Abstract
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is widely used to map histone marks and transcription factor binding throughout the genome. Here we present ChIPmentation, a method that combines chromatin immunoprecipitation with sequencing library preparation by Tn5 transposase (“tagmentation”). ChIPmentation introduces sequencing-compatible adapters in a single-step reaction directly on bead-bound chromatin, which reduces time, cost, and input requirements, thus providing a convenient and broadly useful alternative to existing ChIP-seq protocols.
ChIP-seq combines chromatin immunoprecipitation with next generation sequencing in order to measure the genome-wide distribution of chromatin proteins and their posttranslational modifications1. Despite recent efforts to optimize and improve ChIP-seq2, current protocols are still quite tedious, time-consuming, and costly. The development of a hyperactive Tn5 transposase that enables simultaneous DNA fragmentation and adapter tagging (“tagmentation”) presents an opportunity for faster and more sensitive library preparation from purified DNA3. Furthermore, tagmentation of intact chromatin followed by sequencing (ATAC-seq) was shown to yield open chromatin profiles comparable to those obtained by DNase-seq4. In contrast, this technique has not yet been adapted for ChIP-seq sample preparation.
Here, we demonstrate tagmentation of immunoprecipitated chromatin in a robust one-step reaction performed directly on bead-bound chromatin. This method – which we call ChIPmentation – provides a rapid, cost-effective, low-input ChIP-seq workflow and yields excellent results for both histone marks and transcription factors. Compared to recent ChIP-seq protocol variants that are optimized for minimal cell numbers5-11 or maximum resolution12, 13, which come at the expense of increased complexity and/or high reagent costs (Supplementary Table 1), ChIPmentation is a convenient general-purpose protocol that is well-suited for a broad range of applications.
We initially tested an approach that combines standard ChIP with subsequent tagmentation of the purified ChIP DNA (Supplementary Fig. 1). This “ChIP-tagmentation” protocol gave acceptable results (Supplementary Fig. 2), but was difficult to standardize across samples and across antibodies. ChIP-tagmentation was particularly sensitive to the ratio of DNA to transposase, which is problematic because DNA concentrations obtained by ChIP can be highly variable and too low to quantify. Furthermore, purified ChIP DNA is already fragmented, and excess transposase can result in small fragments that are difficult to sequence.
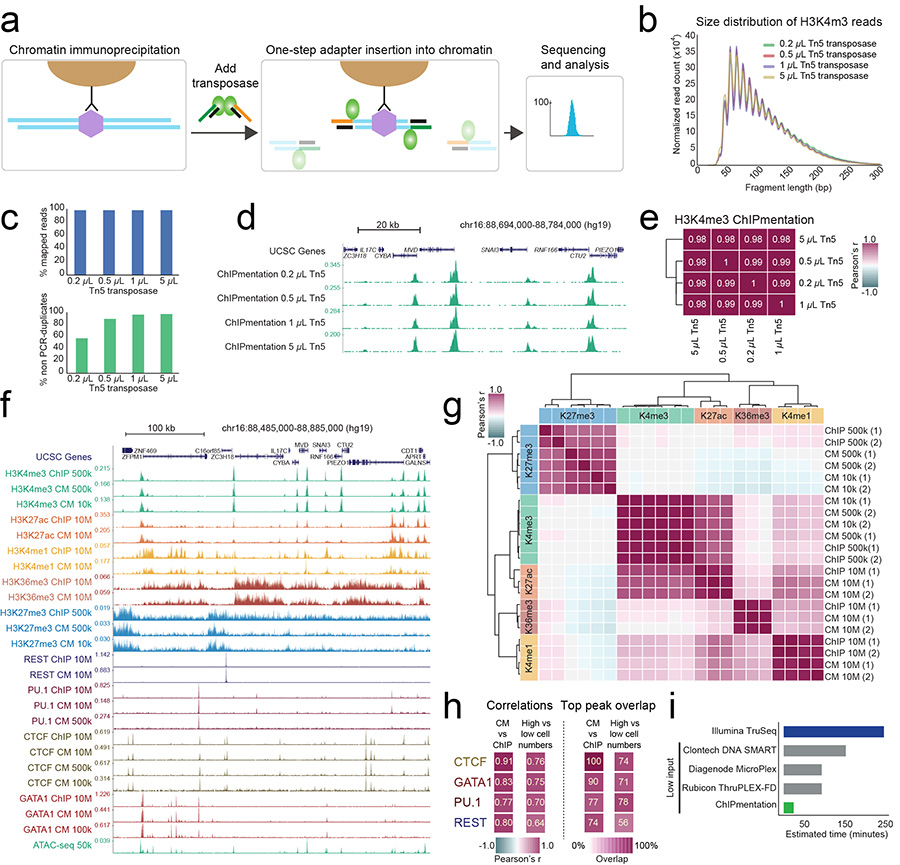
We reasoned that performing tagmentation directly on the immunoprecipitated and bead-bound chromatin would allow chromatin proteins to protect the DNA from excessive tagmentation. Our ChIPmentation protocol (Fig. 1a, Supplementary Fig. 1 and Online Methods) was indeed robust over a 25-fold difference in transposase concentrations according to five different metrics: measured size distribution of ChIPmentation libraries (Supplementary Fig. 3), size distribution inferred from paired-end sequencing reads (Fig. 1b), read mapping performance (Fig. 1c), concordance between sequencing profiles (Fig. 1d), and signal correlations (Fig. 1e). Furthermore, the ChIPmentation protocol is fast and convenient, does not give rise to sequencing adapter dimers, and requires only a single DNA purification step prior to library amplification.
