Abstract
In Clostridium difficile, erm(B) genes are located on mobile elements like Tn5398 and Tn6215. In previous studies, some of these elements were transferred by conjugation-like mechanisms, mobilized in trans by helper conjugative systems. In this study, we analyzed the genomes of several recipient strains that acquired either Tn5398 or Tn6215-like elements. We demonstrated that the integration of the transposons in the genome of the recipient cell was always due to homologous recombination events, involving exchange of large chromosomal segments. We did not observed transposon transfer to a C. difficile strain in presence of DNAse, suggesting that a possible transformation-like mechanism occurred in this recipient.
Keywords: antibiotic resistance, Clostridium difficile, erythromycin, mobile elements
Introduction
In Clostridium difficile, resistance to the macrolide-lincosamide-streptogramin B (MLSB) group of antibiotics is generally conferred by erm(B) genes. In strain 630, 2 copies of this gene are located on the mobilizable non-conjugative Tn5398, previously shown to be transferable by a conjugation-like mechanism.1-3 Even though this element has been extensively studied, its transfer mechanism is still not fully understood.
Several other erm(B)-containing elements showing important genetic diversity have been detected in clinical isolates of C. difficile.1,4,5 In particular, the conjugative transposon Tn6194 has recently been detected in the genome of PCR-ribotype 027 clinical isolates.6,7 Even more recently, the transposon Tn6215 has been described in a PCR-ribotype 010 isolate. Transfer of this element has been shown to be mediated by a bacteriophage, even if its conjugation-like transfer has also been observed.8
The aim of this study was to investigate the transfer mechanisms of the Tn5398 from C. difficile 630, the Tn6194-like element from C. difficile CII7, and the Tn6215-like element from C. difficile F17. Two different non-toxigenic MLSB-susceptible strains of C. difficile, CD37 and CD13, were used as recipient strains.
Results and Discussion
A 13015bp erm(B)-containing transposon was identified in the genome of the C. difficile F17 strain. This element shared 99.9% identity with Tn6215 from C. difficile CD80, another PCR-ribotype 010 strain, and had the same genomic localization. Analysis of the GC% content suggests that this element is the result of the integration of an erm(B)-containing cassette, almost identical to part of pAMβ1 from E. faecalis, within a transposon already integrated in the genome (Fig. 1).
Figure 1.
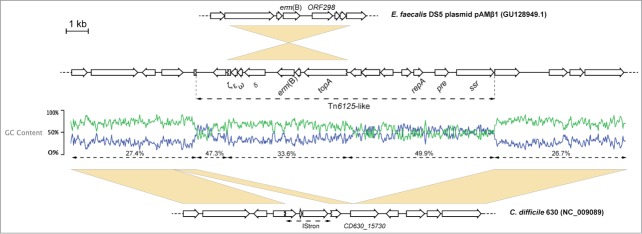
Tn6215-like from C. difficile F17. Top to bottom: position 3244 to 10038 from pAMβ1 from E. faecalis DS5; Tn6215-like and flanking regions in C. difficile F17; position 1814421 to 1827426 from C. difficile 630. Accession numbers of deposited sequences are indicated in brackets. Yellow boxes indicate regions of homology.
