/L xylose+20 g/L glucose (D) at 42 °C with initial OD600=1. The values are the means of three biological replicates±standard deviation (n=3).
1.5. XR and XDH activities of K. marxianus strains growth at 37 °C were higher than those at 42 °C
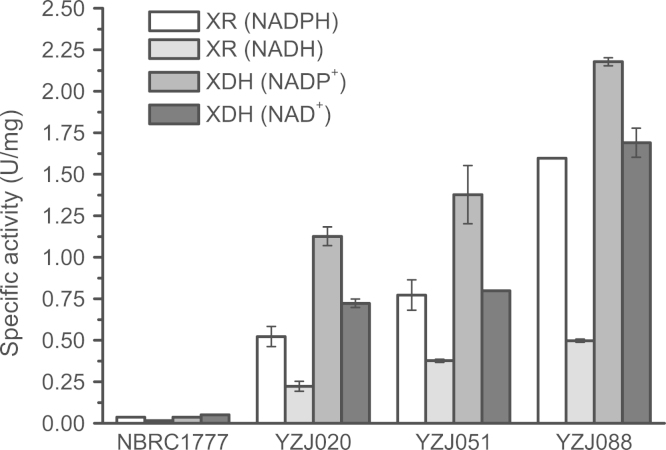
XR and XDH activities were determined for NBRC1777, YZJ020, YZJ051, and YZJ088 cells cultured with YP medium contained 20 g/L xylose at 37 °C. The cells were harvested by centrifugation at 10,000×g for 10 min at room temperature and washed with 100 mM potassium phosphate buffer (pH 7.4). The cells were resuspended in the same buffer and then lysed by sonication (Vibra-Cell VC505, Connecticut, USA) for 20 min at 40% power in an ice–water bath. The cell debris was removed by centrifugation at 10,000×g for 10 min, and the supernatant was used to measure enzyme activity. The assay mixture (1.0 mL) for the XR enzyme reaction contained 100 mM of phosphate buffer (pH 7.4), 200 μM NAD(P)H, 200 mM xylose, and crude enzyme solution (0.1 mL). The assay mixture (1.0 mL) for the XDH enzyme reaction contained 50 mM MgCl2, 50 mM Tris–HCl buffer (pH 9.0), 20 mM NAD(P)+, 300 mM xylitol, and crude enzyme solution (0.1 mL). The reaction was started by adding 0.1 mL of crude enzyme. One unit of enzyme activity is defined as the amount of enzyme required to oxidize/reduce 1 μmol of NAD(P)H/NAD(P)+ per min under the specified conditions [22].
XR and XDH activities in these strains growth at 37 °C were higher than those at 42 °C (Fig. 3) [25]. The XR (NADPH) and XDH (NADP+) activities of YZJ088 cultured at 37 °C were 3.69- and 3.91-fold higher, respectively, than those at 42 °C. Although the enzymatic activities at 37 °C were higher than those at 42 °C, they did not yield higher productivity. More xylitol accumulation at 37 °C with high xylose concentration could reflect lower efficiency of the downstream enzymes at 37 °C [25].
Fig. 3.
Comparison of the XR and XDH activities of NBRC1777, YZJ020, YZJ051 and YZJ088 cultured in YP medium with 20 g/L xylose at 37 °C for 24 h. The values are the means of three biological replicates±standard deviation (n=3).
Acknowledgments
This work was supported by a Grant-in-Aid from the National High Technology Research and Development Program (2012AA02A708), the National Natural Science Foundation of China (31070028 and 31270149), and the National Basic Research Program of China (2011CBA00801). This work also received technical support from the Core Facility Center for Life Sciences, University of Science and Technology of China. The authors do not have any possible conflicts of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.08.038.
Appendix A. Supplementary material
Supplementary material
References
- 1.Diao L., Liu Y., Qian F., Yang J., Jiang Y., Yang S. Construction of fast xylose-fermenting yeast based on industrial ethanol-producing diploid Saccharomyces cerevisiae by rational design and adaptive evolution. BMC Biotechnol. 2013;13:110. doi: 10.1186/1472-6750-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dmytruk O.V., Dmytruk K.V., Abbas C.A., Voronovsky A.Y., Sibirny A.A. Engineering of xylose reductase and overexpression of xylitol dehydrogenase and xylulokinase improves xylose alcoholic fermentation in the thermotolerant yeast Hansenula polymorpha. Microb. Cell Fact. 2008;7:21. doi: 10.1186/1475-2859-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha S.J., Galazka J.M., Kim S.R., Choi J.H., Yang X., Seo J.H., Glass N.L., Cate J.H., Jin Y.S. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation , Proc. Natl. Acad. Sci. USA. 2011;2011;108108:504–509. 504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karhumaa K., Sanchez R.G., Hahn-Hagerdal B., Gorwa-Grauslund M.F. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2007;6:5. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khattab S., Kodaki T. Efficient bioethanol production by overexpression of endogenous Saccharomyces cerevisiae xylulokinase and NADPH-dependent aldose reductase with mutated strictly NADP+-dependent Pichia stipitis xylitol dehydrogenase Sadat Mohammad Rezq Khattab. Process Biochem. 2014;49:1838–1842. [Google Scholar]
- 6.Khattab S.M.R., Saimura M., Kodaki T. Boost in bioethanol production using recombinant Saccharomyces cerevisiae with mutated strictly NADPH-dependent xylose reductase and NADP(+)-dependent xylitol dehydrogenase. J. Biotechnol. 2013;165:153–156. doi: 10.1016/j.jbiotec.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.R., Kwee N.R., Kim H., Jin Y.S. Feasibility of xylose fermentation by engineered Saccharomyces cerevisiae overexpressing endogenous aldose reductase (GRE3), xylitol dehydrogenase (XYL2), and xylulokinase (XYL3) from Scheffersomyces stipitis. FEMS Yeast Res. 2013;13:312–321. doi: 10.1111/1567-1364.12036. [DOI] [PubMed] [Google Scholar]
- 8.Kotter P., Ciriacy M. Xylose Fermentation by Saccharomyces Cerevisiae. Appl. Microbiol. Biotechnol. 1993;38:776–783. [Google Scholar]