Figure 1.
Rapid and robust analysis of histone marks and transcription factors by ChIPmentation.
(a) Schematic overview of ChIPmentation (see Supplementary Fig. 1 for a graphical comparison of standard ChIP-seq, ChIP-tagmentation with purified ChIP DNA, and ChIPmentation).
(b) Size distribution of fragment lengths measured by paired-end sequencing of ChIPmentation libraries for H3K4me3 at different Tn5 transposase concentrations.
(c) Percentages of aligned (mapped) reads and unique (non PCR-duplicate) fragments for ChIPmentation of H3K4me3 at different Tn5 transposase concentrations.
(d) ChIPmentation signal for H3K4me3 at different Tn5 transposase concentrations.
(e) Genome-wide correlation heatmap (1,000 bp windows) for ChIPmentation of H3K4me3 at different Tn5 transposase concentrations.
(f) Genome browser screenshot showing ChIP-seq (“ChIP”) and ChIPmentation (“CM”) data with different cell numbers as input for five histone marks and four transcription factors. Data from two biological replicates were combined.
(g) Genome-wide correlation heatmap (1,000 bp windows) for standard ChIP-seq and ChIPmentation data across different histone marks and different cell numbers.
(h) Genome-wide correlation values (1,000 bp windows) and top peak overlap percentage for standard ChIP-seq and ChIPmentation across different transcription factors and different cell numbers (high cell numbers: 10 million cells; low cell numbers: 100,000 or 500,000 cells). Overlap percentages indicate the proportion of top 50% of peaks from one experiment that were also present among all peaks in a second experiment.
(i) Comparison of library preparation time for standard ChIP-seq (dark blue), commercially available library preparation kits for low-input samples (grey), and ChIPmentation (green). Library preparation time was measured up to the point when sequencing-compatible adapters are introduced, excluding the final library amplification by PCR that is similar for all methods.
We validated ChIPmentation for five histone marks (H3K4me1, H3K4me3, H3K27ac, H3K27me3, and H3K36me3) and four transcription factors (CTCF, GATA1, PU.1, and REST). All ChIPmentation profiles were of high quality and in agreement with those obtained by standard ChIP-seq (Fig. 1f), which we confirmed by the following metrics: correlations in 1-kilobase tiling regions across the genome (Fig. 1g, 1h, Supplementary Fig. 4, 5a), overlap of transcription factor binding peaks (Fig. 1h, Supplementary Fig. 5b), signal distributions at annotated genes (Supplementary Fig. 5c), fractions of reads in peaks as a measure of specific enrichment (Supplementary Fig. 5d), sequencing statistics such as alignment and unique read rates (Supplementary Table 2), and concordance between biological replicates (Fig. 1g, Supplementary Fig. 4).
