Abstract
MicroRNAs and chromatin remodeling complexes represent powerful epigenetic mechanisms that regulate the pluripotent state. miR-302 is a strong inducer of pluripotency, which is characterized by a distinct chromatin architecture. This suggests that miR-302 regulates global chromatin structure; however, a direct relationship between miR-302 and chromatin remodelers has not been established. Here, we provide data to show that miR-302 regulates Brg1 chromatin remodeling complex composition in human embryonic stem (hESs) cells through direct repression of the BAF53a and BAF170 subunits. With the subsequent overexpression of BAF170 in hESCs, we show that miR-302’s inhibition of BAF170 protein levels can affect the expression of genes involved in cell proliferation. Furthermore, miR-302-mediated repression of BAF170 regulates pluripotency by positively influencing mesendodermal differentiation. Overexpression of BAF170 in hESCs led to biased differentiation toward the ectoderm lineage during EB formation and severely hindered directed definitive endoderm differentiation. Taken together, these data uncover a direct regulatory relationship between miR-302 and the Brg1 chromatin remodeling complex that controls gene expression and cell fate decisions in hESCs and suggests that similar mechanisms are at play during early human development.
Keywords: MicroRNAs, Chromatin, Cell Differentiation, Embryonic Stem Cells, BAF170
Introduction
Embryonic stem (ES) cells hold great promise for regenerative medicine, drug screening, and toxicity testing. A necessary prerequisite to realizing this potential is a sophisticated understanding of human ES cell (hESC) biology and early cell fate decisions [1]. ES cells are defined by their characteristics of self-renewal and pluripotency, which are maintained by a complex regulatory network underlying a unique gene expression profile and chromatin landscape [2–5]. This robust yet flexible network includes epigenetic regulators, such as chromatin modifying complexes and microRNAs (miRNAs) [reviewed in 6, 7]. These factors have well-defined relationships with the core transcriptional regulators responsible for ES cell identity; however, little is known about direct regulation between miRNAs and chromatin modifiers and how these relationships dictate cell fate decisions.
MiRNAs repress gene expression through base pairing with target mRNAs, generally within the 3′ untranslated region (UTR), which leads to translational repression or mRNA destabilization. Many miRNAs are key regulators of the ES cell state [8]. For example, the cluster of miRNAs containing miR-302, which is highly expressed in ES cells, controls ES cell self-renewal and proliferation [9, 10] and acts by targeting cell cycle regulators [11, 12] and critical signaling pathways [13, 14]. MiR-302 is directly regulated by the core pluripotency factors Oct4 and Sox2 [11]. Interestingly, it has been shown that miR-302 itself can reprogram fibroblasts to induced pluripotent stem cells (iPCs), even in the absence of exogenous Oct4 and Sox2 [15, 16]. Both reprogramming and differentiation require extensive changes in chromatin structure [17], suggesting that miR-302 expression leads to large-scale chromatin changes. However, whether miR-302 directly regulates chromatin modifiers or specific differentiation events remains unknown.
A number of chromatin modifiers are critical to ES cell biology, iPS formation, and lineage commitment, including the Brg1 ATP-dependent chromatin remodeling complex [reviewed in 18]. This complex regulates gene expression through the opening of chromatin and repositioning of nucleosomes [19]. It is comprised of a central ATPase (Brm or Brg1) and multiple Brg1-associated factors (BAFs), which are assembled in a combinatorial fashion to dictate functional specificity. Overall, complex stoichiometry is influenced by individual BAFs that can regulate the expression of other subunits. For example, the core subunits BAF155 and BAF170 dictate the incorporation and stability of BAF57 [20, 21]. BAFs exhibit different expression patterns throughout development and across cell types [22, 23]. In mouse ES cells (mESCs), the Brg1 complex, termed esBAF, is characterized by the presence of Brg1 and BAF155 and a lack of their respective homologs Brm and BAF170 [24, 25]. This complex regulates stem cell- and lineage-specific factors and is essential for self-renewal and pluripotency [24, 26–28]. Until recently, most studies were performed in mESCs, and little was known of the Brg1 complex function in hESCs. Brg1 and BAF170 have recently been implicated in self-renewal in hESCs [29]; however, the regulation and roles of complex composition during early differentiation events remain unclear.
Here, we have identified BAF53a and BAF170 as developmentally regulated targets of miR-302. We have also identified sites in the 3′UTR of the BAF53a and BAF170 genes that are directly repressed by miR-302. Changes in BAF53a and BAF170 expression during in vitro differentiation mirrored changes in miR-302, suggesting that miR-302 regulates these subunits in a developmental context. Furthermore, we explored the role of BAF170 repression in hESC gene expression and differentiation potential. Overexpression of BAF170 revealed no obvious biological phenotype in hESCs; however, gene expression changes suggested that miR-302-mediated BAF170 repression may contribute to miR-302-dependent effects on cell cycle regulators and cell proliferation genes. Strikingly, we find that BAF170 overexpression severely limited the ability of hESCs to induce mesodermal and endodermal markers during EB formation and directed differentiation, suggesting that miR-302-mediated BAF170 repression is critical for mesendodermal differentiation. Taken together, these data provide mechanistic and biological insights into miR-302-mediated chromatin regulation and reveal a complex relationship between miR-302 and the Brg1 complex that regulates hESC gene expression and early cell fate decisions.
Materials and Methods
ES cell growth and differentiation
H1 cells were maintained on Matrigel (BD Biosciences)-coated plates in mTeSR medium (Stem Cell Technologies). Retinoic acid-induced differentiation was performed by addition of 1 μM retinoic acid. Definitive endoderm differentiation was performed using the STEMdiff Definitive Endoderm Differentiation Kit, with minor variations to the manufacturer’s instructions (StemCell Technologies). Specifically, cells were plated as aggregates without the use of ROCK inhibitor.
