Abstract
Schizophrenia (SCZ) is a common complex disorder with poorly understood mechanisms and no effective drug treatments. Despite the high prevalence and vast unmet medical need represented by the disease, many drug companies have moved away from the development of drugs for SCZ. Therefore, alternative strategies are needed for the discovery of truly innovative drug treatments for SCZ. Here, we present a disease phenome-driven computational drug repositioning approach for SCZ. We developed a novel drug repositioning system, PhenoPredict, by inferring drug treatments for SCZ from diseases that are phenotypically related to SCZ. The key to PhenoPredict is the availability of a comprehensive drug treatment knowledge base that we recently constructed. PhenoPredict retrieved all 18 FDA-approved SCZ drugs and ranked them highly (recall = 1.0, and average ranking of 8.49%). When compared to PREDICT, one of the most comprehensive drug repositioning systems currently available, in novel predictions, PhenoPredict represented clear improvements over PREDICT in Precision-Recall (PR) curves, with a significant 98.8% improvement in the area under curve (AUC) of the PR curves. In addition, we discovered many drug candidates with mechanisms of action fundamentally different from traditional antipsychotics, some of which had published literature evidence indicating their treatment benefits in SCZ patients. In summary, although the fundamental pathophysiological mechanisms of SCZ remain unknown, integrated systems approaches to studying phenotypic connections among diseases may facilitate the discovery of innovative SCZ drugs.
Keywords: drug repositioning, drug discovery, systems biology, disease phenotype, schizophrenia
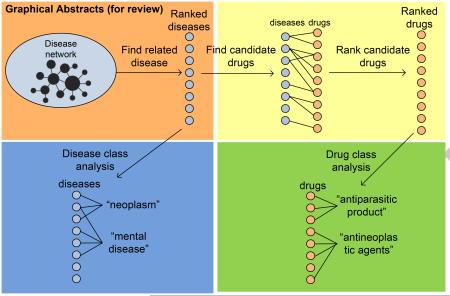
Graphical Abstract
1. Introduction
Mental illness causes enormous personal and societal burdens [1]. In fact, mental illnesses, such as schizophrenia (SCZ), bipolar disease, depression and other psychiatric disorders, is the leading cause of impairment and disability in the United States and world-wide, accounting for around one-third of the disabilities in the world [2, 3]. SCZ is arguably the most intractable among all psychiatric disorders [4]. SCZ has a life-time prevalence of 1%, typically beginning before age 25 years and persisting throughout the life of the individual [3]. Currently, effective drugs do not exist for treating SCZ [5]. Despite the vast unmet medical need, many drug companies have moved away from SCZ drug development, in part because of the high costs, high failure rates in clinical trials, lengthy development processes, and a poor understanding of underlying mechanisms of the disease [5, 6, 7].
In this study, we present a computational drug repositioning approach towards discovering innovative drug candidates for the treatment of SCZ. Psychiatric drug discovery has traditionally come from repositioning existing drugs based on serendipitous clinical observations [8]. For example, lithium, originally approved as a sedating agent, is now used in the treatment of mania [9]. Chlorpromazine, originally approved as an antihistamine, is now used in the treatment of schizophrenia [10]. Iproniazid, originally approved as an anti-tuberculosis agent, is now used in the treatment of depression [11]. Ketamine, originally approved as an anesthetic agent, has rapid antidepressant effects in patients with major depression [12]. Computation-based repositioning approaches that automatically reason over vast amounts of genetic, genomic, chemical, and phenotypic data can greatly speed up the timeline of traditional serendipity-based psychiatric drug discovery process and facilitate the identification of truly innovative drug treatments for SCZ and other psychiatric disorders [13, 14, 15]. However, computational drug repositioning approaches for identifying novel drug candidates for SCZ has not been fully explored.
Computational drug repositioning approaches can be classified as either drug-based or disease-based [14, 15]. Drug-based approaches leverage on the known molecular structures or functions of drugs, such as chemical structures and properties, molecular docking, gene expression, drug treatment indications, and drug side effects [16, 17, 18, 19, 20, 21, 22, 23, 24]. In the past 50 years, psychiatric drug discovery has been largely drug-based and has focused on identifying molecules with which existing drugs interact. Consequently, all current antidepressants, antipsychotics, and anti-anxiety drugs developed and marketed from the 1950s to the current day have targeted the same molecular pathways in the brain as their prototypes [5]. it has been recognized that drug-based discovery, with its focus on finding drug candidates based on existing drugs, might by definition fail to identify new therapeutic mechanisms [25]. An alternative approach is disease-based discovery, which puts less emphasis on existing drugs and focuses more on disease mechanisms and interrelationships. Because disease-based approaches look for similarities and interrelationships among diseases, these approaches are able to identify innovative drugs. Compared to drug-based repositioning approaches, disease-based approaches are surprisingly less explored and mainly used disease gene expression data [19, 20].
We hypothesize that higher-level phenotypic overlaps among diseases reflect underlying biological commonalities and that insights from one disease may be used to inform our developing knowledge of others. We developed a phenotype-driven drug repositioning system, PhenoPredict, to exploit drug repositioning opportunities rendered by disease phenotype data captured in the Human Phenotype Ontology (HPO) and a comprehensive drug-disease treatment relationship knowledge base (TreatKB) that we recently constructed [26, 27, 28]. HPO is a standardized vocabulary of phenotypic abnormalities encountered in human disease [29]. HPO contains phenotypic descriptions of 7,529 diseases, the majority of which were derived from the Online Mendelian Inheritance in Man (OMIM) [30]. Studies of phenotypic abnormalities in HPO have advanced our understanding of the genetic bases of diseases [31, 32, 33]. In a recent study, Gottlieb et al. used disease phenotypic similarities defined in HPO and drug-drug similarities from other databases to construct a classifier (PREDICT) and then used it to determine treatment associations between 593 drugs and 313 diseases, including SCZ [34]. Different from PREDICT, PhenoPredict used a network-based approach to systematically exploit phenotypic interrelationships among diseases as defined in HPO. More importantly, PhenoPredict used a novel drug prioritization algorithm to exploit treatment connections among diseases as defined in TreatKB, which is a key component of PhenoPredict. Compared to PREDICT, our study included significantly more drugs and diseases (2,482 drugs and 24,511 unique disease concepts). We compared PhenoPredict to PREDICT in novel drug predictions using multiple evaluation datasets and demonstrated that PhenoPredict achieved consistently better performances.