Compared to standard ChIP-seq, ChIPmentation also allowed us to substantially reduce the number of cells required for obtaining high-quality data. We generated accurate ChIPmentation profiles for H3K4me3 and H3K27me3 from 10,000 cells, and we obtained good results from 100,000 cells for GATA1 and CTCF (Fig. 1f). The signal quality obtained for these low-input samples was confirmed by genome-wide correlations (Fig. 1g, 1h, Supplementary Fig. 4), agreement of sequencing profiles (Supplementary Fig. 6a), and peak overlap (Fig. 1h, Supplementary Fig. 6b). Nevertheless, as with every ChIP approach, low-input ChIPmentation depends on antibody quality and may not be feasible for all histone marks and transcription factors.
Given that ChIPmentation involves tagmentation of fragmented chromatin instead of purified ChIP DNA, we also investigated whether the tagmentation patterns correlate with local chromatin structure as described for ATAC-seq4. We observed characteristic signal intensity patterns surrounding the consensus binding motifs of the respective transcription factor in the ChIPmentation data (Supplementary Fig. 7a). However, some of these patterns were also present in genome sequencing libraries prepared by tagmentation of purified genomic DNA (Supplementary Fig. 7a), suggesting that they originate in part from DNA sequence biases of the transposase. After bioinformatic correction we observed footprints for CTCF and REST, which resembled those found in ATAC-seq and DNase-seq data, but none of these methods detected footprints for GATA1 and PU.1 (Supplementary Fig. 7b and Online Methods).
Furthermore, using ChIPmentation we observed a striking periodicity of approximately 10 basepairs in the average distance between neighboring tagmentation events (Supplementary Fig. 7c). Similar patterns detected in DNase-seq data appear to be related to the wrapping of DNA around nucleosomes and have been used to predict nucleosome stability14. We also observed characteristic ChIPmentation patterns surrounding the center of nucleosomes (Supplementary Fig. 7d), indicating increased Tn5 insertion frequency at those positions along the nucleosome where the DNA is most accessible15. These observations suggest that ChIPmentation data may be useful for inferring transcription factor footprints, nucleosome stability, and nucleosome positioning.
Our results establish ChIPmentation as a convenient general-purpose alternative to standard ChIP-seq, providing a fast (Fig. 1i) and cost-effective (Supplementary Fig. 8) method that works well for low-input samples (Fig. 1f). ChIPmentation is compatible with numerous variations of the ChIP protocol (Supplementary Note), which facilitates its use with a broad range of different antibodies. While the ChIP part of the protocol can profit from optimization for new antibodies and cell types, the remainder of the protocol is highly robust and standardized. We sequenced 52 ChIPmentation libraries using nine different antibodies and 10,000 to 10 million cells without having to individually optimize the tagmentation conditions. An interesting additional aspect of ChIPmentation is our observation of high-resolution patterns in ChIPmentation data (Supplementary Fig. 7), which may become useful for inferring chromatin structure at the immunoprecipitated target regions. For example, with suitable bioinformatic algorithms that correct for the inherent sequence bias of tagmentation, it may be possible to use ChIPmentation of histone marks as a cost-effective approach to positioning nucleosomes in transcribed regions or in repressive chromatin. We expect that the simplicity and robustness of ChIPmentation will make it attractive for small-scale studies; in addition, its speed and cost-effectiveness will facilitate high-throughput research in the context of large epigenome projects16-18.
Online Methods
Cell culture and sample collection
K562 cells were cultured in RPMI medium supplemented with 10% FCS and antibiotics. Cells were routinely checked for mycoplasma contamination, and K562 cell line identity was recently confirmed by sequencing-based analysis of copy number aberrations19. A CASY cell counter (Roche) was used to determine cell numbers. Peripheral blood was obtained from healthy donors as approved by the ethics commission of the Medical University of Vienna and the Vienna General Hospital. All donors provided written informed consent. Coagulation was prevented with EDTA or heparin, peripheral blood was diluted 1:1-1:3 in PBS, and peripheral blood mononuclear cells (PBMCs) were isolated with Lymphoprep density gradient (Axis-Shield) following manufacturer’s instructions. Purified cells were suspended in RPMI supplemented with 10% FBS and penicillin-streptomycin.