Luciferase reporter assays
Wild-type and mutant fragments of the BAF170 and BAF53a 3′UTR were cloned into the pMIR-Report vector (Stratagene). The reporter was cotransfected with 20 nM pre-miR-302a precursor or negative control precursor (Ambion) and pRL-CMV for normalization (Promega) into HeLa cells using Lipofectamine 2000 (Invitrogen). Cells were harvested 48 hours after transfection, and luciferase activity was assayed with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was calculated as firefly luciferase/Renilla luciferase and expressed relative to controls.
Transfections
For ectopic miR-302 expression, HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) with 50 nM negative control or pre-miR-302a (Ambion). H1 cells were transfected using Dharmafect 1 (Thermo Scientific) with 100 nM total Miridian miR-302 hairpin inhibitors (25 nM each miR-302a, b, c and d), 100nM BAF170 siRNA or 100 nM NC1 inhibitors.
Western blot analysis
Cells were lysed for Western blotting in whole cell extract lysis buffer (100mM Tris-HCl, 250mM NaCl, 1mM EDTA, 1% NP-40) containing protease inhibitor cocktail (Roche). Proteins were separated by SDS-PAGE and subjected to western blotting with the following antibodies: BAF170 H-116 (Santa Cruz), BAF155 H-76 (Santa Cruz), BAF53a (Bethyl Labs), BAF180 (Millipore), BAF60a (Transduction Labs), and Oct4 C-10 (Santa Cruz).
The Brg1 polyclonal antibody was generated by injecting BRG1 fragments, aa437–678, purified from E. coli into rabbits housed at Covance Laboratories and collecting serum at intervals using standard methods. Serum was tested by western blot for detection of BRG1. Antiserum was purified using Nab Protein A Spin purification Kit (Pierce). Protein concentration of resulting fractions was determined by absorbance at 280 nm using a standard curve of purified rabbit IgG (Santa Cruz) and pooled. Specificity was determined using western blots of BRG1 protein expressed in SW-13 cells probed with antisera.
Quantitative RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) or a Total RNA Purification Plus Kit (Norgen). For mRNA detection, cDNA was produced using the SuperScript III First Strand Synthesis System (Invitrogen) or the iScript cDNA Synthesis Kit (BioRad). Real-time PCR was performed using Brilliant III Ultra-Fast SYBR Green QPCR Mix (Agilent) or SsoAdvanced™ Universal SYBR® Green Supermix (BioRad). Ct values were normalized to the geometric mean of four control genes (GAPDH, ACTIN, 18S rRNA, and CYPB1). For miRNA detection, qRT-PCR was performed using TaqMan MicroRNA Assays (Life Technologies). Ct values were normalized to the geometric mean of two controls (U6 snoRNA and RPL21). Fold change was calculated relative to control samples. Statistical analyses (Student’s T-tests) were performed on normalized Ct values from at least three independent replicates.
Microarray analysis
Total RNA was isolated from biological triplicates of transfected H1 cells using the Qiagen RNeasy kit with on-column DNase treatment. RNA quality was analyzed on a Bioanalyzer (Agilent). Gene expression analysis was conducted using Affymetrix Human Genome U133 Plus 2.0 GeneChip® arrays in the NIEHS Microarray Core Laboratory. Briefly, 100 ng of total RNA was amplified as directed in the Affymetrix 3′ IVT Express kit protocol. Amplified biotin-aRNAs were fragmented and hybridized to arrays using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Array slides were stained and washed according to the GeneChip Hybridization, Wash and Stain Kit and user manual. Arrays were scanned in an Affymetrix Scanner 3000, and data obtained using the GeneChip® Command Console Software (AGCC; Version 1.1) using the MAS5 algorithm to generate .CHP files.
The data were analyzed using Bioconductor in R. Probe set intensities were extracted and normalized using the GCRMA method from Bioconductor R package ‘gcrma’ version 2.36.0. Gene annotations were assigned to probe sets using the Bioconductor R package ‘hgu133plus2.db’ version 2.14.0, supplemented using the Bioconductor R package ‘org.Hs.eg.db’ version 2.14.0 to resolve accession numbers into corresponding Entrez genes, where possible. Statistical comparisons were modeled and fit using the Bioconductor R package ‘limma’ version 3.20.9, using n=3 biological replicates of miR-302-inhibitor or BAF170 overexpression (BAF170OE) versus empty vector (EV) control samples for fold change calculations and Benjamini-Hochberg (BH) adjusted P-values for statistical significance. Statistical hits were defined as probes having an adjusted P-value less than 0.05 and fold change magnitude greater than 1.5. For gene-wise comparisons, per-gene data were generated by taking the mean log10 P-value and mean log2 fold change for the representative probe sets for each gene. For genes with multiple probe sets, probes with intensities at or below the chip background were excluded. For genes with statistically significant probe sets, only those with adjusted p-values < 0.05 were included. In cases of significant probes in both directions for a given gene, the direction with the most statistical significance was used and probes having that direction were used to define the representative probes. Functional analysis was performed using the Ingenuity Pathway Analysis software.
Flow Cytometry
Undifferentiated H1 cells or Day 4 definitive endoderm-differentiated cells were washed in PBS and treated with trypsin for 3–5 minutes. The cells were then resuspended and washed three times in mTeSR to obtain single-cell suspensions. Live cells were stained with either a PE-CXCR4 antibody or a FITC-TRA-1-60 antibody (BD Pharmingen) according to the manufacturer’s instructions. Cells were treated with propidium iodide or Pacific Blue prior to flow cytometry to identify viable cells. Stained cells were analyzed in the NIEHS Flow Cytometry Center on a Becton Dickinson LSR II Flow Cytometer.
Lentiviral expression
The lentiviral construct used for overexpression of BAF170 was constructed by cloning the SMARCC2 coding region into the pCDH-EF1-MCS-IRES-Puro lentiviral expression vector (System Biosciences) using standard molecular cloning techniques. The sequence of the PCR-amplified SMARCC2 insert was confirmed by primer walking and Sanger sequencing performed at Eton Bioscience. Empty pCDH-CMV-MCS-EF1-Puro vector was used a control. The viral particles were produced at the NIEHS Viral Vector Core Laboratory according to a previously established protocol[30]. The polyclonal BAF170-overexpressing cell line and control cell line were produced by treating H1 hESCs with lentivirus at an MOI of 50 and culturing the treated cells in mTeSR containing 1 μg/ml puromycin for 9 days to select for cells with lentiviral integration.