Previously, it has been demonstrated that both the Tn5398 from C. difficile 630 and the Tn6215-like element from C. difficile F17 integrate into the genome of the recipient strain, at the same target site of their respective donors.1,8,9 In this study, SNPs occurring in the region containing the transposon and flanking its integration sites were used to distinguish the genomes of the progeny and those of the recipient and donor strains. The SNPs comparative analysis demonstrated that large genomic fragments containing the transposons from the donors have substituted, likely by homologous recombination events, the respective orthologous chromosomal segments in the recipients. In fact, the nucleotide sequences flanking the newly acquired elements in the progeny genomes were indistinguishable from those of their respective donors (Fig. 2). The length of the transferred fragments in each genome analyzed was determined by calculating the distance between the first SNP upstream and the last SNP downstream of the transferred element, as performed for PaLoc transfer.10 Notably, a segment with a size comprised between 254458 and 256507 bp from C. difficile 630 was identified in the genome of 630xCD37A. In 630xCD13A, 2 segments of donor DNA covering a region comprised between 44869 and 46115 bp in length were detected, separated by a fragment with a length comprised between 10775 and 12064 bp corresponding to the recipient's sequence, suggesting that multiple events of homologous recombination occurred (Fig. 2A). Likewise, the acquired C. difficile F17 genome fragments were between 22048 and 26338 bp in length in F17xCD37A and between 22098 and 22306 bp in length in F17xCD13A (Fig. 2B). Therefore, no site-specific transposition but chromosomal recombination events occurred in the progeny acquiring the erm(B)-containing transposon. These results are in accordance with previously reported evidences suggesting that in C. difficile large chromosomal region exchange can occur.6 Moreover, a recent study has shown that the pathogenicity locus (PaLoc) can be transferred through a conjugation-like mechanism and integrate the genome of CD37 by homologous recombination involving flanking regions.10
Figure 2.
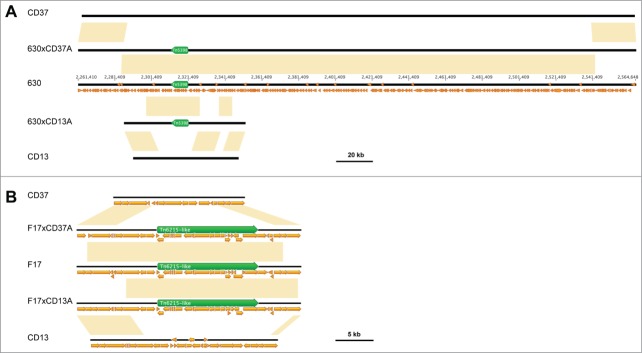
Donor-specific DNA fragments present in progeny obtained after transfer of (A) Tn5398 from C. difficile 630 and (B) Tn6215-like from C. difficile F17 to C. difficile CD37 and CD13, respectively. Yellow boxes indicate identity between sequences of progeny and those of donor or recipient strains. Small orange arrows represent CDS, and green arrows represent the erm(B)-containing elements. Sequence of 630 is numerated according to the Genbank accession number NC_009089.
Filter mating assays were performed to evaluate the effect of DNase on transfer frequencies of Tn5398 and Tn6215-like to both CD37 and CD13. Transfer of the conjugative Tn6194-like from strain CII7 9,11 was also performed, as control. Each transfer was repeated in at least 3 independent experiments. Addition of DNAse to the mix of donor and recipient lowered the transfer frequencies of Tn5398 to the CD13 recipient strain but not to CD37. Moreover, transfer of Tn6194-like or Tn6215-like was not detected when only CD13 was used as recipient in the presence of DNase (Table 2). The different behavior of the 2 strains suggested that a transformation-based transfer of the transposons could be excluded when CD37 is used as the recipient strain. The large size of the transferred fragment (>250 kb) identified in 630 × CD37A makes it also improbable that generalized transduction occurred in this progeny, while conjugation cannot be excluded, as already hypothesized for the PaLoc transfer.10
Table 2.
