Abstract
Two recent studies by Van Wynsberghe et al. and Perales et al. in the nematode C. elegans have demonstrated a new function of the Period protein homolog LIN-42 in negatively regulating microRNA (miRNA) biogenesis at the transcriptional level. LIN-42 is a complex gene with 4 isoforms and multiple functions including the regulation of molting, developmental timing and entry into dauer. These recent studies uncover an additional function of LIN-42 as a negative regulator of miRNA transcription. Approximately 95% of miRNAs present in eggs and 33% of miRNAs present in L4 stage worms were upregulated in lin-42 mutant worms relative to wild type (WT) worms, suggesting that LIN-42 globally regulates miRNA biogenesis. Expression from both a let-7 miRNA and a lin-4 miRNA transcriptional reporter were enhanced in the absence of lin-42. Additionally, chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) of late larval stage worms showed that LIN-42 bound the let-7 promoter, suggesting that LIN-42 affects mature miRNA levels by inhibiting their transcription. In addition to miRNAs, LIN-42 also predominantly bound to the promoters of many diverse protein-coding genes. These findings support the action of LIN-42 at multiple points within the heterochronic and other regulatory pathways to impact a multitude of functions including developmental timing.
Keywords: C. elegans, developmental timing, heterochronic, let-7, LIN-42, miRNA, Period
Abbreviations
- bp
base pair
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation followed by high throughput sequencing
- gf
gain of function
- lf
loss of function
- miRNA
microRNA
- pre-miRNA
precursor microRNA
- pri-miRNA
primary microRNA
- RISC
RNA induced silencing complex
- TF
transcription factor
- TSS
transcription start site
- UTR
untranslated region
- WT
wild type
LIN-42, a Complicated Protein
Proper timing of molecular events is essential for appropriate development and behaviors. In C. elegans, LIN-42 acts as a linker of developmental timing and circadian rhythmic behaviors through its homology with period “clock” proteins and its role in development. The multiple isoforms, bipartite function and oscillatory pattern of LIN-42 expression throughout development suggest that it is a complicated protein with many different functions throughout development.
Mutations in LIN-42 cause a variety of phenotypes related to development. As described in more detail below, many cell types develop precociously in the absence of LIN-42. In addition to hypodermal seam cell development, gonad migration, vulval precursor cell development and sex myoblast development occurs precociously in lin-42 mutant worms.1 Lin-42 mutant worms are egg laying defective.1 They also have difficulty shedding cuticles and are thus slightly dumpy; this phenotype is exasperated in later stage worms.2,3 LIN-42 is important for negatively regulating entry into dauer.4 By acting in opposition to the nuclear receptor and heterochronic gene, daf-12, LIN-42 promotes continuation through the molting cycle under normal or mild stress conditions.4 Like other circadian rhythm behaviors, locomoter activity in C. elegans is entrainable by both light-dark cycles and low-amplitude temperature cycles.5 Since lin-42 mutant worms display altered locomotor activity rhythms,5 in addition to regulating many developmental events, LIN-42 also regulates circadian rhythmic behavior in C. elegans.
Multiple isoforms of LIN-42 have been identified (Fig. 1).1,2 LIN-42B and LIN-42C are transcribed from the same promoter, and alternative splicing within exon 5 determines whether LIN-42B or LIN-42C is ultimately translated.1 In the case of LIN-42B, the truncated exon 5 is spliced to exons 6 through 9 that are located approximately 3 kb downstream.1 LIN-42A is transcribed from a second promoter and overlaps with exons 6 through 9 of LIN-42B.1 Thus, LIN-42C and LIN-42A sequences are mostly encompassed within LIN-42B sequence (Fig. 1). Homology to the defining protein-interaction PAS domain of Period proteins, which contains a cytoplasmic localization domain, is found within LIN-42B and LIN-42C.2 In addition, the SYQ and LT domains (named after their conserved amino acids) found in LIN-42A and LIN-42B are also found within Period proteins.1 The LT domain is predicted to contain a nuclear localization sequence.1
Figure 1.