Results
MiR-302 directly represses BAF170 in hESCs
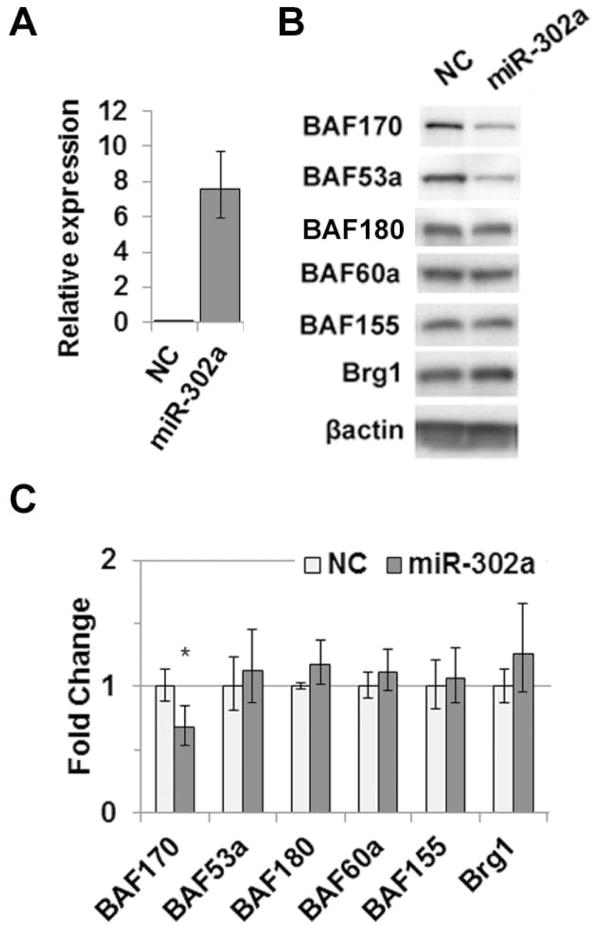
MiR-302 expression can direct reprogramming of human somatic cells to an ES cell state, which requires changes in chromatin structure. We therefore sought to identify miR-302 targets in human cells that affect chromatin structure with a focus on a chromatin modifier critical for self-renewal and pluripotency, the Brg1 chromatin remodeling complex. We searched for miR-302 binding sites in the 3′UTR of all known genes encoding BAF subunits. MiR-302 seed site matches were discovered in genes encoding BAF53a, BAF170, BAF45c and BAF180 (Table S1). Site prediction was confirmed by Miranda (www.microrna.org), and two sites were found to be conserved in mouse -- BAF53a and BAF170. A number of miRNAs share the miR-302 AAGUGC seed sequence, including miR-372, miR-373, and the miR-520 family. It is likely that these miRs can target BAF53a and BAF170 as well; however, we focused our study on miR-302 because it is highly expressed in both hESC and iPSC lines (Figure S1) [Figure S1; 10, 31] and is an important factor in cellular reprogramming [15]. To evaluate the functionality of predicted sites, we ectopically expressed miR-302a in HeLa cells, which lack endogenous miR-302, and assayed BAF expression. Overexpression of miR-302a (Figure 1A) led to repression of BAF53a at the protein level and BAF170 at both the protein and mRNA levels (Figure 1B and C). However, miR-302a overexpression had no effect on BAF180 or other complex members lacking target sites (Figure 1B and C). These data demonstrate that miR-302a can repress BAF53a and BAF170 expression.
Figure 1.
Ectopic miR-302 expression leads to BAF170 and BAF53a repression. (A) Ectopic expression of miR-302a in HeLa cells. (B) Protein levels of Brg1 complex subunits in HeLa cells transfected with pre-miR-302a or negative control (NC). β-actin was used as a loading control. (C) mRNA levels of Brg1 complex subunits measured by qRT-PCR. In all panels, error bars represent standard deviation of at least three independent replicates. * p < 0.05
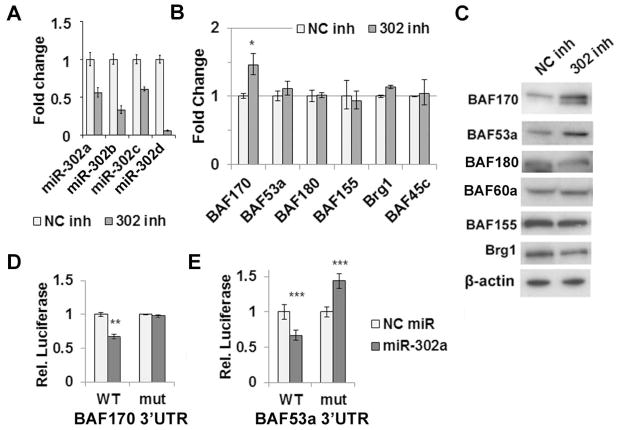
We next evaluated the effect of miR-302 on BAF expression in H1 hESCs. MiR-302 is a family of miRNAs consisting of miR-302a, b, c and d. We inhibited the entire miR-302 family by cotransfecting miRNA inhibitors against miR-302a, b, c and d into H1 cells. Sequestration of the miRs was evident by the inability to amplify the mature forms, with 50% of miR-302a and c, 30% of miR-302b, and only 5% of miR-302d available for amplification in the qRT-PCR reaction (Figure 2A). Other miRs that share a seed sequence with miR-302 were largely unaffected by these inhibitors, except for miR-373, which also showed a 75% decrease in mature levels by qRT-PCR (Figure S1B). We consider it unlikely that the loss of miR-373 expression is responsible for the observed effects given its very low expression level (Fig. S1B), although we cannot rule out this possibility. Loss of miR-302 function was confirmed by increased expression of known miR-302 targets TWF1 and p21 (Figure S2). MiR-302 inhibition had no effect on other BAF subunits but led to increased expression of both BAF53a and BAF170 (Figure 2B and C). We confirmed that derepression was not due to differentiation given that Oct4 and Sox2 expression remained at baseline levels (Figure S2). As with ectopic expression in HeLa cells, repression occurred only at the protein level for BAF53a and at both the mRNA and protein level for BAF170 (Figure 2B and C). These data show that miR-302 represses both BAF53a and BAF170 in hESCs. Furthermore, the differential effects on BAF53a and BAF170 protein and mRNA levels observed in both the loss-of-function and gain-of-function experiments suggest distinct mechanisms of regulation by miR-302.