Transfer frequencies observed in this study
| Transfer frequency (per donor ±SD )a |
|||
|---|---|---|---|
| Donor | Recipient | no DNAse | DNase (50U) |
| 630 | CD37 | 2.99 ± 1.56 x 10–8 | 1.22 ± 0.57 x 10–8 |
| CII7 | CD37 | 6.94 ± 2.66 x 10–9 | 1.04 ± 0.47 x 10–8 |
| F17 | CD37 | 2.92 ± 0.58 x 10–9 | 4.61 ± 1.16 x 10–9 |
| 630 | CD13 | 3.87 ± 0.20 x 10–7 | 9.66 ± 7.46 x 10–9 |
| CII7 | CD13 | 1.04 ± 0.47 x 10–8 | <1.0 x10–9 |
| F17 | CD13 | 4.61 ± 1.16 x 10–9 | <1.0 x10–9 |
SD, standard deviation
On the other hand, competence for transformation can be hypothesized for the CD13 recipient strain, because the transfer of the transposon was impaired by the treatment with DNAse. However, erythromycin-resistant transformants were not obtained when CD13 cells were mixed to 2–4 μg of genomic DNA from the donors and placed on filter for 24 hours, as performed in filter-mating assays (data not shown). This result suggests that the transformation-like event is not supported by naked DNA, but physical contact between donor and recipient cells is still required, likely involving undefined and unknown DNA uptake systems. The genes predicted to code products putatively involved in competence observed in the genome of strain 630 (i.e: comE, cinA and ftsK) were also found in the genome of strains CD37 and CD13. Unique genes coding for proteins involved in transformation were not identified in CD13. Nevertheless, the genome of this strain contains many genes encoding transporters, in particular ABC transporters, which could have a role in DNA uptake.
Interestingly, DNase also affected the transfer of the Tn6194-like conjugative transposon from strain CII7 to CD13. Genome analysis revealed that integration in the recipient genome occurred through recombination events involving homologous flanking regions, instead of transposition (Fig. 3), whereas Tn6194-like from CII7 integrates the genome of CD37 by a transposition mechanism.11 A transposon sharing sequence homology with the Tn6194-like from CII7, already present in the genome of CD13, was also involved in the recombination events which led the generation of erm(B)-containing chimerical elements (Fig. 3).
Figure 3.
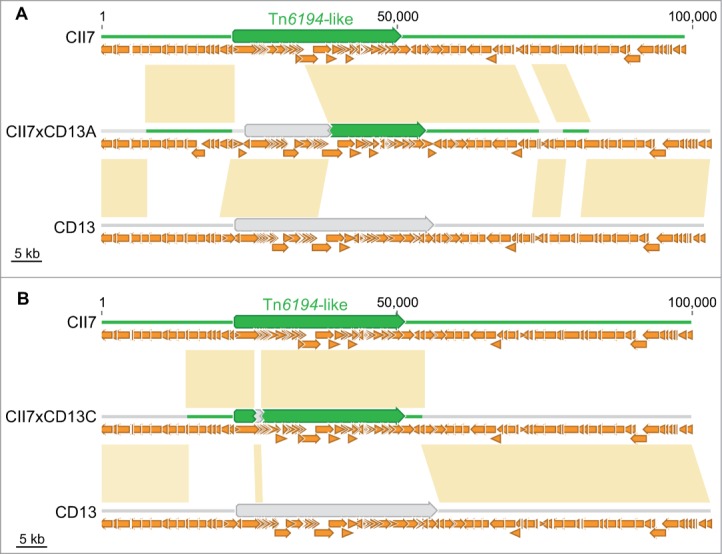
Insertion of the Tn6194-like element in (A) CII7xCD13A and (B) CII7xCD13C. Homologous recombination events involving either orthologous sequences flanking both elements and internal regions of the 2 transposons resulted in the generation of chimeric elements. Donor and recipient specific DNA sequences are shown in green and light gray, respectively. Yellow boxes indicate identity between sequences of progeny and those of donor CII7 or recipient CD13. Small orange arrows represent CDS, and big arrows represent conjugative transposons.
In conclusion, we demonstrated that Tn5398 and Tn6215 integrate the genome of C. difficile through exchange of large genomic fragments. This is in accordance with observations done recently during PaLoc's transfer between C. difficile strains10 suggesting that the mechanism involved can mobilise other regions of the genome. We also reported the peculiar in vitro behavior of the recipient strain CD13 that acquired genetic material through a transformation-like mechanism not yet observed in C. difficile, to our knowledge. Although further studies are necessary, the results obtained support the importance of homologous recombination events in both the spread of antibiotic resistance and in C. difficile genome evolution.