Chromatin immunoprecipitation
ChIPmentation was tested in combination with three different protocols for performing the chromatin immunoprecipitation, which are described in detail in the Supplementary Note.
Standard ChIP-seq
Purified ChIP DNA was end-repaired using the NEBNext End Repair Module (NEB) according to manufacturer’s instructions. Clean-up was done using SPRI Ampure XP beads (Agencourt) according to manufacturer’s instructions. Fragments were A-tailed using Klenow (3′ → 5′ exo-) polymerase (Enzymatics), and TruSeq-compatible adapters were ligated using T4 DNA Ligase (Enzymatics). The final library was size-selected using SPRI Ampure XP beads to remove adapter dimers.
ChIPmentation
ChIPmentation is compatible with various different protocols for ChIP, which makes it easy to apply ChIP-mentation to antibodies that work best with a certain ChIP protocol. The chosen ChIP protocol was followed until the beads carrying immunoprecipitated chromatin were washed with LiCl-containing wash buffer (WBIII for ChIP version 1, RIPA-LiCl for ChIP version 2, and TF-WBIII for ChIP version 3, as described in the Supplementary Note). Beads were then washed twice with 10 mM cold Tris-Cl pH 8.0 to remove detergent, salts, and EDTA. Subsequently, beads were resuspended in 30 μl of the tagmentation reaction buffer (10 mM Tris, pH 8.0, 5 mM MgCl2) containing 1 μl Tagment DNA Enzyme from the Nextera DNA Sample Prep Kit (Illumina) and incubated at 37°C for 10 minutes in a thermocycler. Following tagmentation, the beads were washed twice with 150 μl cold WBI (ChIP version 1), RIPA (ChIP version 2), or TF-WBI (ChIP version 3). Afterward, the chosen ChIP protocol was resumed with the final bead wash, elution from beads, reverse-crosslinking, and DNA purification. A detailed protocol can be found at http://chipmentation.computational-epigenetics.org/.
ChIP-tagmentation with purified ChIP DNA
Purified ChIP DNA from a standard ChIP for H3K4me3 on PBMCs was measured using a Qubit fluorometer (Life Technologies) and then diluted in 10mM Tris-Cl pH 8.5 supplemented with 0.1% Tween-20 to 100 pg, 10 pg, or 2 pg total DNA. The tagmentation reaction was performed for 5 minutes at 55°C in a 10 μl reaction containing diluted DNA, 5 μl 2x tagmentation buffer (Illumina) and 1 μl (for 100 pg DNA) or 0.5 μl (for 10 pg and 2 pg) 1:10 diluted Nextera Tag DNA Enzyme (diluted in precooled TE/50% glycerol). The tagmented DNA was amplified with the Nextera DNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions with the following program: 72°C 5 min, 98°C 30 s, 14 cycles of 98°C 10 s 63°C 30 s 72°C 30 s, and a final elongation at 72°C for 1 min. Libraries were purified using SPRI AMPure XP beads with a beads-to-sample ratio of 1.5:1. Purified ChIP DNA or deproteinized input DNA from K562 ChIP was prepared as for PBMCs with slight modifications: 5 ng of ChIP DNA was taken for the tagmentation reaction using 0.5 μl of a 1:10 diluted Tn5 transposase in a 5 μl reaction at 55°C for 5 minutes. DNA was purified with the MinElute kit (Qiagen) and amplified with the Kapa HiFi HotStart ReadyMix (Kapa Biosystems).