2. Data and Methods
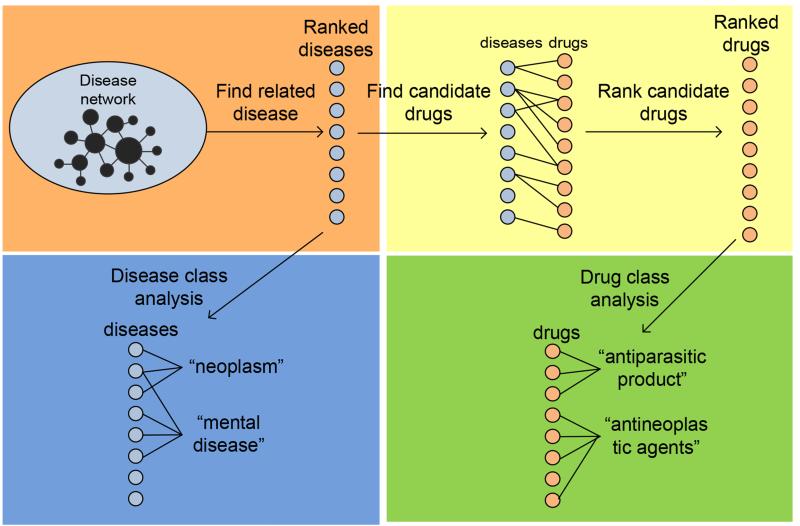
The experiment framework for PhenoPredict is depicted in Fig. 1 and consists of four phases: (1) We constructed a phenotypic disease network (PDN) using disease-disease similarity measures from HPO. We then developed a network-based ranking algorithm to find diseases that are phenotypically related to SCZ; (2) In order to validate the network construction and ranking algorithms of PhenoPredict and to better understand SCZ-related diseases, we analyzed disease class distributions among diseases at different ranking cutoffs and investigated what kinds of diseases were enriched among top-ranked SCZ-related diseases; (3) We developed a novel drug repositioning algorithm to systematically identify drug repositioning candidates from SCZ-related diseases. We evaluated PhenoPredict using FDA-approved SCZ drugs. We compared PhenoPredict to PREDICT in novel predictions; and (4) In order to better understand top-ranked drug candidates, we examined drug class distributions among both top- and intermediate-ranked drug candidates.
Figure 1.
The overall experimental flow chart for PhenoPredict.
2.1. Construct the phenotypic disease network (PDN) and find SCZ-related diseases from PDN
2.1.1. Construct phenotypic disease network (PDN)
PDN was constructed by directly using the disease-disease similarity matrix obtained from HPO. In HPO, individual diseases are often associated with multiple phenotypic terms. Similarity measures for any two given phenotypic terms were calculated based upon shared information content (frequency among annotations of all diseases) in the set of their common-ancestor nodes. The similarity between two diseases was then calculated by matching each phenotypic term of one disease with the most similar term of the other disease; the average was taken over all pairs of phenotypic terms [29]. We downloaded the HPO disease-disease similarity matrix and mapped disease terms to the Unified Medical Language System (UMLS) [35] Concept Unique Identifiers (CUIs) in order to facilitate the subsequent linking to drug-disease treatment pairs in TreatKBs that were constructed from other data sources. A total of 5708 out of 7529 disease terms in HPO were mapped to UMLS CUIs. Instead of excluding unmapped terms, we used the term names as their unique identifiers. In total, we obtained 17,523,509 disease-disease pairs, representing 7210 unique disease concepts. The similarity scores from the matrix were used as the edge weights of PDN. We also generated ten random PDNs by randomly shuffling edges of the real PDN.
2.1.2. Develop network-based ranking algorithm for finding SCZ-related diseases
Recently, we developed network-based approaches to prioritize genes for a given disease [36] and to prioritize diseases for a given microbial metabolite [37]. In this study, we applied these network-based algorithms to prioritize diseases for SCZ. The iterative network-based ranking algorithm is defined as: pt+1 = (1 − r)Mpt + rp0, wherein M is the column-normalized adjacency matrix of PDN, γ is a preset probability of restarting from the initial seed node (γ=0.1 in this study), and pt is a vector in which the ith element holds the normalized ranking score of disease i at tth iteration. The initial probability vector p0 contains normalized probability values for input. In our study, p0 contains SCZ, with a probability of 1.0. Diseases are ranked according to values in the steady-state probability vector, which is obtained by iterating the algorithm until the change between pt+1 and pt is less than 10−6.
2.2. Analyze disease class distribution at different ranking cutoffs
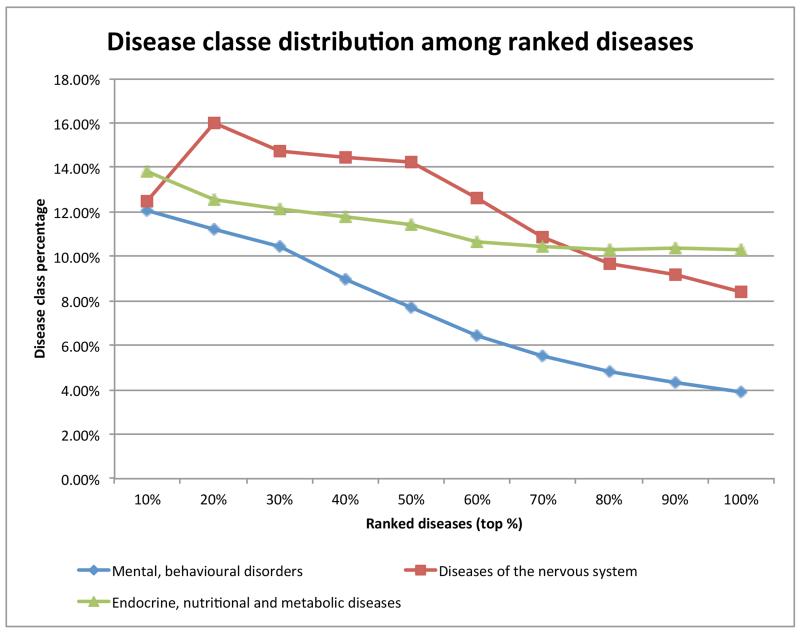
To better understand ranked diseases, we analyzed disease class distribution at ten different ranking cutoffs. Using SCZ as the seed, we retrieved a ranked list of 7204 diseases from PDN. We classified these diseases into sixteen categories using the 10th revision of the International Statistical Classification of Diseases and Related Health Problems (ICD10), a disease classification scheme designated by the World Health Organization (WHO) [38]. The ICD10 includes 22 highest-level disease classes (or chapters) such as “Neoplasms” and “Diseases of the nervous system.” We used sixteen chapters and excluded the other six non-specific disease classes such as “Codes for special purposes” and “Injury, poisoning and certain other consequences of external causes.” Because the terms used in ICD10 may differ from those in PDN, we expanded disease terms in ICD10 to their synonyms through UMLS CUIs. Disease chapters and the numbers of diseases in each chapter are listed in Table 1.