Figure 2.
miR-302 directly regulates BAF170 and BAF53a in hESCs. (A) Inhibition of the miR-302 family in H1 hESCs. Sequestration of mature miR-302 is evident by inability to amplify miR-302a and miR-302c by qRT-PCR. (B) mRNA levels of Brg1 subunits measured by qRT-PCR following inhibition of miR-302 in hESCs compared to negative control. (C) Protein level of Brg1 complex subunits upon miR-302 inhibition in hESCs compared to negative control. (D) Luciferase activity of BAF170 WT and mutant 3′UTR reporter constructs cotransfected into HeLa cells with pre-miR-302a or negative control precursor. (E) Luciferase activity of BAF53a WT and mutant 3′UTR reporter constructs cotransfected into HeLa cells with pre-miR-302a or negative control precursor. In all panels, error bars represent standard deviation of at least three independent replicates. * p < 0.001, ** p < 0.01, *** p < 0.05
The presence of miR-302 target sites within the 3′UTRs of BAF170 and BAF53a suggested direct regulation by miR-302. To test this hypothesis, luciferase reporter constructs were generated with a fragment of the BAF53a or BAF170 3′UTR harboring the miR-302 binding site (Figure S3). Reporter constructs were cotransfected into HeLa cells along with miR-302a or a negative control miR. MiR-302a repressed the luciferase activity of both the BAF53a and BAF170 constructs (Figure 2D and E). Importantly, repression was relieved by mutation of the miR-302 binding site (Figure 2D and E). These results confirm that miR-302 can directly regulate BAF53a and BAF170 through the identified sites in their respective 3′UTRs.
Changes in BAF expression mirror miR-302 repression during differentiation
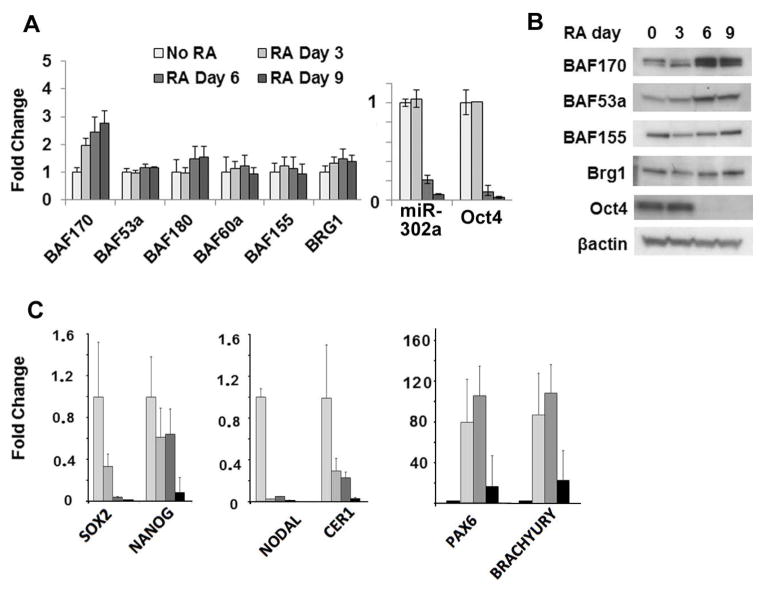
To put this regulatory event into a biological context, we characterized changes in miR-302 and BAF expression during retinoic acid (RA)-induced differentiation of H1 hESCs. After six days of RA treatment, differentiation was evident from the loss of Oct4, Sox2, Nanog and miR-302 expression (Figure 3). To determine which lineage our RA-treated cells acquired, we analyzed the expression of differentiation markers. As expected based on previous literature[32], the neuroectodermal marker Pax6 was strongly up-regulated, whereas the endodermal marker Cer1 was down-regulated; mesodermal markers showed a mixed response, with Nodal being down-regulated and Brachyury up-regulated (Figure 3C). The decrease observed in miR-302 expression occurred at the same time as an increase in both BAF53a and BAF170 at the protein level (Figure 3B). BAF170 was regulated at the mRNA level, as well (Figure 3A). The change in BAF170, but not BAF53a, mRNA is in agreement with the miR-302 inhibition data and further supports differential mechanisms of regulation. Expression of Brg1 and other BAFs remained unchanged (Figure 3A and B). Unexpectedly, an increase of BAF170 mRNA occurred prior to miR-302 repression, suggesting a role for additional transcriptional regulatory mechanisms during differentiation. Importantly, the increase in BAF170 and BAF53a protein occurred later and kinetically mirrored repression of miR-302 levels (Figure 3A and B). These results further support miR-302-mediated regulation of BAF170 and BAF53a translation and suggest that this regulation is important for developmentally relevant changes in Brg1 complex composition.
Figure 3.
Differentiation-induced upregulation of BAF170 and BAF53a mirrors miR-302 downregulation. (A) mRNA levels of miR302a, Oct4 and Brg1 complex subunits measured by qRT-PCR in H1 hESCs treated with 1 μM retinoic acid for the indicated time periods. (B) Protein levels of Brg1 complex subunits and Oct4 during RA-induced differentiation. (C) An assessment of the effects of RA-induced differentiation on H1 ESCs, using pluripotency markers and lineage markers for each of the three germ layers. mRNA levels of the indicated pluripotency genes and lineage markers were measured by qRT-PCR in H1 hESCs treated with 1 μM retinoic acid for the indicated time periods.