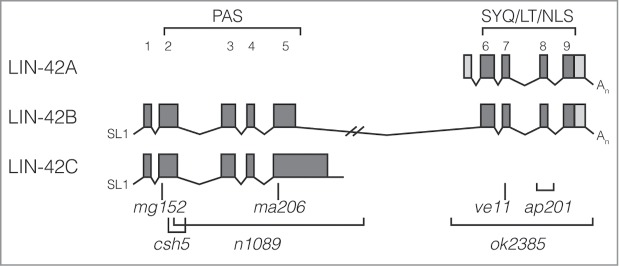
The lin-42 gene has a complex structure. Depiction of the 3 isoforms of LIN-42 as described by WormBase. These isoforms were previously described as lin-42d, lin-42c, and lin-42a, respectively.1 In addition, a fourth isoform (previously described as lin-42b) that overlaps exons 1–7 of LIN-42B and contains a final exon far downstream of exon 9, has also been described.1 Exons and untranslated regions are respectively shaded dark and light gray. Conserved domains and amino acids are marked above the gene diagrams, while select alleles are marked below the gene diagrams.
Because of its homology to the PAS domain of Period proteins, LIN-42C was initially thought to encompass the most important region of LIN-42, however the importance of the region encompassed by LIN-42A was later recognized. Tennessen et al. found that expression of the LIN-42A region could rescue the precocious alae phenotype in either lin-42(mg152) or lin-42(ve11) worms that contain mutations in the LIN-42C and LIN-42A regions, respectively.1 In contrast, expression of the LIN-42C region of LIN-42 could only rescue this phenotype in lin-42(mg152) worms that contain mutations in the LIN-42C region.1 Intriguingly, overexpression of LIN-42C isoform can antagonize phenotypes of lin-42(ok2385), which contains a mutation in the LIN-42A region.3 Thus in addition to maintaining some overlap of function, the different LIN-42 isoforms may also regulate one another to ultimately dictate proper LIN-42 expression and function.
Period proteins are members of the core circadian oscillator that regulates circadian rhythms.6 As part of an autoregulatory feedback loop that is crucial to the time-keeping ability of the circadian oscillator, Period levels cycle over an approximate 24 hour period.6 Both lin-42 mRNA and protein levels also cycle.2-4 Instead of cycling every day, however, LIN-42 oscillates in accordance with the molt throughout the larval stages.2-4 This suggests that LIN-42 plays multiple or continued roles throughout development. LIN-42 is enriched in the nuclei relative to the cytoplasm, but is present in both locations.2 The expression pattern of LIN-42 varies in different cell types, and the timing of expression of the different LIN-42 isoforms also varies.2-4 LIN-42B and C peak during the intermolt, while LIN-42A peaks at the molt.2-4 In fact, LIN-42 is necessary for proper molting.3 Lin-42(ok2385) worms, which contain a deletion that eliminates all of LIN-42A and LIN-42B exons 6–9, molt asynchronously and spend more time in the lethargic stage prior to ecdysis.3
In summary, the complicated structure, diverse temporal and spatial expression patterns, multiple phenotypes and homology to the essential circadian rhythm Period protein, places LIN-42 in a unique situation to regulate diverse pathways. Such ability suggests that LIN-42 uses a common mechanism to regulate multiple genes that ultimately affect C. elegans development and behavior.
LIN-42 and the Heterochronic Pathway
A series of highly regulated molecular interactions ultimately controls C. elegans development through 4 larval stages into adulthood. Genes associated with this pathway have been identified by their heterochronic phenotypes that display developmental events in the correct cell lineages, but at the wrong developmental time.7 At its core, the heterochronic pathway is composed of a series of genes whose timing of expression occurs during specific larval stages.7 Placement of these genes in the heterochronic pathway has been established through their genetic interactions with other members of the pathway. Complicating this analysis, however, is the fact that the pathway does not act simply in a linear fashion (Fig. 2). Instead, some downstream genes repress genes upstream in the pathway, while other genes have multiple targets.7,8
Figure 2.