Table 1.
Sixteen disease chapters (classes) and the number of diseases (synonym expanded) in each chapter.
| Disease Class | Diseases (n) |
Disease Classes | Diseases (n) |
|---|---|---|---|
| Certain infectious and parasitic diseases | 11,598 | Diseases of the circulatory system | 5544 |
| Neoplasms | 14,158 | Diseases of the respiratory system | 3156 |
| Diseases of the blood and blood-forming organs and certain disorders involving the immune mechanism |
3264 | Diseases of the digestive system | 5960 |
| Endocrine, nutritional and metabolic diseases | 5438 | Diseases of the skin and subcutaneous tissue | 4390 |
| Mental and behavioural disorders | 6162 | Diseases of the musculoskeletal system and connective tissue |
11520 |
| Diseases of the nervous system | 5258 | Diseases of the genitourinary system | 5247 |
| Diseases of the eye and adnexa | 3735 | Congenital malformations, deformations and chromosomal abnormalities |
9064 |
| Diseases of the ear and mastoid process | 1815 | Certain conditions originating in the perinatal period |
3454 |
At ten ranking cutoffs (10%, 20%, … 100%), we calculated percentages of these sixteen disease classes among retrieved diseases. For example, at the 100% cut-off (all 7204 retrieved diseases), 3.89% of the diseases were classified as “Mental, behavioural disorders.” At the 10% cutoff (top 720 diseases), 87 out of the 720 diseases (12.05%) were classified as “Mental, behavioural disorders,” representing a 209.8% increase as compared to the 100% cutoff ((12.05-3.89)/3.89 = 209.8%). This means that top-ranked diseases on average included more “Mental, behavioural disorders” than lower-ranked diseases. While this was expected and demonstrates the validity of the disease ranking algorithm, we found that certain other disease classes such as “Endocrine, nutritional and metabolic diseases” were enriched among top-ranked diseases.
2.3. Reposition drugs
2.3.1. Drug repositioning algorithm
We developed an approach to systematically identify drug repositioning candidates from SCZ-related diseases. We ranked drugs based on the number of SCZ-related diseases that they are currently approved to treat as well as the ranking scores of these diseases. For example, if drug 1 treats 25 top-ranked diseases, it would be ranked higher than drug 2, which treats only one or two lower-ranked diseases. The drug ranking algorithm is defined as: , wherein n is the number of SCZ-related diseases that are currently approved to treat and R_disease_i is the disease ranking score (output from the network-based disease ranking algorithm). During the experiment, we found that certain drugs were consistently ranked highly for both the real PDN and random PDNs. For example, the drug “chlordiazepoxide” was ranked at top 0.32% for the real PDN and on average at top 0.36% for andom PDNs. We designed our reprioritization strategy by accounting for rankings of a drug for random PDNs. A drug was ranked highly if and only if it was ranked highly based on the real PDN and the ratio of its ranking for the real PDN to that for random PDNs is at least 2 fold.
2.3.2. Comparison of four TreatKBs in a de-novo validation setting using 18 known SCZ drugs as evaluation dataset
In order to systematically reposition drug treatments from one disease to another, it is critical to have a comprehensive drug treatment knowledge base. In our recent studies, we constructed four large-scale drug-disease treatment knowledge bases (TreatKBs) from multiple heterogeneous and complementary data sources using advanced computational techniques including natural language processing, text mining, and data mining [26, 27, 28]. The databases included 9,216 drug-disease treatment pairs extracted from FDA drug labels, 111,862 pairs extracted from the FDA Adverse Event Reporting System (FAERS), a database supporting the FDA’s post-marketing drug safety surveillance efforts, 34,306 pairs extracted from 22 million published biomedical literature abstracts, and 69,724 pairs extracted from 171,805 clinical trials. The combined TreatKB consists of 208,330 unique drug-disease treatment pairs, representing 2484 drugs and 24,511 unique disease concepts.
We evaluated PhenoPredict using all 18 FDA-approved SCZ drugs by comparing its performance across four TreatKBs. Since SCZ and its associated drug treatment pairs were removed from the inputs to the repositioning algorithm (SCZ-related diseases and drug-disease treatment pairs), the evaluation is in fact a de-novo validation. We calculated the rankings of the 18 FDA-approved SCZ drugs among all retrieved drugs and used them as our gold standard. We assumed that the higher these gold standard drugs were ranked, the better the ranking algorithm was. We compared the performances (recall and average rankings) across four TreatKBs separately and in combination.
2.3.3. Compare PhenoPredict to PREDICT in novel predictions
We compared PhenoPredict to PREDICT in novel predictions using three evaluation datasets: (1) 195 drugs that had been tested in SCZ clinical trials; (2) 50 drugs that were in ongoing SCZ clinical trials initiated in 2012 and after. These drugs may represent newer SCZ drugs; and (3) 114 drugs that the literature implies have been used to treat varying symptoms of SCZ. These three evaluation datasets were derived from TreatKBs, which was constructed from multiple data resources including 22 million published biomedical literature abstracts and 171,805 clinical trials [26, 27, 28]. The 18 FDA-approved drugs were removed from these three evaluation datasets. Note that all SCZ-related drug treatment information were removed from TreatKB before PhenoPredict made predictions for SCZ.
We used Precision-Recall (PR) curves instead of Receiver Operator Characteristic (ROC) curves to evaluate and compare PhenoPredict to PREDICT. PR curves are often used to evaluate ranked classification results in information retrieval [39]. ROC curves are commonly used to evaluate binary classification problems in machine learning and data mining [40]. A PR space is defined as precision (fraction of examples classified as positive that are truly positive) and recall (true positive rate) as x and y axes, respectively. An ROC space is defined by FPR (false positive rate) and TPR (the same as recall) as x and y axes, respectively. Studies have shown that in domains where the number of negatives greatly exceeds the number of positives, such as in drug repositioning and most other biomedical classification domains, ROC curves can present an overly optimistic view of an algorithm’s performance as compared to PR curves [41, 42]. Davis et al. proved that a curve dominates in ROC space if and only if it dominates in PR space and algorithms that optimize the ROC curve are not guaranteed to optimize the PR curve [42]. Therefore, in our study, we used PR curves even though most biomedical classification studies use ROC curves.