Amplification and sequencing of standard ChIP-seq, ChIPmentation, and ChIP-tagmentation libraries
1 μl of each library was amplified in a 10-μl qPCR reaction containing 0.15 μM primers, 1x SYBR green and 5 μl 2x KAPA HiFi HotStart ReadyMix (Kapa Biosystems) to estimate the optimum number of enrichment cycles with the following program: 72°C 5 min, 98°C 30 s, 24 cycles of 98°C 10 s 63°C 30 s 72°C 30 s, and a final elongation at 72°C for 1 min. Kapa HiFi HotStart ReadyMix was incubated at 98°C for 45 s prior to preparation of the PCR reaction to activate the hot-start enzyme for successful nick translation in the first PCR step. Final enrichment of the libraries was performed in a 50 μl reaction using 0.75 μM primers and 25 μl 2x KAPA HiFi HotStart ReadyMix. Libraries were amplified for N cycles, where N is equal to the rounded-up Cq value determined in the qPCR reaction. Enriched libraries were purified with size-selection using SPRI AMPure XP beads at a beads-to-sample ratio of 0.7:1 to remove long fragments (>600 bp), recovering the remaining DNA in the reaction using a beads-to-sample ratio of 2:1. Sequencing was performed by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 2000/2500 platform (see Supplementary Table 2 for details). Library preparation was performed using custom Nextera primers as described for ATAC-seq4.
ATAC-seq
Open chromatin mapping was performed using the ATAC-seq method as described4 with minor adaptations for K562 cells. In each experiment, 1 × 105 cells were washed once in 50 μl PBS, resuspended in 50 μl ATAC-seq lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.01% IGEPAL CA-630), and centrifuged for 10 min at 4°C. Upon centrifugation, the pellet was washed briefly in 50 μl MgCl2 buffer (10 mM Tris pH 8.0 and 5 mM MgCl2) before incubating in the transposase reaction mix (12.5 μL 2× TD buffer, 2 μL transposase (Illumina), and 10.5 μL nuclease-free water) for 30 min at 37°C. After DNA purification with the MinElute kit, 1 μl of the eluted DNA was used in a qPCR reaction to estimate the optimum number of amplification cycles. Library amplification was followed by SPRI size-selection to exclude fragments larger than 1,200 bp. DNA concentration was measured with a Qubit fluorometer (Life Technologies).
Sequencing data processing
Reads were trimmed using skewer20. Trimmed reads were aligned to the hg19/GRCh37 assembly of the human genome using Bowtie221 with the “--very-sensitive” parameter and allowing for multi-mapper reads according to the aligner’s default. Duplicate reads were marked and removed using Picard. Unless otherwise stated, all downstream analyses were performed on non-duplicate reads. For ChIPmentation and ATAC-seq data, we adjusted the read start positions to represent the center of the transposition event. To that end, reads aligning to the plus strand were offset by +4 bp, and reads aligning to the minus strand were offset by −5 bp as described4. Genome browser tracks were created with the genomeCoverageBed command in bedtools22 and normalized such that each value represents the read count per basepair per million reads. Finally, the UCSC Genome Browser’s bedGraphToBigWig tool was used to produce a bigWig file. Peak calling was performed with MACS223. For both ChIP-seq and ChIPmentation data, MACS2 was run independently for biological replicates using a bandwidth of 200 bp and the matched IgG control as background. For broad his-tone marks (H3K27me3, H3K36me3) the “--broad”, “--nomodel”, “--extsize 73”, and “--pvalue 1e-3” flags and arguments were provided. After ensuring consistency among replicates, downstream analysis was performed on peaks called from merged biological replicates in the same way as described. Comparisons between replicates were done by calculating the fraction of top 1%, 2%, 5%, 10%, 25%, 50%, and 100% peaks (based on a ranking by increasing q-value) that overlap peaks from the other replicate. The same comparison was performed between ChIP-seq and ChIPmentation data, and between ChIPmentation samples produced with different number of cells (with both replicates combined).