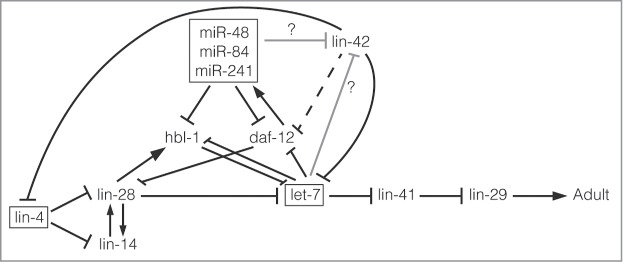
LIN-42 and the heterochronic pathway. A simplified depiction of the heterochronic pathway in C. elegans. miRNAs that act in the pathway are boxed. The 3′UTR of LIN-42C has a potential let-7 miRNA binding site, suggesting that LIN-42 could be regulated by let-7 or its sisters (miR-48, miR-84 and miR-241).17 Recent work shows that LIN-42 directly inhibits primary lin-4 and let-7 transcription.15,16 In addition, LIN-42 antagonizes the ligand-free form of DAF-12 to inhibit entry into dauer.4
Heterochronic defects can be analyzed in any cell lineage that displays specific, methodical phenotypes throughout development. For example, most lateral hypodermal cells, otherwise known as seam cells, divide before each molt. One daughter cell fuses with the hypodermis while the other daughter cell continues to divide. However, at the beginning of the L2 stage, both daughter cells undergo an extra cell division and at the L4-to-adult molt the seam cells exit the cell cycle, fuse and secrete a cuticular structure called alae. Thus precocious heterochronic mutants exhibit alae formation prior to the adult stage due to skipping of an earlier developmental event, while seam cells in retarded heterochronic mutants may never exit the cell cycle. Consequently depending on the larval stage affected, heterochronic mutants may vary in the number of seam cells and the timing of seam cell fusion and alae production.8
Central to the heterochronic pathway are the first discovered microRNAs (miRNAs), lin-4 and let-7. miRNAs act post-transcriptionally to inhibit gene expression by imperfectly binding to target gene mRNA and, in association with the RNA induced silencing complex (RISC), causing mRNA degradation and/or translation inhibition.9 Though miRNAs function as ∼22 nt RNAs they are encoded in the genome and transcribed by RNA polymerase II into primary miRNAs (pri-miRNA) that can be several kilobases in length.10 Each pri-miRNA is capped and polyadenylated before being processed by the Drosha/Pasha complex into the ∼70 nt precursor miRNA (pre-miRNA).10 The pre-miRNA is exported to the cytoplasm before being further processed by Dicer into the mature miRNA.10 Each of these steps in miRNA biogenesis is highly regulated to ensure the correct amount of miRNA is produced in the right cells at the right time.10 Accordingly, improper amounts of miRNAs have been associated with multiple human cancers, various diseases and developmental defects.11,12 Though some regulators of miRNA biogenesis have been identified, many more likely remain to be uncovered.10
LIN-42 was identified as a member of the heterochronic pathway in C. elegans since loss of function mutations in lin-42 cause precocious alae production.2 However, precise placement of LIN-42 in the heterochronic pathway has been hindered by the numerous genetic interactions exhibited by lin-42 mutants (Table 1).1,2,13 LIN-42 functions upstream of lin-29 in the heterochronic pathway, since mutations in lin-42 have no effect on the retarded phenotype of lin-29 mutants.1,13 However, mutations in lin-42 suppress the retarded phenotypes found in lin-14 gain-of-function mutants or lin-4, let-7 or alg-1 loss-of-function mutants, and mutations in the latter also suppress the precocious phenotypes of lin-42 mutants.1,13-16 LIN-42 acts synergistically with hbl-1, miR-48 or lin-41, since mutations in lin-42 and any of these genes causes enhanced precocious phenotypes.13,14 Lin-42 and daf-12 mutations mutually suppress each other.1 In contrast, mutations in lin-46 have no effect on lin-42 precocious phenotypes.1
Table 1.