PREDICT utilizes multiple drug-drug and disease-disease similarity measures for the prediction task [34]. PREDICT first trains a logistic regression classifier using known drug-disease associations. It then classifies additional drug-disease associations based on their similarity to the known associations. We compared PhenoPredict to PREDICT in novel predictions. A total of 593 drugs were included in PREDICT, among which 79 drugs were classified as positives for SCZ. The 79 drugs along with their corresponding probabilities (ranging from 0.543–0.994) are publicly available [34]. The remaining 524 drugs were predicted by PREDICT as negatives for treating SCZ. We assigned each negative prediction a value that was randomly picked from 0.0 to 0.499. We repeated this process of assigning values to negatives for ten iterations and generated ten datasets for PREDICT. PR curves for these ten datasets were similar, therefore we did not generate more datasets for PREDICT. The PR curves for PREDICT were then averaged across the ten datasets that we generated. The output from PhenoPredict is a ranked list of 2484 drugs. Using each of the three evaluation datasets as gold standard, we calculated precisions at 10 different recall cutoffs (0.1, 0.2, …; 1.0) for both PhenoPredict and PREDICT and plotted the PR curves. The area under curves (AUC) was used to compare the two approaches.
2.4. Analyze repositioned drug candidates
It is important to the current study, as well as to future work in the fields of computational drug repositioning, to better understand the nature of identified drug repositioning candidates. In order to facilitate such an understanding, we examined the class distributions of drug repositioning candidates. Drug classes were defined by the Anatomical Therapeutic Chemical (ATC) classification system [43]. The ATC system consists of 13 first-level codes, 94 second-level codes, 267 third-level codes, 882 fourth-level codes, and 4580 fifth-level codes. The fifth-level codes are individual drugs. In our study, we used the third level ATC codes for the analysis. We examined top ranked drug classes for drug candidates ranked in the range of top 0-15% and in the range of top16-30% separately.
3. Results
3.1. Disease class analysis
Using SCZ as the seed, we retrieved a ranked list of 7204 diseases from PDN. We calculated percentages of sixteen disease classes among these retrieved diseases at ten different ranking cutoffs (10%, 20%, … 100%). Among the sixteen disease classes, three disease classes were enriched among top-ranked diseases: “Mental, behavioural disorders,” “Diseases of the nervous system,” and “Endocrine, nutritional and metabolic diseases” (Fig. 2). The increase for the disease class “Mental, behavioural disorders” was particularly pronounced, with a 209.8% increase for the top 10% diseases as compared to all retrieved diseases. The increases for the other two classes are similar but less prominent. In summary, the enrichment of “Mental, behavioural disorders,” to which SCZ belongs, among top-ranked diseases, demonstrated the validity of our phenotype-driven network-based disease ranking algorithm.
Figure 2.
Percentages of three disease classes among 7204 diseases retrieved from HPON at ten ranking cutoffs (top 10%, 20%, … 100% (all diseases)). For example, ranked diseases at top 10% cuto (top 720 diseases) contain 12.05% diseases from the class “Mental, behavioural disorders”. Thirteen unenriched disease classes are not shown.
3.2. FDA-approved SCZ drugs were ranked highly
When the TreatKB containing only FDA-approved drug-disease treatments was used, PhenoPredict achieved a recall of 0.33 and an average ranking of 30.9%. When the other three TreatKBs were used, PhenoPredict achieved a significantly better performance in terms of both recalls and rankings (Table 2). Significantly, when all four TreatKB were combined, PhenoPredict achieved a recall of 1.00 and an average ranking of 8.49%. These results demonstrate that a comprehensive TreatKB is critical component of PhenoPredict.
Table 2.
Comparing recalls and average rankings of 18 FDA-approved SCZ drugs for four TreatKBs individually and combined.
| TreatKB | Recall | Average ranking |
|---|---|---|
| FDA-approved | 0.33 | 30.9% |
| Post-market | 1.00 | 10.48% |
| ClinicalTrials | 0.67 | 21.65% |
| Literature | 0.83 | 10.97% |
| Combined | 1.00 | 8.49% |
3.3. PhenoPredict performed better than PREDICT in novel predictions
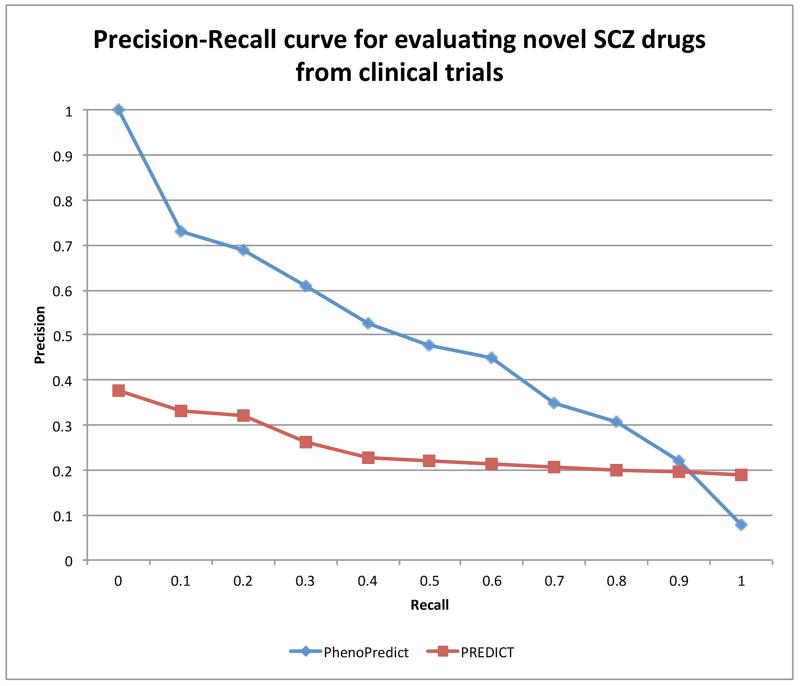
We plotted PR curves for PhenoPredict and PREDICT using the 195 drugs extracted from SCZ clinical trials as the evaluation set. The PR curve for PhenoPredict clearly dominates that for PREDICT. The area under the curve (AUC) for PhenoPredict is 0.489, representing a 98.8% improvement as compared to the AUC of 0.246 for PREDICT (Fig. 3).