As mentioned above, we observed upregulation of BAF170 mRNA prior to miR-302 repression during RA-induced differentiation (Figure 3A), suggesting additional mechanisms of transcriptional regulation for BAF170. Given the pleiotropic nature of RA-induced differentiation, it is unlikely that miR-302 is the only factor impinging on Brg1 complex composition; however, it may influence the kinetics of BAF170 protein induction. To test this hypothesis, we assayed BAF170 levels during RA-induced differentiation while overexpressing miR-302. Transient transfection of H1 cells with miR-302a led to a 16-fold increase in expression over the endogenous miR-302a levels. Although the overall levels decreased at a rate similar to the endogenous miR-302a gene, levels of miR-302a expression remained higher in the miR-302a-transfected cells than in negative controls throughout the differentiation process (Figure S4A). No effect on BAF170 mRNA was observed (Figure S4B), yet BAF170 protein expression was significantly decreased at 0, 3 and 6 days of RA treatment in miR-302 overexpressing cells compared to the negative control (Figure S4C). This result is consistent with the view that miR-302 regulates the kinetics of BAF170 induction during differentiation.
BAF170 repression contributes to miR-302-mediated changes in gene expression
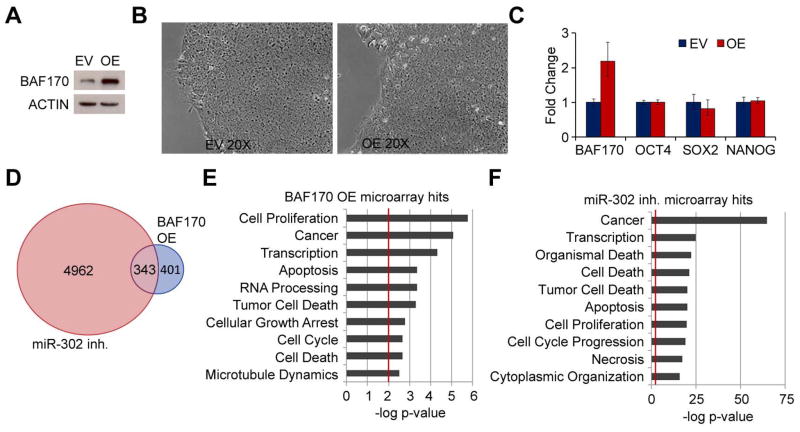
While the role of BAF53a during early development has been studied previously [33], no specific biological function in early development has been described for the BAF170 core subunit. We therefore sought to determine the biological role of BAF170 repression in hESC gene expression and pluripotency. To this end, we overexpressed BAF170 in H1 hESCs using a lentiviral expression system. Polyclonal BAF170-overexpressing cell lines (OE) and negative controls transduced with empty vector (EV) were isolated following puromycin selection. Robust and stable overexpression of BAF170 was observed in the BAF170 OE cell line (Figure 4A). No obvious difference in phenotype was observed between these cells and EV cells. The cells retained their ES cell-like appearance, showed no signs of differentiation and expressed normal levels of pluripotency genes such as Oct4, Sox2, and Nanog (Figure 4B and C). These results suggest that BAF170 repression is not required for ES cell self-renewal.
Figure 4.
BAF170 overexpression and miR-302 inhibition both affect cell cycle and proliferation genes. (A) Western blot of BAF170 levels in cells lines stably transduced with empty lentiviral vector (EV) or lentiviral vector containing the BAF170 ORF (OE). Actin was used as a loading control. (B) Phase contrast microscopy of EV and OE cell lines three days after passaging showing characteristic hESC morphology. (C) mRNA levels of BAF170, OCT4, SOX2, and NANOG in the EV and OE cell lines. (D) Venn diagram demonstrating the overlap in gene sets misregulated upon miR-302 inhibition or BAF170 overexpression based on microarray gene expression analysis. (E) Top ten functional categories enriched in the set of genes misregulated in the BAF170 OE cell line. (F) Top ten functional categories enriched in the set of miR-302-regulated genes.
Microarray analysis of the BAF170 OE cells revealed 744 genes that were misregulated compared to controls (adjusted p-value < 0.05); however, only 169 genes exhibited fold changes greater than 1.5 in the BAF170 OE cells compared with EV (Figure S5). This represents relatively few genes compared to the gene expression changes that occur 48 hours after inhibition of miR-302 in H1 cells, which led to greater than 1.5 fold misregulation of 2594 genes (Figure S5). This observation is not surprising, as miR-302 has potentially hundreds of targets in hESCs and pleotropic effects on gene expression and cellular processes. Nevertheless, given the direct regulation of BAF170 by miR-302, it is likely that BAF170 repression contributes to the gene expression changes observed upon miR-302 inhibition. Indeed, almost half of the genes significantly affected by BAF170 overexpression were also misregulated by miR-302 inhibition (Figure 4D). Although many genes affecting the cell cycle and pluripotency are likely to be independently regulated by mir-302 or BAF170, this observation suggests that miR-302-mediated BAF170 repression may contribute somewhat to the gene expression changes observed upon miR-302 inhibition. Furthermore, functional analysis of the genes misregulated by either miR-302 inhibition or BAF170 overexpression revealed enrichment of very similar functional categories, including cell proliferation, cell cycle regulation, cell death and apoptosis, and transcription (Figure 4E and F). For example, cell cycle-related genes with significantly altered expression due to both BAF170 overexpression and miR-302 inhibition included cyclins C and G2, cdc25a, cdc27, and aurora kinase A, and many other cell cycle-related genes were significantly regulated by either BAF170 or miR-302. The effects on cell cycle genes upon miR-302 inhibition corroborate the well-known role of miR-302 in cell cycle regulation and proliferation in hESCs. These data further suggest that in addition to directly regulating cell cycle regulators such as p21 and cyclin D1 [11, 12], miR-302 may also affect cell proliferation indirectly through BAF170 repression.