Genetic interactions of lin-42 in the heterochronic pathway
| Worm Strain | Alae Phenotype | References |
|---|---|---|
| lin-42(lf) | precocious | 1,14,16 |
| lin-4(e912) | retarded | 1,13,16, |
| lin-4(e912);lin-42(lf) | wt | 1,13,16 |
| lin-14(gf) | retarded | 13 |
| lin-14(gf);lin-42(lf) | wt | 13 |
| lin-46(ma174) | wt | 1 |
| lin-46(ma174);lin-42(lf) | precocious | 1 |
| mir-48(ve33) | precocious | 13 |
| mir-48(ve33);lin-42(lf) | enhanced precocious | 13 |
| hbl-1(mg285) | precocious | 14 |
| hbl-1(mg285);lin-42(lf) | enhanced precocious | 14 |
| daf-12(lf) | wt | 1 |
| daf-12(lf);lin-42(lf) | precocious | 1 |
| let-7(n2853) | retarded | 3,14,16 |
| let-7(n2853);lin-42(lf) | wt | 3,14,16 |
| lin-41(ma104) | precocious | 14 |
| lin-41(ma104);lin-42(lf) | enhanced precocious | 14 |
| lin-29(n546) | retarded | 1,13 |
| lin-29(n546);lin-42(lf) | retarded | 1,13 |
| alg-1(ma192) | retarded | 16 |
| alg-1(ma192);lin-42(lf) | wt | 16 |
gf – gain of function; lf – loss of function refers to one or more alleles or RNAi experiments.
Though a lin-42 mutation suppresses the vulvaless phenotype of lin-4(ma161) worms or the bursting phenotype of let-7(n2853) worms, these phenotypes are not suppressed in lin-4(e912) or let-7(mn112) worms (Table 2).14-16 The former alleles reduce, but do not eliminate lin-4 or let-7 expression while the latter alleles are both null. Thus, at a minimum, LIN-42 functions through lin-4 and let-7 miRNAs to impact the heterochronic pathway (Fig. 2). Intriguingly, since the 3′untranslated region (UTR) of lin-42 has a putative let-7 binding site,17 LIN-42 may also be subject to regulation by miRNAs like let-7 or the let-7 sisters (miR-48, miR-84, and miR-241).
Table 2.
Vulva phenotypes in lin-42 mutant worms
| Category | Worm Strain | Phenotype | References |
|---|---|---|---|
| Vulva Development | lin-42(lf) | Wt | 16 |
| lin-4(e912) | Vulvaless | 16 | |
| lin-4(e912);lin-42(lf) | Vulvaless | 16 | |
| lin-4(ma161) | Vulvaless | 16 | |
| lin-4(ma161);lin-42(lf) | Wt | 16 | |
| Vulva Bursting | lin-42(lf) | None | 15,16 |
| let-7(mn112) | Burst | 16 | |
| let-7(mn112);lin-42(lf) | Burst | 16 | |
| let-7(n2853) | Burst | 14-16 | |
| let-7(n2853);lin-42(lf) | None | 14-16 |
lf – loss of function refers to one or more alleles or RNAi experiments.
The mechanism by which LIN-42 affects these miRNAs is beginning to become clear based on 2 recent papers. Both Van Wynsberghe et al. and Perales et al. found that levels of mature lin-4 and let-7 are increased in lin-42 mutant worms at multiple stages throughout development.15,16 This effect of LIN-42 on miRNAs extended far beyond the heterochronic pathway however, as 95% of miRNAs in eggs and ∼33% of miRNAs in L4 stage worms were upregulated in the absence of lin-42.15 This increase in mature miRNA levels was accompanied by an increase in pri-let-7 and pri-lin-4 levels.15 Surprisingly, the natural oscillatory patterns of primary let-7 and lin-4 levels were not affected by LIN-42.15,18,19 These results suggested that LIN-42 regulates the transcription of miRNAs, but not the timing or oscillatory nature of primary miRNA expression. This was further verified by the use of transcriptional reporters for let-7 and lin-4.15,16 GFP mRNA and protein levels expressed from these reporters was increased in the absence of lin-42.15,16
Thus LIN-42 regulates a multitude of miRNAs by inhibiting primary miRNA transcription. Such regulation is likely carefully spatially and temporally controlled, however, as LIN-42 is expressed in multiple cell types throughout development. Indeed, this regulation has clear effects on cell functions beyond seam cells and the heterochronic pathway. For example, mutations in lin-42 suppress lsy-6 miRNA neuronal cell fate specification phenotypes,16 likely by increasing lsy-6 levels above a required threshold. Additionally, the finding that LIN-42 also regulates primary miRNA transcription in embryos suggests that LIN-42 also has important functions prior to larval development.15
Gene Regulation by Period Proteins
Period proteins of diverse organisms use varying mechanisms to inhibit gene expression and thus influence the circadian clock. In Drosophila, the period protein acts indirectly to regulate circadian rhythms. Drosophila Period binds Clock to inhibit dimerization of the transcriptional activator of Clock-Cycle, which once activated binds to enhancer sequences associated with the Period and Timeless genes, thereby completing the core circadian feedback clock loop.20,21 However, in mammalian systems the period protein binds to the Clock-Cycle homologs CLOCK-BMAL1 while bound to the enhancer regions (E-boxes) of their target genes.6 An alternative mechanism by which Period could regulate target genes is to directly bind to target gene regulatory sequences. Binding to target gene enhancer sequences could inhibit transcription by blocking activator binding, while binding to target gene silencer sequences could recruit a transcriptional repressor. Though all of these potential mechanisms result in inhibition of gene transcription, the Drosophila mechanism does not involve target gene binding while the remaining mechanisms do. Recent work by Perales et al. shows that LIN-42 targets can be isolated by chromatin immunoprecipitation (ChIP).16 Thus, LIN-42 must bind target genes either directly or through interactions with a bound transcription factor, as in mammalian systems.