Figure 3.
Precision-Recall curves for PhenoPredict and PREDICT using 195 drugs from SCZ clinical trials as gold standard.
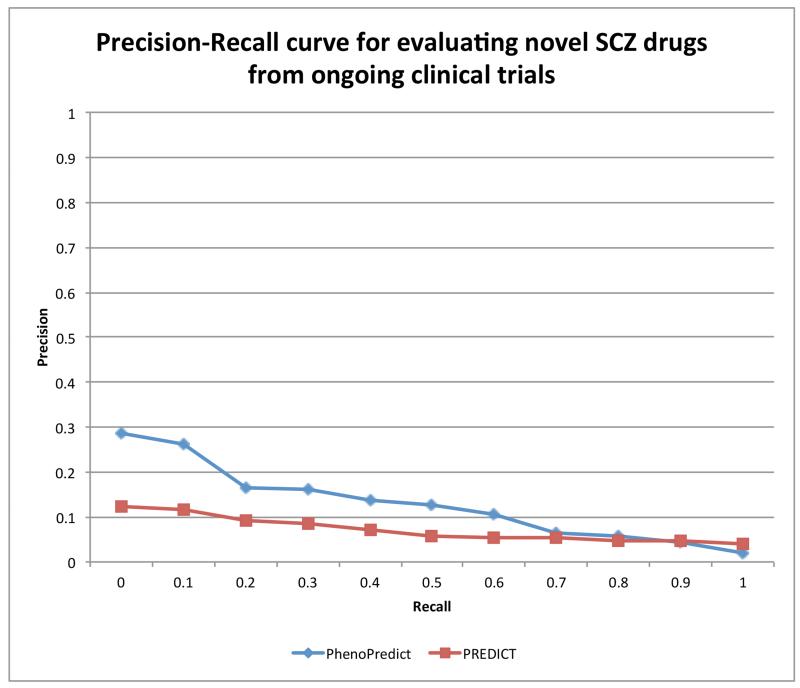
When evaluated with 50 drugs extracted from ongoing SCZ clinical trials, PhenoPredic achieved an AUC of 0.128, representing an 81.1% improvement as compared to the AUC of 0.071 for PREDICT (Fig. 4).
Figure 4.
Precision-Recall curves for PhenoPredict and PREDICT using 50 drugs from ongoing SCZ clinical trials as gold standard.
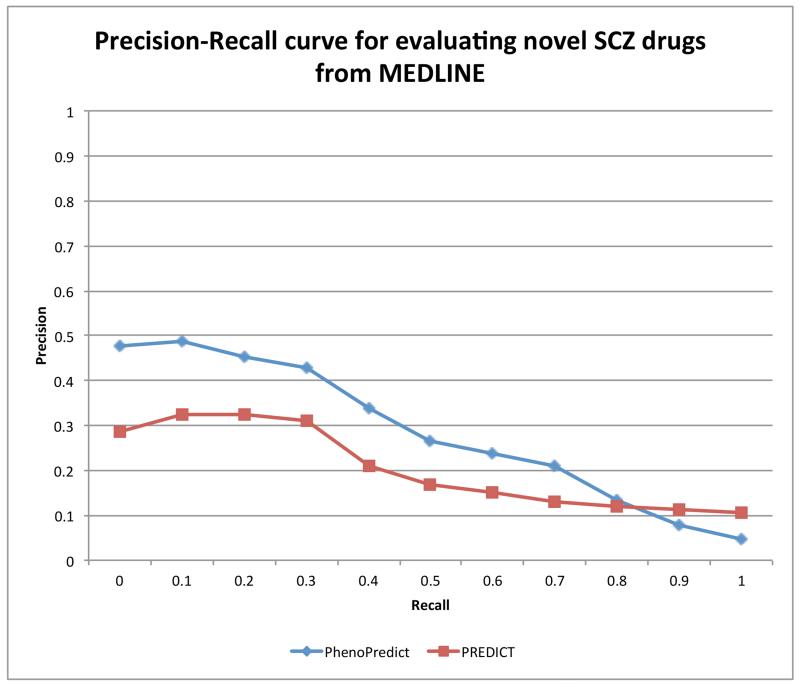
The PR curves determined using the 114 drugs that the literature implies have been used to treat varying symptoms of SCZ as the evaluation set are shown in Fig. 5. PhenoPredict achieved of an AUC of 0.289, representing a 41.2% improvement as compared to the AUC of 0.208 for PREDICT. In summary, PhenoPredict consistently showed improved PR curves compared to those for PREDICT across three different evaluation datasets.
Figure 5.
Precision-Recall curves for PhenoPredict and PREDICT using 114 drugs extracted from biomedical literature (MEDLINE) as gold standard.
Table 3 shows the top 20 repositioned drug candidates, all of which are implicated as promising candidates through evidence from sources other than our experiment, such as FDA drug labels, clinical trials, or biomedical literature. Among these 20 drugs, 8 are FDA-approved drugs. These specific examples further demonstrate the potential of PhenoPredict in identifying promising drug repositioning candidates for SCZ.
Table 3.
Top 20-ranked repositioned drug candidates. NCT**: SCZ drugs from clinical trials. PMID**: SCZ drugs from biomedical literature.