BAF170 overexpression biases cells toward ectodermal differentiation
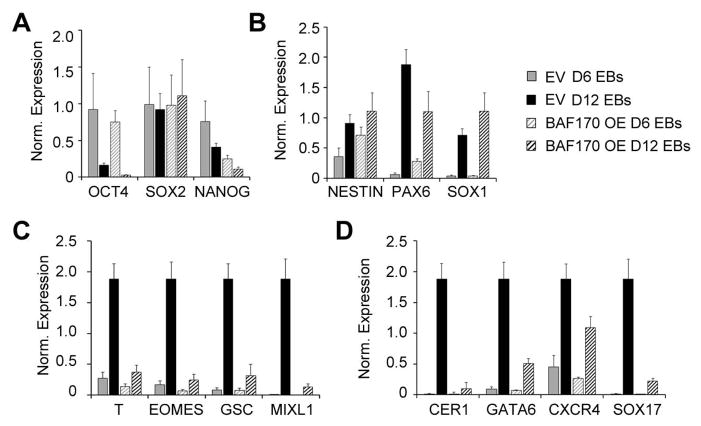
Because BAF170 OE cells exhibited no self-renewal defect, we next performed in vitro differentiation experiments to assess the pluripotency of these cells. Embryoid bodies were formed from EV and BAF170 OE cell lines and analyzed for gene expression on days 6 and 12. Sox2 expression was unaffected by BAF170 OE; however, the levels of OCT4 and NANOG were lower in BAF170 OE EBs compared with controls (Figure 5). These results indicate that BAF170 OE cells are capable of downregulating pluripotency markers during EB formation. Expression of the neural progenitor markers NESTIN, PAX6 and SOX1 were generally increased in day 6 & 12 BAF170 OE EBs compared with controls, with PAX6 expression showing a slight decline at day 12 (Figure 5B), suggesting that BAF170 overexpression does not impede ectodermal differentiation. On the other hand, the induction of mesodermal (T, EOMES, GSC, and MIXL1) and endodermal markers (CER1, GATA6, CXCR4, and SOX17) was decreased by 50–90% in BAF170 OE EBs compared with controls (Figure 5C and D). These results demonstrate that BAF170 overexpression biases cells toward a neuroectodermal fate and may block mesendodermal differentiation. Oct4, Sox2, and Nanog have differential roles in the differentiation of the three germ layers [34]. Given that high levels of OCT4 and NANOG are associated with repression of ectodermal differentiation, the decreased levels of OCT4 and NANOG in the BAF170 OE EBs are consistent with the idea that these cells are biased toward ectodermal differentiation during EB formation.
Figure 5.
BAF170 overexpression biases EB differentiation toward the ectodermal lineage. (A–D) mRNA expression analysis of Day 6 and Day 12 embryoid bodies obtained from the EV and BAF170 OE cell lines. The genes analyzed included typical pluripotency (A), ectoderm (B), mesoderm (C), and endoderm (D) markers. The qRT-PCR data are expressed as the median-centered normalized expression for each gene.
BAF170 repression is required for directed induction of mesendodermal differentiation
The observation that ectopic BAF170 expression biases cells toward an ectodermal fate is consistent with the repression of miR-302 and upregulation of BAF170 observed during RA-induced differentiation (see Figure 3). To determine if the opposite expression pattern may occur during endodermal differentiation, we differentiated H1 hESCs using the STEMdiff Definitive Endoderm Differentiation Kit (Stemcell Technologies), which leads to a primitive streak-like mesendodermal intermediate state before inducing differentiation into definitive endoderm. Protein and RNA changes were measured every day for four days of differentiation. During this time, BAF170 exhibited biphasic repression and remained at levels lower than those in hESCs throughout the differentiation process (Figure 6A). Strikingly, miR-302 exhibited a parallel biphasic induction, consistent with the idea that miR-302 is at least partially responsible for the repression of BAF170 during endodermal differentiation (Figure 6B).
Figure 6.
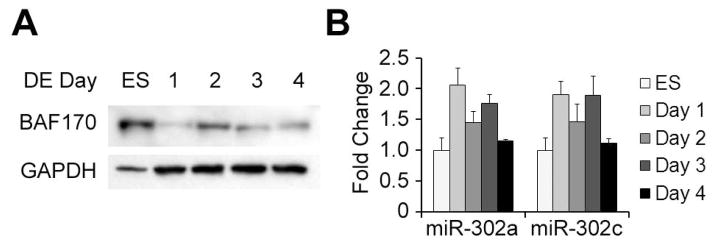
BAF170 is repressed as miR-302 is upregulated during definitive endoderm differentiation. (A) Western blot analysis of BAF170 expression during definitive endoderm differentiation. GAPDH was used as a loading control. (B) miRNA expression of miR-302a and miR-302c during definitive endoderm differentiation.
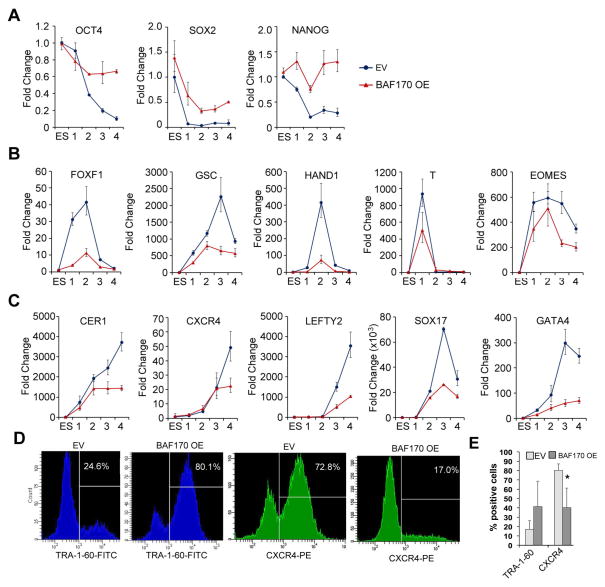
To determine if the observed repression of BAF170 is important for directed endodermal differentiation, we subjected BAF170 OE and EV hESCs cells to the same definitive endoderm differentiation protocol. BAF170 OE cells were unable to downregulate the pluripotency factors OCT4, SOX2, and NANOG to the same degree as EV cells (Figure 7A), suggesting that BAF170 overexpression confers a differentiation defect. Further gene expression analysis demonstrated that BAF170 OE cells were unable to fully induce either mesodermal or endodermal markers during directed differentiation (Figure 7B and C, respectively). The largest defects were observed for the mesodermal markers FOXF1, GSC, and HAND1 and the endodermal markers CER1, LEFTY2, and SOX17, which were induced to less than 25% of control levels in the BAF170 OE cells. Approximately 30–50% induction was observed in BAF170 OE cells for T, EOMES, CXCR4, and GATA4. These results demonstrate that ectopic BAF170 expression inhibits the induction of mesodermal and endodermal genes in response to differentiation cues.