- 9.Kumar S., Singh S.P., Mishra I.M., Adhikari D.K. Ethanol and xylitol production from glucose and xylose at high temperature by Kluyveromyces sp IIPE453. J. Ind. Microbiol. Biotechnol. 2009;36:1483–1489. doi: 10.1007/s10295-009-0636-6. [DOI] [PubMed] [Google Scholar]
- 10.Kurylenko O.O., Ruchala J., Hryniv O.B., Abbas C.A., Dmytruk K.V., Sibirny A.A. Metabolic engineering and classical selection of the methylotrophic thermotolerant yeast Hansenula polymorpha for improvement of high-temperature xylose alcoholic fermentation. Microb. Cell Fact. 2014;13:122. doi: 10.1186/s12934-014-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.M., Jellison T., Alper H.S. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol. Biofuels. 2014;7:122. doi: 10.1186/s13068-014-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margaritis A., Bajpai P. Direct fermentation of d-Xylose to ethanol by Kluyveromyces marxianus strains. Appl. Environ. Microbiol. 1982;44:1039–1041. doi: 10.1128/aem.44.5.1039-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsushika A., Watanabe S., Kodaki T., Makino K., Inoue H., Murakami K., Takimura O., Sawayama S. Expression of protein engineered NADP+-dependent xylitol dehydrogenase increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2008;81:243–255. doi: 10.1007/s00253-008-1649-1. [DOI] [PubMed] [Google Scholar]
- 14.Matsushika A., Watanabe S., Kodaki T., Makino K., Sawayama S. Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP(+)-dependent xylitol dehydrogenase, and xylulokinase. J. Biosci. Bioeng. 2008;105:296–299. doi: 10.1263/jbb.105.296. [DOI] [PubMed] [Google Scholar]
- 15.Rodrussamee N., Lertwattanasakul N., Hirata K., Suprayogi S., Limtong T., Kosaka, Yamada M. Growth and ethanol fermentation ability on hexose and pentose sugars and glucose effect under various conditions in thermotolerant yeast Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 2011;90:1573–1586. doi: 10.1007/s00253-011-3218-2. [DOI] [PubMed] [Google Scholar]
- 16.Su Y.K., Willis L.B., Jeffries T.W. Effects of aeration on growth, ethanol and polyol accumulation by Spathaspora passalidarum NRRL Y-27907 and Scheffersomyces stipitis NRRL Y-7124. Biotechnol. Bioeng. 2015;112:457–469. doi: 10.1002/bit.25445. [DOI] [PubMed] [Google Scholar]
- 17.Wang R., Li L., Zhang B., Gao X., Wang D., Hong J. Improved xylose fermentation of Kluyveromyces marxianus at elevated temperature through construction of a xylose isomerase pathway. J. Ind. Microbiol. Biotechnol. 2013;40:841–854. doi: 10.1007/s10295-013-1282-6. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe S., Abu Saleh A., Pack S.P., Annaluru N., Kodaki T., Makino K. Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP(+)-dependent xylitol dehydrogenase. J. Biotechnol. 2007;130:316–319. doi: 10.1016/j.jbiotec.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Wilkins M.R., Mueller M., Eichling S., Banat I.M. Fermentation of xylose by the thermotolerant yeast strains Kluyveromyces marxianus IMB2, IMB4, and IMB5 under anaerobic conditions. Process Biochem. 2008;43:346–350. [Google Scholar]
- 20.Xiong M., Chen G., Barford J. Alteration of xylose reductase coenzyme preference to improve ethanol production by Saccharomyces cerevisiae from high xylose concentrations. Bioresour. Technol. 2011;102:9206–9215. doi: 10.1016/j.biortech.2011.06.058. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B., Li L.L., Zhang J., Gao X.L., Wang D.M., Hong J. Improving ethanol and xylitol fermentation at elevated temperature through substitution of xylose reductase in Kluyveromyces marxianus. J. Ind. Microbiol. Biotechnol. 2013;40:305–316. doi: 10.1007/s10295-013-1230-5. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B., Zhang L., Wang D., Gao X., Hong J. Identification of a xylose reductase gene in the xylose metabolic pathway of Kluyveromyces marxianus NBRC1777. J. Ind. Microbiol. Biotechnol. 2011;38:2001–2010. doi: 10.1007/s10295-011-0990-z. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Zhang B., Wang D.M., Gao X.L., Hong J. Xylitol production at high temperature by engineered Kluyveromyces marxianus. Bioresour. Technol. 2014;152:192–201. doi: 10.1016/j.biortech.2013.10.109. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., Zhang B., Wang D.M., Gao X.L., Hong J. Improving xylitol production at elevated temperature with engineered Kluyveromyces marxianus through over-expressing transporters. Bioresour. Technol. 2015;175:642–645. doi: 10.1016/j.biortech.2014.10.150. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Zhang B., Wang D.M., Gao X.L., Sun L.H., Hong J. Rapid ethanol production at elevated temperatures by engineered thermotolerant Kluyveromyces marxianus via the NADP(H)-preferring xylose reductase–xylitol dehydrogenase pathway. Metab. Eng. 2015;31:140–152. doi: 10.1016/j.ymben.2015.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material