Bioinformatic analysis
For the correlation analysis of ChIP-seq and ChIPmentation samples, we excluded ENCODE blacklisted regions and regions with average mappability score equal or lower than 0.2 to minimize bias. We counted reads in 1,000 bp windows genome-wide and normalized the counts to the total non-duplicate reads per sample. We then calculated Pearson correlation coefficients and plotted the base-2 logarithm of the signal, comparing biological replicates, different techniques (ChIP-seq versus ChIPmentation), and different numbers of cells, in the latter two cases based on merged reads from biological replicates. To compare signal-to-noise ratio, we calculated the fraction of reads in peaks (FRiP), defined as the number of non-duplicate reads that overlap called peaks for the same sample over the total number of non-duplicate reads. To investigate the insertions patterns created by the Tn5 transposase around transcription factors, we used HOMER24 for de novo motif discovery in ChIP-seq peaks for CTCF, GATA1, PU.1, and REST with a masked genome background. We retrieved a 400 bp window around each motif and used these windows to count four signals: Tn5 insertion events for ChIPmentation of the respective factor, ATAC-seq hits (5′ position of overlapping reads), DNase-seq cut sites in K562 cells from ENCODE25, and Tn5 insertion sites (5′ position of overlapping reads) for a Nextera whole genome sequencing sample prepared from genomic DNA. To normalize the ChIPmentation and ATAC-seq signals, we divided the observed value at each position by the exponential of the Nextera background signal at the same position. For visualization purposes, normalized signal was averaged over all peaks, smoothed with a 20 bp Hanning window, and Z score transformed. To assess the periodicity between Tn5 insertion sites, after removing duplicate reads we calculated the distances between each pair of Tn5 insertion sites (5′ position) of a single biological replicate of H3K4me3 ChIPmentation. We also analyzed Tn5 insertion patterns around nucleosomes. Using the NucleoATAC software15 we inferred center-of-nucleosome (dyad) positions based on ATAC-seq samples for the GM12878 cell line4, and we counted insertion events for ChIPmentation of H3K4me1 in a 300 bp window centered on the GM12878 dyads.
Supplementary Material
Acknowledgements
We would like to thank the Biomedical Sequencing Facility at CeMM for assistance with next generation sequencing, the Superti-Furga lab for contributing cells, and all members of the Bock lab for their help and advice. This work was performed in the context of the BLUEPRINT project (European Union’s Seventh Framework Programme grant agreement No. 282510) and funded in part by the ERA-NET project CINOCA (FWF grant agreement no. I 1626-B22). C.S. was supported by a Feodor Lynen Fellowship of the Alexander von Humboldt Foundation. N.C.S. was supported by a long-term fellowship of the Human Frontier Science Program (LT000211/2014). C.B. was supported by a New Frontiers Group award of the Austrian Academy of Sciences.
Footnotes
Accession Codes
In total, 52 libraries were sequenced for ChIPmentation, 24 libraries for standard ChIP-seq, nine libraries for ChIP-tagmentation, and two libraries for ATAC-seq. All sequencing data reported in this paper have been submitted to the GEO database and are available under accession number GSE70482. A detailed protocol and interactive browser tracks can be found at http://chipmentation.computational-epigenetics.org/.
Competing Financial Interests
C.S. and C.B. declare competing financial interests in the form of a pending patent application for ChIPmentation.
References
- 1.Park PJ. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furey TS. Nat Rev Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adey A, et al. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adli M, Zhu J, Bernstein BE. Nat Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobsen JS, et al. BMC Genomics. 2015;16:46. doi: 10.1186/s12864-014-1195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lara-Astiaso D, et al. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng JH, et al. Dev Cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Sachs M, et al. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankaranarayanan P, et al. Nat Methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 11.Zwart W, et al. BMC Genomics. 2013;14:232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Q, Johnston J, Zeitlinger J. Nat Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HS, Pugh BF. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter DR, Song L, Mukherjee S, Furey TS, Crawford GE. Genome Res. 2013;23:1118–1129. doi: 10.1101/gr.150482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schep A, et al. bioRxiv. 2015 doi: 10.1101/016642. [Google Scholar]
- 16.Adams D, et al. Nat Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 17.Bock C. Genome Med. 2014;6:41. doi: 10.1186/gm557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satterlee JS, Schübeler D, Ng HH. Nat Biotechnol. 2010;28:1039–1044. doi: 10.1038/nbt1010-1039. [DOI] [PubMed] [Google Scholar]
Methods-only References
- 19.Farlik M, et al. Cell Reports. 2015;10:1386–1397. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Lei R, Ding SW, Zhu S. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Salzberg SL. Nature Methods. 2012;9:357–U354. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinlan AR, Hall IM. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinz S, et al. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurman RE, et al. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.