Figure 7.
BAF170 overexpression blocks mesendodermal marker expression and definitive endodermal differentiation. (A–C) mRNA expression of untreated BAF170 OE and EV hESCs and cells undergoing definitive endoderm differentiation. Samples were taken on days 1, 2, 3 and 4 during the differentiation process. The qRT-PCR data are expressed as fold change over expression in untreated EV hESCs. (D) Flow cytometry showing the percentage of BAF170 OE and EV cells expressing the pluripotency marker TRA-1-60 or the endodermal marker CXCR4 after 4 days of definitive endoderm differentiation. The data shown are representative results from three independent replicates. (E) Histogram showing collected data from at least three measurements of TRA-1-60 and CXCR4 expression in both the EV and BAF170 OE cell lines following the directed definitive endoderm protocol. * p < 0.05
To determine the percentage of cells that were able to successfully undergo differentiation to definitive endoderm, pluripotency and endodermal surface markers were analyzed by flow cytometry on day four of the differentiation protocol. Eighty percent of BAF170 OE cells remained positive for the pluripotency cell surface marker TRA-1-60 after four days of differentiation, a dramatic increase over the 17% of positive-staining cells observed in the EV population. Conversely, only 40% of the BAF170 OE cells expressed the endodermal marker CXCR4 after four days of differentiation, while 80% of the EV cells were CXCR4 positive. Together, these data suggest that BAF170 repression is critical for efficient endodermal differentiation and required for induction of mesoderm- and endoderm-specific gene expression programs.
Discussion
It is well established that miRNAs and chromatin modifiers are critical for ES cell biology and development; however, how these players interact remains largely unexplored. Here, we have identified a novel regulatory mechanism between miR-302 and the Brg1 complex subunits BAF53a and BAF170; specifically, our results indicate that miR-302 binds the 3′UTRs and directly regulates BAF53a and BAF170. Previous studies on murine neural development have demonstrated regulation of BAF53a by other miRNAs during mouse development [33]. Similarly, more recent work has revealed miRNA-mediated regulation of BAF60 isoforms during myogenesis in chick embryos [35]. Our data from hESCs, models of early human embryonic development, extend the miRNA-mediated regulation of BAF subunits to an earlier developmental window and suggest that similar regulation occurs during human development. Furthermore, these data provide, to our knowledge, the first mechanistic insight into BAF regulation in ES cells. Combined with the additional recent work in mouse and chick development, our data in hESCs suggests that miRNA-mediated regulation may be a general mechanistic approach used during development to dictate functional changes in multisubunit complexes such as the Brg1 chromatin remodeling complex. More in-depth studies matching tissue-specific miRNA expression with mRNA expression and miRNA target prediction of tissue-specific isoforms of dynamic multisubunit complexes may reveal additional regulatory events important for development or other cellular transformations.
Recent work has shown that Brg1 and BAF170 are important for self-renewal in hESCs [29]. Specifically, knock down of Brg1 or BAF170, but not BAF155, led to loss of the stem cell-like phenotype in hESCs. Given that BAF170 is required for self-renewal, it is perhaps not surprising that we observed no loss of self-renewal in BAF170 OE cells. In hESCs, BAF170 overexpression does not appear to vitally affect complex function; i.e., whereas too little BAF170 impairs self-renewal [29], too much BAF170 has no affect in this context (Figure 4B and C). On the other hand, Zhang et al., also showed that embryoid bodies formed from Brg1-deficient cells are biased toward ectodermal differentiation and impaired for mesendodermal differentiation. These results are similar to those seen here for BAF170-overexpressing embryoid bodies, suggesting that overexpression of BAF170 negatively impacts the overall function of the Brg1 complex in the context of embryoid body formation and mesendodermal differentiation. This seemingly paradoxical result is not surprising given the dual role of the complex in both stem cell self-renewal and lineage determination. Overall, these results support a role for BAF170 in dictating context-dependent Brg1 complex function.
Interestingly, BAF170 and its homolog BAF155 are core subunits of the Brg1 complex that are generally thought to be involved solely in scaffolding and complex stoichiometry[36]. The importance of BAF170 regulation in development described here suggests that BAF170 may have more specific roles and play a larger part in dictating functional specificity of the complex than previously appreciated. Some evidence of this exists from work on mESCs [24] and murine neural development [37], which has shown distinct expression patterns of BAF170 and BAF155 in specific cell types during development. Interestingly, recent results showed that manipulation of BAF170 levels affected chromatin regulation and differentiation patterns in neocortex development[37]. In this context, BAF170 overexpression led to decreased neocortical volume and changes in cellular differentiation patterns. Along with the data presented here, these results suggest that Brg1 core subunits such as BAF170 and BAF155 play complex roles in regulating cellular differentiation at many levels of development and have distinct functions that are dependent on the cellular and developmental context.
The context-dependent roles of different core subunits may also be integral to species-specific differences in ES cell biology. Our current data support a Brg1 core complex composition in hESCs that is distinct from that observed in mESCs, as recently reported [29], [24, 25]. Most notably, compared with mESCs, we observed significant expression of BAF170 in hESCs (Figure S6), which we have demonstrated to have a key biological role in hESC pluripotency and differentiation. Given the potentially far-reaching applications of ES cell-derived technologies for clinical biology, it is critical to fully understand the differences between mESCs and hESCs. There is currently a large body of work outlining many of the intricacies of the networks controlling mESC function; however, applying this knowledge to clinically relevant hESCs remains a challenge given the differences between these cell types. The present work provides additional insight into those differences and elucidates mechanisms that can be further explored and exploited to understand the complexities of both mouse and human embryonic stem cell biology.