| R | Drug | Evidence | R | Drug | Evidence |
|---|---|---|---|---|---|
| 1 | risperidone | FDA-approved | 11 | memantine |
NCT02001103 NCT00757978 NCT00097942 |
| 2 | methylphenidate | NCT00794040 | 12 | buspirone | NCT00178971 |
| 3 | quetiapine | FDA-approved | 13 | paliperidone | FDA-approved |
| 4 | citalopram |
NCT00893256 NCT00047450 NCT01032083 NCT01032083 |
14 | haloperidol | FDA-approved |
| 5 | olanzapine | FDA-approved | 15 | lithium |
NCT00202306 NCT00183443 NCT00202293 |
| 6 | sertraline |
NCT00169988, NCT00531518 |
16 | amantadine |
NCT00999505 NCT00975611 NCT00401973 |
| 7 | aripiprazole | FDA-approved | 17 | levodopa | NCT01636037 |
| 8 | ziprasidone | FDA-approved | 18 | atomoxetine |
NCT00420498 NCT00222794 NCT00488163 NCT00628394 NCT00161031, NCT00089869 |
| 9 | clozapine | FDA-approved | 19 | clomipramine | PMID9659874 PMID7635998 PMID7903293 |
| 10 | valproic acid |
NCT00194025 NCT01094249 NCT02011750 |
20 | prednisone | PMID17245324 PMID23738211 |
3.4. Analysis of repositioned drug candidates offers insights to common mechanisms of action
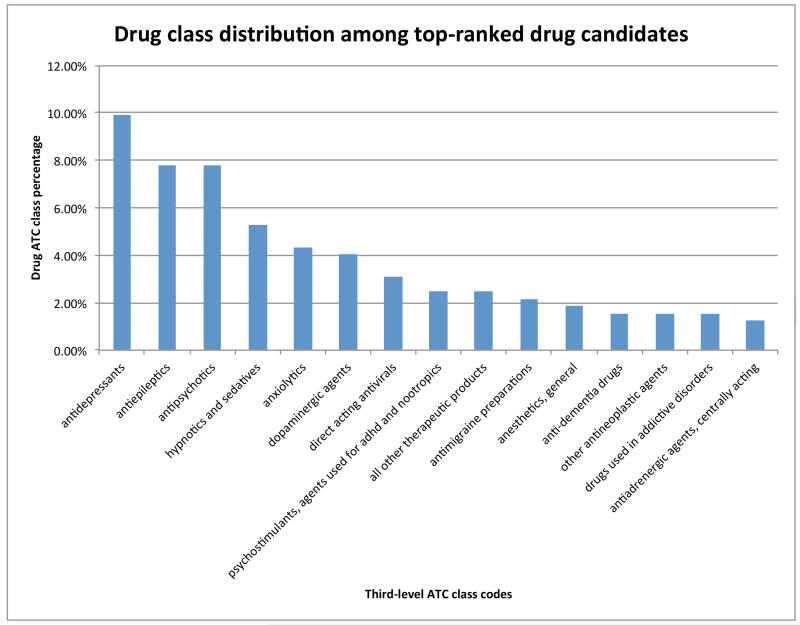
The top drug candidates, those ranked in the 0-15% range, were associated with a total of 95 third-level ATC codes. Figure 6 shows the top 15 drug classes, among which 13 classes are related to antipsychotics, including antidepressants, antiepileptics, and dopaminergic agents. We have shown in the disease class analysis that mental diseases were highly enriched among top-ranked diseases; therefore, it is not surprising that most of the top ranked drug candidates are typical antipsychotics. This result also demonstrates that common pathophysiologic mechanisms are shared among phenotypically related psychiatric disorders and that traditional psychiatric drug discovery may have fully exploited this commonality (i.g. the same drugs are used among related diseases).
Figure 6.
Top 15 third level ATC codes (out of 95 codes) and their percentages for drug candidates ranked in the range of 0-15%. For example, 9.94% of drugs ranked in the range of 0-15% belong to the class “antidepressants”.
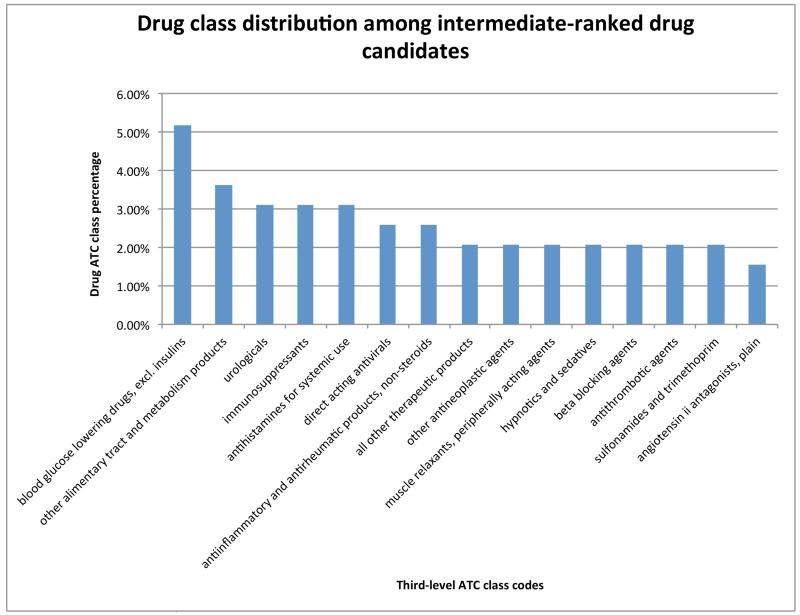
While top ranked drugs are mainly antipsychotics, drugs with intermediate rankings may provide opportunities for discovering innovative drugs. Figure 7 shows the top 15 drug ATC codes for drugs ranked in the range of 16-30%. The majority of these top ATC codes are not related to antipsychotics. Evidence gleaned from the published biomedical literature shows that these drug classes may have treatment potential in SCZ patients. For example, two ATC codes “immunodepressants” and “antiinflammatory and antirheumatic” were ranked highly. Studies have shown that immune dysfunction and inflammation are involved in patients with SCZ [44, 45]. Therefore, anti-inflammatory drugs may represent promising treatments for SCZ. In a randomized controlled study, celecoxib, a widely used anti-inflammatory agent, was shown to improve symptoms experienced by SCZ patients without major side effects [46]. Recent genetic findings from genome-wide association studies (GWAS) also point to possible common genetic connections between SCZ and immune disorders [47]. Beta-blockers were also ranked highly. Beta blockers are commonly used to treat hypertension and cardiovascular diseases. Studies have shown that they may reduce anxiety and extrapyramidal symptoms in SCZ and have been suggested as adjunctive therapies to antipsychotics in SCZ or similar severe mental disorders [48, 49]. The output of PhenoPredict also suggests the potential use of angiotensin antagonists as an atypical SCZ treatment. Angiotensin antagonists are primarily used in the treatment of hypertension, congestive heart failure, and heart attacks. Interestingly, angiotensin has been shown to regulate the central nervous system activity [50, 51]. Neurochemical and anecdotal reports suggest that angiotensin antagonists may have mood-elevating and cognitive enhancing functions in patients, however mechanisms of actions by which these inhibitors modify cognitive performance remain unknown [52].
Figure 7.
Top 15 third level ATC codes (out of 93 codes) and their percentages for repositioned drug candidates ranked in the range of 16-30%. For example, 3.31% of drugs ranked in the range of 16-30% belong to the class “immunosuppressants.”