In addition to associating with non-protein-coding genes like miRNAs, LIN-42 also interacted with many protein-coding genes.16 Chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) analysis showed that though LIN-42 bound intronic and exonic sequences of protein-coding genes, LIN-42 predominantly associated with both protein-coding and non-protein-coding target genes at either the transcription start site (TSS) or approximately 750 base pairs upstream of the TSS.16 LIN-42 target genes fit into multiple different categories including those related to transport, signal transduction, growth, locomotion, cell cycle, and stress response.16 Corroborating the finding that LIN-42 regulates almost all miRNAs in eggs, many L4 stage LIN-42 target genes are involved in reproduction and embryo development.15,16 Thus throughout development, LIN-42 associates with target genes to regulate their transcription and thus influence multiple developmental and behavioral phenotypes.
Conclusions
Altogether these new findings further confirm the complicated nature of the period protein homolog LIN-42, while illuminating some mechanisms by which LIN-42 carries out diverse functions throughout the course of development. Period proteins are known to inhibit gene regulation by acting as inhibitors of target gene transcription.6,20 The finding that LIN-42 also performs this function further supports its identity as a Period protein homolog. Period proteins are thought to predominantly control circadian rhythms by acting on protein-coding gene targets, though circadian clocks have been shown to impact miRNA biogenesis pathways at multiple steps and miRNAs have been found to post-transcriptionally regulate core circadian clock feedback loops.22-24 Studies have also found that miRNAs can be influenced by core circadian oscillator components in Drosophila and mammals.24 For example, miR-279 has recently been shown to impact JAK/STAT signaling and thus rest/activity circadian rhythms in Drosophila,25 and miR-219–1 was found to be a direct transcriptional target of the oscillator protein CLOCK.24 The new findings by Van Wynsberghe et al. and Perales et al. nicely show that the effects of the circadian oscillator on miRNA expression encompass more than just a few miRNAs, but instead transcriptionally regulate a large group of miRNAs and other mRNAs to ultimately affect developmental timing and circadian rhythms (Fig. 3). Thus miRNAs may be used more than previously thought by core circadian proteins to impact circadian rhythmic behavior.
Figure 3.
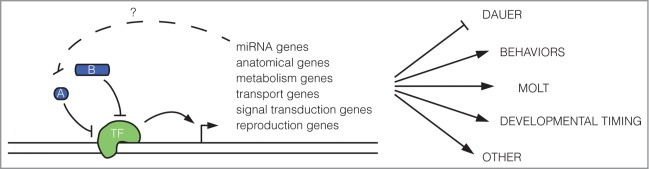
The role of LIN-42 in regulating target gene transcription. LIN-42 inhibits transcription of multiple primary miRNAs and physically associates with target miRNAs and mRNAs at the transcription start site (TSS) and ∼750 bp upstream of the TSS.15,16 Based on the mechanism by which Period inhibits target gene transcription in mammalian cells,6 it is likely that LIN-42 (A & B) bind a transcriptional activator (TF) to inhibit gene expression. LIN-42 targets a diverse set of genes including those involved in forming anatomical structures, metabolism, transport, signal transduction, reproduction and other functions. By inhibiting expression of these targets, LIN-42 ultimately impacts developmental timing, entrance into dauer, behaviors including locomotion, and molting throughout development.