Materials and Methods
Bacterial strains, culture conditions and filter mating assays
Strains used in this study, listed in Table 1, were grown on Brain Heart Infusion (BHI) agar plates or in BHI broth (Oxoid Ltd, Basingstoke, UK) supplemented with 0.5% yeast extract and 0.1% L-cysteine (BHIS), at 35°C under anaerobic conditions (85% N2, 10% H2, 5% CO2). Filter-mating experiments were performed as previously described.1 When specified, 50U of DNAseI (New England Biolabs) were added to the mix of cells. Resistant colonies obtained were confirmed by PCR-ribotyping 16 and PCR detection of erm(B) using primers E5 and E6.17
Table 1.
Bacterial strains and plasmids used in this study
| Strains | Characteristics a | Source/Reference |
|---|---|---|
| CII7 | EryR CliR RifS | 12 |
| 630 | EryR CliR RifS | 13 |
| F17 | EryR CliR RifS | 14 |
| CD13 | EryS CliS RifR | 9 |
| CD37 | EryS CliS RifR | 15 |
| progeny of filter-mating assays | ||
| 630xCD37 A | EryR CliR RifR | 9 |
| 630xCD13 A | EryR CliR RifR | 9 |
| F17xCD37 A | EryR CliR RifR | 9 |
| F17xCD13 A | EryR CliR RifR | 9 |
| CII7xCD13 A | EryR CliR RifR | 9 |
| CII7xCD13 C | EryR CliR RifR | 9 |
Ery, erythromycin; Cli, clindamycin; Rif, rifampicin; R, resistant; S, susceptible.
Genomic DNA extraction
Genomic DNA extraction was performed using the NucleoBond® AXG columns and NucleoBond® Buffer Set III (Macherey-Nagel) according to the manufacturer's instructions.
Genome sequencing
The 110-bp single read genomic libraries of C. difficile CD13 and CII7xCD13C were sequenced on the Illumina HiSeq 2000 platform. The CII7xCD13A and F17xCD13A genome sequences were obtained using the 454-Genome Sequencer FLX procedure (Roche Diagnostic, Monza, Milan). The 51bp-single read genomic libraries of F17, 630xCD37A, 630xCD13A and F17xCD37A were sequenced using the Illumina HiSeq 2000 platform (GATC, Konstanz, Germany). Read-mapping analysis, gene predictions and pairwise alignment were performed with the Geneious software (Biomatters Ltd, New Zealand).
Nucleotide sequence accession number
The whole genome contigs of strains CII7xCD13A and F17xCD13A have been deposited in the European Nucleotide Archive (ENA) under accession number ‘PRJEB5538’ and can be accessed online (http://www.ebi.ac.uk/ena/data/view/PRJEB5538).
The short reads genomic data of all remaining strains sequenced in this study have been deposited in the ENA under accession number ‘PRJEB5926’ and can be accessed online (http://www.ebi.ac.uk/ena/data/view/PRJEB5926).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The research leading to these results has received funding from the European Community's Seventh Framework Program FP7/2007–2013 under grant agreement No. 237942; A.C. has received funding from MIUR-CNR for the Italian FLAGSHIP “InterOmics” project (PB.P05).