4. Discussion
We developed a drug repositioning system, PhenoPredict, to exploit the phenotypic connections among diseases and applied it to identify drug repositioning candidates for the treatment of SCZ. PhenoPredict ranked many traditional antipsychotic drugs highly, demonstrating the validity of the algorithms. In addition, we discovered many drug repositioning candidates with mechanisms of action fundamentally different from traditional antipsychotics, each of which has substantial literature-based evidence implicating its potential benefits in the treatment of SCZ patients. However, PR curves for PhenoPredict are not optimal and can certainly be improved upon with future research efforts.
First, it will be interesting to test the generalizability of PhenoPredict for other diseases. Currently, PhenoPredict included drug-disease treatment relationships for a total of 24,511 diseases and 2,484 drugs. In theory, PhenoPredict can rank the 2,484 drugs for each of the 24,511 diseases or vice versa.
Second, it will be interesting to investigate why PhenoPredict outperformed PREDICT. Such knowledge can offer insight into how to further improve both systems. Since the algorithms as well as the datasets included in both PhenoPredict and PREDICT are integral parts of these two systems, it is unclear which (algorithms or datasets or both) contributed to the PhenoPredict’s advantage over PREDICT in finding drug candidates for schizophrenia. It will be interesting to investigate whether integrating datasets from both PhenoPredict and PREDICT can further improve the performances for each system.
Third, a limitation of using disease phenotypes in HPO for drug repositioning is that HPO mainly includes rare Mendelian disorders, the majority of which themselves have no available drug treatments. Therefore, the success of PhenoPredict in identifying drug repositioning candidates from similar diseases to a given input disease largely depends on the input disease as well as the treatment availability for top-ranked diseases.
Fourth, disease genetics and genomics, in combination with disease phenotypes, may further facilitate the discovery of truly innovative drug candidates for SCZ. Psychiatric disorders are among the most heritable of all common complex diseases. Human genomics and genetics studies have recently identified a large number of genetic risk factors for psychiatric disorders [47]. Although nearly all of the identified SCZ loci are nonspecific and not fully penetrant, recent GWAS studies have demonstrated shared genetic loci among phenotypically related psychiatric disorders including SCZ and bipolar disorder. While this justifies our approach of using disease phenotype data for drug repositioning, disease genetics and genomics may provide additional information not captured by disease phenotypes. However, the task of how to combining different level of evidence, including genetics, genomics, and phenomics, in order to build compassing predictive models for drug repositioning is challenging. We are actively exploring options for how to best accomplish this task.
Last but not least, incorporating other types of disease-phenotype relationships such as disease comorbidities and disease risk factors may o er additional drug repositioning opportunities for SCZ. Recently, we constructed three large-scale disease phenotypic knowledge bases, including a disease comorbidity knowledge base, a disease-risk relationship knowledge base, and a disease-manifestation knowledge base [27, 53, 54]. Unlike HPO, which includes exclusively Mendelian genetic disorders, these disease phenotype knowledge bases contain not only Mendelian disorders but also many common complex diseases. Currently, we are developing approaches to integrate disease phenotype knowledge from these complementary and heterogeneous data resources in an e ort to further improve PhenoPredict.
Highlights (for review).
Schizophrenia (SCZ) is among the most enigmatic disorders without curative drugs.
Systems approaches to study phenotypic interconnections among diseases can lead novel drug discovery.
We developed a phenome-wide systems drug repositioning approach for SCZ.
Our approach is effective in finding FDA-approved SCZ drugs as well as in novel predictions.
Our algorithm performed significantly better than one of most comprehensive drug repositioning system.
Acknowledgement
Xu and Wang have jointly conceived the idea, designed and implemented the algorithms. All the authors have participated in study discussion and manuscript preparation.
Funding
RX was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under the NIH Director’s New Innovator Award number DP2HD084068 and the Training grant in Computational Genomic Epidemiology of Cancer (CoGEC) (R25 CA094186-06). QW was partially supported by ThinTek LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Collins PY, Patel V, Joestl SS, March D, Insel TR, Daar AS, Walport M. Grand challenges in global mental health. Nature. 2011;475(7354):27–30. doi: 10.1038/475027a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kessler RC, Chiu WT, Demler O, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;762(6):617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mathers C, Fat DM, Boerma JT. The global burden of disease: 2004 update. World Health Organization; 2008. [Google Scholar]
- [4].Sullivan PF. Puzzling over schizophrenia: schizophrenia as a pathway disease. Nature medicine. 2012;18(2):210–211. doi: 10.1038/nm.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hyman SE. Time for New Schizophrenia Rx. Science. 2014;343(6176):1177–1177. doi: 10.1126/science.1252603. [DOI] [PubMed] [Google Scholar]
- [6].Hyman SE. Revolution stalled. Science translational medicine. 2012;4(155):155cm11–155cm11. doi: 10.1126/scitranslmed.3003142. [DOI] [PubMed] [Google Scholar]
- [7].Miller G. Is pharma running out of brainy ideas. Science. 2010;329(5991):502–504. doi: 10.1126/science.329.5991.502. [DOI] [PubMed] [Google Scholar]
- [8].Hyman SE. Cerebrum: the Dana forum on brain science. Vol. 2013. Dana Foundation; Psychiatric Drug Development: Diagnosing a Crisis. [PMC free article] [PubMed] [Google Scholar]
- [9].Mitchell PB, Hadzi-Pavlovic D. Lithium treatment for bipolar disorder. Bulletin of the World Health Organization. 2000;78(4):515–517. [PMC free article] [PubMed] [Google Scholar]
- [10].Lopez-Munoz F, Alamo C, Cuenca E, Shen WW, Clervoy P, Rubio G. History of the discovery and clinical introduction of chlorpromazine. Annals of Clinical Psychiatry. 2005;17(3):113–135. doi: 10.1080/10401230591002002. [DOI] [PubMed] [Google Scholar]
- [11].Maxwell RA, Eckhardt SB. Drug discovery: a casebook and analysis. Humana Press; Clifton, NJ: p. xvii. [Google Scholar]
- [12].Dolgin E. Rapid antidepressant effects of ketamine ignite drug discovery. Nature medicine. 2013;19(1):8–8. doi: 10.1038/nm0113-8. [DOI] [PubMed] [Google Scholar]