While miR-302 has largely been studied for its role in ES cell self-renewal and somatic cell reprogramming, evidence suggests that it also has a role in early differentiation events. Previous studies have implicated the miR-302 family and the related miR-373 in mesendodermal differentiation [38, 39]. These findings are consistent with the increase in miR-302 observed in the present study during definitive endoderm differentiation. This role of miR-302 and miR-373, however, has until now largely been attributed to direct repression of the TGFβ/Nodal inhibitor Lefty [13, 38, 39]. While Lefty repression is undoubtedly important in this process, our work has identified an additional novel target of miR-302 whose repression is required for endodermal differentiation – the Brg1 complex subunit BAF170. Importantly, this finding may provide clues regarding, and direct further studies to elucidate, the role of miRNAs, such as miR-302, and chromatin remodeling complex composition in the genome-wide chromatin changes that occur during differentiation or cellular reprogramming.
Summary
By identifying BAF170 as a miR-302 target required for mesendodermal differentiation, we have expanded the role of the Brg1 complex, and BAF170 in particular, in critical aspects of hESC biology and differentiation. The Brg1 complex is among a number of chromatin modifiers critical to maintain the distinct chromatin architecture of ES cells [23, 40]. These chromatin modifiers, along with core transcription factors like Oct4 and Sox2, miRNAs and signaling pathways, are critical components of the core regulatory circuitry underlying ES cell self-renewal and pluripotency [41]. This work contributes substantially to our understanding of the network of chromatin modifiers, miRNAs, transcription factors and signaling pathways underlying hESC self-renewal and pluripotency. Importantly, it underscores the critical nature of interplay between epigenetic factors such as miRNAs and chromatin remodeling complexes in driving cell fate decisions in hESCs and suggests that similar mechanisms are at play in human embryonic development.
Supplementary Material
SIGNIFICANCE STATEMENT.
This work establishes a regulatory connection between microRNAs and chromatin remodeling protein complexes critical for pluripotency in human ES cells. Our data demonstrate show that specific micro RNAs can regulate chromatin remodeling complex composition in human embryonic stem cells through direct repression of the BAF53a and BAF170 protein expression. We show that this altered protein expression contributes to miR-302-mediated regulation of genes involved in cell proliferation. We are confident that our identification of novel regulatory mechanisms involving key epigenetic factors in embryonic stem cell biology may allow for the development of novel therapeutic strategies for human diseases.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (project #Z01 ES071006-14).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors have no conflict of interest to disclose.
Staton Wade: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing
Lee Langer: collection and assembly of data, data analysis and interpretation, manuscript writing
James Ward: Data analysis
Trevor Archer: Conception and design, financial support, provision of study material, assembly of data, data interpretation, manuscript writing, final approval of manuscript.
References
- 1.Daley GQ. Prospects for stem cell therapeutics: myths and medicines. Curr Opin Genet Dev. 2002;12:607–613. doi: 10.1016/s0959-437x(02)00346-5. [DOI] [PubMed] [Google Scholar]
- 2.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 4.Ramalho-Santos M, Yoon S, Matsuzaki Y, et al. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 5.Meshorer E, Yellajoshula D, George E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao M. Conserved and divergent paths that regulate self-renewal in mouse and human embryonic stem cells. Dev Biol. 2004;275:269–286. doi: 10.1016/j.ydbio.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & development. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 10.Suh MR, Lee Y, Kim JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 11.Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Baskerville S, Shenoy A, et al. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barroso-delJesus A, Lucena-Aguilar G, Sanchez L, et al. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. Faseb J. 2011;25:1497–1508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- 14.Lipchina I, Elkabetz Y, Hafner M, et al. Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes & development. 2011;25:2173–2186. doi: 10.1101/gad.17221311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyoshi N, Ishii H, Nagano H, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koche RP, Smith ZD, Adli M, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–532. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon H, Imbalzano AN, Khavari PA, et al. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keppler BR, Archer TK. Ubiquitin-dependent and ubiquitin-independent control of subunit stoichiometry in the SWI/SNF complex. The Journal of biological chemistry. 2010;285:35665–35674. doi: 10.1074/jbc.M110.173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Xue Y, Zhou S, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 23.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaeser MD, Aslanian A, Dong MQ, et al. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaniel C, Ang YS, Ratnakumar K, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 28.Ho L, Jothi R, Ronan JL, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Li B, Li W, et al. Transcriptional Repression by the BRG1-SWI/SNF Complex Affects the Pluripotency of Human Embryonic Stem Cells. Stem Cell Reports. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barde I, Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2010;Chapter 4(Unit 4):21. doi: 10.1002/0471142301.ns0421s53. [DOI] [PubMed] [Google Scholar]
- 31.Wilson KD, Venkatasubrahmanyam S, Jia F, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baharvand H, Mehrjardi N-Z, Hatami M, et al. Neural differentiation from human embryonic stem cells in a defined adherent culture condition. The International Journal of Developmental Biology. 2007;51:371–378. doi: 10.1387/ijdb.72280hb. [DOI] [PubMed] [Google Scholar]
- 33.Yoo AS, Staahl BT, Chen L, et al. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Oron E, Nelson B, et al. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 35.Goljanek-Whysall K, Mok GF, Fahad Alrefaei A, et al. myomiR-dependent switching of BAF60 variant incorporation into Brg1 chromatin remodeling complexes during embryo myogenesis. Development. 2014;141:3378–3387. doi: 10.1242/dev.108787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung I, Sohn DH, Choi J, et al. SRG3/mBAF155 stabilizes the SWI/SNF-like BAF complex by blocking CHFR mediated ubiquitination and degradation of its major components. Biochem Biophys Res Commun. 2012;418:512–517. doi: 10.1016/j.bbrc.2012.01.057. [DOI] [PubMed] [Google Scholar]
- 37.Tuoc TC, Boretius S, Sansom SN, et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Rosa A, Papaioannou MD, Krzyspiak JE, et al. miR-373 is regulated by TGFbeta signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev Biol. 2014;391:81–88. doi: 10.1016/j.ydbio.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 41.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.