References
- 1. Farrow KA, Lyras D, Rood JI. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 2001. 147:2717-28; PMID:11577151 [DOI] [PubMed] [Google Scholar]
- 2. Mullany P, Wilks M, Tabaqchali S. Transfer of macrolide-lincosamide-streptogramin B (MLS) resistance in Clostridium difficile is linked to a gene homologous with toxin A and is mediated by a conjugative transposon, Tn5398. J. Antimicrob. Chemother 1995. 35:305-315; PMID:7759394; http://dx.doi.org/ 10.1093/jac/35.2.305 [DOI] [PubMed] [Google Scholar]
- 3. Wüst J, Hardegger U. Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother 1983. 23:784-6; PMID:6870225; http://dx.doi.org/ 10.1128/AAC.23.5.784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Spigaglia P, Carucci V, Barbanti F, Mastrantonio P. ErmB determinants and Tn916-like elements from clinical isolates of Clostridium difficile. Antimicrob. Agents. Chemother 2005. 49:2550-3; PMID:15917571; http://dx.doi.org/ 10.1128/AAC.49.6.2550-2553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spigaglia P, Barbanti F, Mastrantonio P. on behalf of the European Study Group on Clostridium difficile (ESGCD) . Multidrug resistance in European Clostridium difficile clinical isolates. J. Antimicrob. Chemother 2011. 66:2227-34; PMID:21771851; http://dx.doi.org/ 10.1093/jac/dkr292 [DOI] [PubMed] [Google Scholar]
- 6. He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holta KE, Seth-Smith HMB, Quail MA, Rance R, et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc. Natl. Acad. Sci. USA 2010. 107:7527-32; http://dx.doi.org/ 10.1073/pnas.0914322107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet 2012. 45:109-13; PMID:23222960; http://dx.doi.org/ 10.1038/ng.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P. Phage φC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio 2013. 4(6):e00840-13. doi: 10.1128/mBio.00840-13; PMID:24255122; http://dx.doi.org/ 10.1128/mBio.00840-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wasels F, Spigaglia P, Barbanti F, Mastrantonio P. Clostridium difficile erm(B)-containing elements and the burden on the in vitro fitness. J. Med. Microbiol 2013. 62:1461-7; PMID:23741023; http://dx.doi.org/ 10.1099/jmm.0.057117-0 [DOI] [PubMed] [Google Scholar]
- 10. Brouwer MSM, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat. Commun 2013. 4:2601; PMID:24131955; http://dx.doi.org/ 10.1038/ncomms3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wasels F, Monot M, Spigaglia P, Barbanti F, Ma L, Bouchier C, Dupuy B, Mastrantonio P. Inter and intra-species transfer of a Clostridium difficile conjugative transposon conferring resistance to MLSB. Microb. Drug Resist 2014. 20(6):555-560; PMID:25055190; http://dx.doi.org/ 10.1089/mdr.2014.0015 [DOI] [PubMed] [Google Scholar]
- 12. Barbut F, Mastrantonio P, Delmée M, Brazier J, Kuijper E, Poxton I, European Study Group on Clostridium difficile (ESGCD) . Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin. Microbiol. Infect 2007. 13:1048-57; PMID:17850341; http://dx.doi.org/ 10.1111/j.1469-0691.2007.01824.x [DOI] [PubMed] [Google Scholar]
- 13. Wüst J, Sullivan NM, Hardegger U, Wilkins TD. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol 1982. 16:1096-101; PMID:7161375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spigaglia P, Mastrantonio P. Analysis of macrolide-lincosamide-streptogramin B (MLSB) resistance determinant in strains of Clostridium difficile. Microb. Drug Resist 2002. 8:45-53; PMID:12002649; http://dx.doi.org/ 10.1089/10766290252913755 [DOI] [PubMed] [Google Scholar]
- 15. Smith CJ, Markowitz SM, Macrina FL. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother 1981. 19:997-1003; PMID:7271279; http://dx.doi.org/ 10.1128/AAC.19.6.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett 1999. 175:261-6; PMID:10386377; http://dx.doi.org/ 10.1111/j.1574-6968.1999.tb13629.x [DOI] [PubMed] [Google Scholar]
- 17. Spigaglia P, Mastrantonio P. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J. Med. Microbiol 2004. 53:1129-36; http://dx.doi.org/ 10.1099/jmm.0.45682-0 [DOI] [PubMed] [Google Scholar]