- [13].Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews Drug discovery. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- [14].Dudley JT, Deshpande T, Butte AJ. Exploiting drugdisease relationships for computational drug repositioning. Briefings in bioinformatics. 2011;12(4):303–311. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: From data to therapeutics. Clinical Pharmacology and Therapeutics. 2013;93(4):335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- [16].Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ. Roth BL: Predicting new molecular targets for known drugs. Nature. 2009;462(7270):175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kinnings SL, Liu N, Buchmeier N, Tonge PJ, Xie L, Bourne PE. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS computational biology. 2009;5(7):e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- [19].Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3(96ra):77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Science translational medicine. 2011;3(96):96ra76–96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chiang AP, Butte AJ. Systematic evaluation of drugdisease relationships to identify leads for novel drug uses. Clinical Pharmacology & Therapeutics. 2009;86(5):507–510. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- [23].Duran-Frigola M, Aloy P. Recycling side-effects into clinical markers for drug repositioning. Genome Med. 2012;4(3) doi: 10.1186/gm302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nature Neurosci. 2010;2010(13):1161–169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu R, Wang Q. Large-scale extraction of drug-disease treatment pairs from biomedical literature for drug repurposing. BMC Bioinformatics. 2013;14(1):181. doi: 10.1186/1471-2105-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu R, Li L, Wang Q. Towards building a disease-phenotype relationship knowledge base: large scale extraction of disease-manifestation relationship from literature. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt359. doi: 10.1093/bioinformatics/btt359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu R, Wang Q. Automatic signal prioritizing and filtering approaches in detecting post-marketing cardiovascular events associated with targeted cancer drugs from the FDA Adverse Event Reporting System (FAERS) Journal of Biomedical Informatics. 2014:171–177. doi: 10.1016/j.jbi.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. The American Journal of Human Genetics. 2008;83(5):610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research. 2005;33(suppl 1):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Robinson PN, Kohler S, Oellrich A, Wang K, Mungall CJ, Lewis SE, Smedley D. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome research. 2014;24(2):340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhou X, Menche J, Barabasi AL, Sharma A. Human symptomsdisease network. Nature communications. 2014:5. doi: 10.1038/ncomms5212. doi:10.1038/ncomms5212. [DOI] [PubMed] [Google Scholar]
- [33].Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nature Reviews Genetics. 2010;13(8):523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- [34].Gottlieb A, Stein GY, Ruppin E, Sharan R. PREDICT: a method for inferring novel drug indications with application to personalized medicine. Molecular systems biology. 2011;7(1) doi: 10.1038/msb.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen Y, Xu R. Network-based Gene Prediction for Plasmodium falciparum Malaria Towards Genetics-based Drug Discovery; International Conference on Intelligent Biology and Medicine (ICIBM 2014); San Antonio, TX. Dec 4-6. [Google Scholar]
- [37].Xu R, Wang Q, Li L. Genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat; International Conference on Intelligent Biology and Medicine (ICIBM 2014); San Antonio, TX. Dec 4-6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].ICD-10: International Statistical Classification of Diseases and Related Health Problems: 10th revision. World Health Organization; 1992. [PubMed] [Google Scholar]
- [39].Manning CD. In: Foundations of statistical natural language processing. Schutze H, editor. MIT press; 1999. [Google Scholar]
- [40].Provost FJ, Fawcett T, Kohavi R. The case against accuracy estimation for comparing induction algorithms; International Conference on Machine Learning (ICML); 1998.pp. 445–453. [Google Scholar]
- [41].Davis J, Burnside ES, de Castro Dutra I, Page D, Ramakrishnan R, Costa VS, Shavlik JW. View Learning for Statistical Relational Learning: With an Application to Mammography; International Joint Conference on Artificial Intelligence (IJCAI); 2006.pp. 677–683. [Google Scholar]
- [42].Davis J, Goadrich M. The relationship between Precision-Recall and ROC curves; Proceedings of the 23rd international conference on Machine learning; 2005; ACM; pp. 233–240. [Google Scholar]
- [43].World Health Organization . The anatomical therapeutic chemical classification system with defined daily doses (ATC/DDD) WHO; Norway: [Google Scholar]
- [44].Dean B. Understanding the role of inflammatory-related pathways in the pathophysiology and treatment of psychiatric disorders: evidence from human peripheral studies and CNS studies. The International Journal of Neuropsychopharmacology. 2011;14(07):997–1012. doi: 10.1017/S1461145710001410. [DOI] [PubMed] [Google Scholar]
- [45].Meyer U. Anti-inflammatory signaling in schizophrenia. Brain, behavior, and immunity. 2011;25(8):1507–1518. doi: 10.1016/j.bbi.2011.05.014. [DOI] [PubMed] [Google Scholar]
- [46].Muller N, Schwarz MJ, Dehning S, Douhe A, Cerovecki A, Goldstein-Muller B, Riedel M. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Molecular psychiatry. 2006;11(7):680–684. doi: 10.1038/sj.mp.4001805. [DOI] [PubMed] [Google Scholar]
- [47].Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics. 2012;13(8):537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wahlbeck K, Cheine MV, Gilbody S, Ahonen J. Efficacy of Beta-blocker supplementation for schizophrenia: a systematic review of randomized trials. Schizophrenia Research. 2000;41(2):341–7. doi: 10.1016/s0920-9964(99)00069-9. [DOI] [PubMed] [Google Scholar]
- [49].Shek E, Bardhan S, Cheine MV, Ahonen J, Wahlbeck K. Beta-blocker supplementation of standard drug treatment for schizophrenia. Schizophrenia bulletin. 2010:sbq089. doi: 10.1093/schbul/sbq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].van den Buuse M, Zheng TW, Walker LL, Denton DA. Angiotensin-converting enzyme (ACE) interacts with dopaminergic mechanisms in the brain to modulate prepulse inhibition in mice. Neuroscience letters. 2005;380(1):6–11. doi: 10.1016/j.neulet.2005.01.009. [DOI] [PubMed] [Google Scholar]
- [51].Phillips IM. Functions of angiotensin in the central nervous system. Annual Review of Physiology. 1987;49(1):413–433. doi: 10.1146/annurev.ph.49.030187.002213. [DOI] [PubMed] [Google Scholar]
- [52].Domeney AM. Angiotensin converting enzyme inhibitors as potential cognitive enhancing agents. Journal of Psychiatry and Neuroscience. 1994;19(1):46. [PMC free article] [PubMed] [Google Scholar]
- [53].Xu R, Li L, Wang Q. dRiskKB: a large-scale disease-disease risk relationship knowledge base constructed from biomedical text. BMC bioinformatics. 2014;15(1):105. doi: 10.1186/1471-2105-15-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen Y, Zhang X, Zhang GQ, Xu R. Creation and Comparative Analysis of a Novel Disease Phenotype Network Based on Clinical Manifestation. Journal of Biomedical Informatics. 2014 doi: 10.1016/j.jbi.2014.09.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]