Once again, the regions of LIN-42 that lack the notorious Period PAS domain are particularly important for this function.15,16 But, numerous questions remain regarding the roles, intra-relationships and clear expression patterns of the different LIN-42 isoforms. Though much attention has been paid to the requirement of LIN-42 for larval stage functions, little has been done to understand its function in embryo stage worms. The new findings that LIN-42 regulates ∼95 percent of miRNAs at this stage and that LIN-42 targets in L4 stage worms include those involved in embryo development suggest that LIN-42 also functions at this early time in development.15,16 Additionally, little is known about how LIN-42 is regulated to cause its oscillatory expression pattern throughout development. Period proteins are subject to phosphorylation-associated degradation pathways,6 and homologs of Period kinases are present in C. elegans.14,21 Thus similar mechanisms may be at work in C. elegans. The 3′UTR of lin-42 also has putative let-7 binding sites,17 suggesting that miRNAs might also post-transcriptionally regulate LIN-42. Future work will continue to address these and other questions to determine how a single protein can impact so many different pathways throughout development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Funding was provided by Colgate University (P.M.V.W) and grants by the US National Institutes of Health (GM071654), Keck Foundation and Peter Gruber Foundation (A.E.P).
References
- 1. Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol [Internet] 2006. [cited 2012 Dec 11]; 289:30-43. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16300753; PMID:16300753; http://dx.doi.org/ 10.1016/j.ydbio.2005.09.044 [DOI] [PubMed] [Google Scholar]
- 2. Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science (80- ) [Internet] 1999. [cited 2012 Dec 19]; 286:1141-6. Available from: http://www.sciencemag.org/cgi/doi/ 10.1126/science.286.5442.1141; http://dx.doi.org/ 10.1126/science.286.5442.1141 [DOI] [PubMed] [Google Scholar]
- 3. Monsalve GC, Van Buskirk C, Frand AR. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr Biol [Internet] 2011. [cited 2012 Nov 27]; 21:2033-45. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22137474; PMID:22137474; http://dx.doi.org/ 10.1016/j.cub.2011.10.054 [DOI] [PubMed] [Google Scholar]
- 4. Tennessen JM, Opperman KJ, Rougvie AE. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development [Internet] 2010. [cited 2012 Dec 5]; 137:3501-11. Available from: Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2947761&tool=pmcentrez&rendertype=abstract; PMID:20843862; http://dx.doi.org/ 10.1242/dev.048850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simonetta SH, Migliori ML, Romanowski A, Golombek D a. Timing of locomotor activity circadian rhythms in Caenorhabditis elegans. PLoS One [Internet] 2009. [cited 2014 Aug 4]; 4:e7571. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2764868&tool=pmcentrez&rendertype=abstract; PMID:19859568; http://dx.doi.org/ 10.1371/journal.pone.0007571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci [Internet] 2006. [cited 2012 Nov 30]; 119:4793-5. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17130292 PMID:17130292; http://dx.doi.org/ 10.1242/jcs.03174 [DOI] [PubMed] [Google Scholar]
- 7. Rougvie A, Moss E. Developmental transitions in C. elegans larval stages. Curr Top Dev Biol [Internet] 2013. [cited 2014 Jul 28]; 105:153-80. Available from: http://books.google.com/books?hl=en&lr=&id=9_Up_WN8vkQC&oi=fnd&pg=PA153&dq=Developmental+Transitions+in+C.+elegans+Larval+Stages&ots=7vWBN-HvWF&sig=GtAZh_EdDdLg0U0LxzTxC1yJMis; PMID:23962842; http://dx.doi.org/ 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- 8. Resnick TD, McCulloch KA, Rougvie AE. miRNAs give worms the time of their lives: small RNAs and temporal control in Caenorhabditis elegans. Dev Dyn [Internet] 2010. [cited 2014 Jul 28]; 239:1477-89. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20232378; PMID:20232378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet [Internet] 2012. [cited 2012 Oct 27]; 13:271-82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22411466; PMID:22411466 [DOI] [PubMed] [Google Scholar]
- 10. Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol [Internet] 2012. [cited 2012 Dec 7]; 48:1-18. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23163351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abbott AL. Uncovering new functions for microRNAs in Caenorhabditis elegans. Curr Biol [Internet] 2011. [cited 2014 Aug 4]; 21:R668-71. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3189681&tool=pmcentrez&rendertype=abstract; PMID:21920301; http://dx.doi.org/ 10.1016/j.cub.2011.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev [Internet] 2011. [cited 2012 Oct 25]; 91:827-87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21742789; PMID:21742789; http://dx.doi.org/ 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- 13. Abrahante J, Miller E, Rougvie A. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen::green fluorescent protein fusion gene. Genetics [Internet] 1998; 149:1335-51. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1460241&tool=pmcentrez&rendertype=abstract; PMID:9649524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee D, Kwok A, Lin S-Y, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell [Internet] 2005. [cited 2012 Nov 20]; 8:287-95. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15691769; PMID:15691769; http://dx.doi.org/ 10.1016/j.devcel.2004.12.006 [DOI] [PubMed] [Google Scholar]
- 15. Van Wynsberghe PM, Finnegan EF, Stark T, Angelus EP, Homan KE, Yeo GW, Pasquinelli AE. The Period protein homolog LIN-42 negatively regulates microRNA biogenesis in C. elegans. Dev Biol [Internet] 2014. [cited 2014 Jul 28]; 390:126-35. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24699545; PMID:24699545; http://dx.doi.org/ 10.1016/j.ydbio.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perales R, King DM, Aguirre-Chen C, Hammell CM. LIN-42, the Caenorhabditis elegans PERIOD homolog, negatively regulates microRNA transcription. PLoS Genet [Internet] 2014. [cited 2014 Jul 18]; 10:e1004486. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25032706; PMID:25032706; http://dx.doi.org/ 10.1371/journal.pgen.1004486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature [Internet] 2000; 403:901-6. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10706289; PMID:10706289; http://dx.doi.org/ 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- 18. Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol [Internet] 2011. [cited 2012 Dec 19]; 18:302-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3077891&tool=pmcentrez&rendertype=abstract; PMID:21297634; http://dx.doi.org/ 10.1038/nsmb.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bracht JR, Van Wynsberghe PM, Mondol V, Pasquinelli AE. Regulation of lin-4 miRNA expression, organismal growth and development by a conserved RNA binding protein in C. elegans. Dev Biol [Internet] 2010. [cited 2012 Dec 19]; 348:210-21. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2982876&tool=pmcentrez&rendertype=abstract; PMID:20937268; http://dx.doi.org/ 10.1016/j.ydbio.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hardin PE. The circadian timekeeping system of Drosophila. Curr Biol [Internet] 2005. [cited 2012 Oct 31]; 15:R714-22. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16139204; PMID:16139204; http://dx.doi.org/ 10.1016/j.cub.2005.08.019 [DOI] [PubMed] [Google Scholar]
- 21. Temmerman L, Meelkop E, Janssen T, Bogaerts A, Lindemans M, Husson SJ, Beets I, Schoofs L. C. elegans homologs of insect clock proteins: a tale of many stories. Ann N Y Acad Sci [Internet] 2011. [cited 2014 Aug 4]; 1220:137-48. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21388411; PMID:21388411; http://dx.doi.org/ 10.1111/j.1749-6632.2010.05927.x [DOI] [PubMed] [Google Scholar]
- 22. Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci [Internet] 2011. [cited 2014 Aug 1]; 124:311-20. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3021995&tool=pmcentrez&rendertype=abstract; PMID:21242310; http://dx.doi.org/ 10.1242/jcs.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu K, Wang R. MicroRNA-mediated regulation in the mammalian circadian rhythm. J Theor Biol [Internet] 2012; 304:103-10. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22554948; PMID:22554948; http://dx.doi.org/ 10.1016/j.jtbi.2012.03.037 [DOI] [PubMed] [Google Scholar]
- 24. Mehta N, Cheng H-YM. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J Mol Biol [Internet] 2013. [cited 2014 Jul 22]; 425:3609-24. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23142644; PMID:23142644; http://dx.doi.org/ 10.1016/j.jmb.2012.10.022 [DOI] [PubMed] [Google Scholar]
- 25. Luo W, Sehgal A. Regulation of a circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell [Internet] 2012; 148:765-79. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22305007; PMID:22305007; http://dx.doi.org/ 10.1016/j.cell.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]


