Abstract
In immune intervention trials, the comprehensive investigation of immunogenicity or T-cell epitope-mapping is challenging especially when a large set of epitopes needs to be screened and limited sample material is available. To this end, T-cell responses are often monitored using peptide pools. Here, we assessed the magnitude and sensitivity of detection of antigen-specific CD8+ and CD4+ T cells using a single peptide alone or mixed into large pools. Interestingly the magnitude of ex vivo anti-viral and anti-tumor T-cell responses was identical irrespective of the presence and number of irrelevant peptides, in different functional assays with PBMCs from healthy donors and cancer patients. Moreover, the presence of up to 300 irrelevant peptides did not affect the threshold of responsiveness of antigen-specific CD8+ T cells to single cognate peptides. These data demonstrate the relevance of using very large peptide pools for the sensitive and specific immune-monitoring of epitope-specific T cells in natural or immune-modulated context.
Keywords: immune monitoring, peptide pools, tumor-associated antigens, tumor-specific T cells, vaccine trials
Abbreviations
- AA
amino acid
- APC
antigen presenting cell
- DMSO
dimethyl sulfoxide
- ELISpot
enzyme-linked immunospot
- HLA
human leukocyte antigen
- ICS
intracellular cytokine staining
- MAGE
melanoma associated antigen
- MHC
major histocompatibility complex
- MoDC
monocyte derived dendritic cell
- PBMC
peripheral blood mononuclear cell
- TCR
T-cell receptor
Introduction
T cells recognize antigenic peptide ligands non-covalently bound to MHC molecules. Naturally processed and presented peptides can be very diverse and the affinity of the interaction is relatively low (10−4 to 10−5 M) but the sensitivity of recognition can be very high. Cells express 105 MHC molecules on their surface and the number of MHC/peptide complexes for any given specificity is estimated to range from 10 to 100 complexes.1 T cells can recognize target cells expressing few complexes and it was recently demonstrated that one complex per cell can fully activate the specific T cell.2 This feature allows the use of pools of synthetic peptides with lengths mimicking those of MHC class II or MHC class I binding peptides in antigen recognition assays for the rapid identification of the cognate antigenic peptide being recognized by T cells of unknown antigen specificity.
Monitoring of antigen-specific T-cell responses is a key step in vaccine and immunotherapeutic clinical trials as well as for T-cell epitope-mapping and delineation of immune correlates of protection in various diseases. Several approaches are available to monitor antigen-specific T-cell mediated immune responses. On the one hand, tetramers are widely used in immunology,3,4 but this technique relies on known peptide-MHC complex molecules. On the other hand, the detection of antigen-specific T cells can be performed by in vitro stimulation with synthetic peptides − typically 9 to 20 amino acids (AA) in length − followed by quantitative and sensitive read-outs such as intracellular-cytokine-staining (ICS)5 and IFNγ-ELISpot.6 However, the comprehensive monitoring of cellular immune responses is complex, in particular when (i) limited sample material is available, (ii) whole protein vaccines are used, and/or (iii) patients are not selected for particular HLA haplotypes. Thus, the use of peptide pools can be mandatory when a broad range of different epitopes needs to be tested.
Overlapping synthetic peptides spanning a whole protein sequence have been used to stimulate T cells for immunomonitoring purposes.7,8 Notably, this allowed comprehensive analyses of both breadth and magnitude of viral-specific responses, such as those targeting the entire HIV genome.9 In order to simplify the assays and minimize the number of tests required, several labs are using peptide pools in a matrix format to identify single epitope specific T cells by a deconvolution process. These “peptide arrays” are usually made of pools containing 78 to 36 peptides in an optimized protocol.10 Nevertheless, as the standard format is pools of 15-mer peptides overlapping by 11 AA, a high number of peptides can be required to span long molecules such as tumor-associated proteins. When limited material is available, especially when screening tumor-infiltrating lymphocytes from cancer patients, it would be helpful to test for the presence of antigen-specific responses using very few if not one stimulation test. Since scientific literature lacks of comprehensive studies indicating how to handle several hundreds of peptides in standardized immunomonitoring assays, we here determined whether, and to what extent, a high number of peptides per pool can affect the detection of viral and tumor antigen-specific T-cell responses in different conditions of stimulation.
Results & Discussion
Detection of virus- and tumor antigen-specific CD8+ T-cell responses in the presence of irrelevant peptide pools
Lyophilized peptide libraries are typically solubilized and stored in dimethyl sulfoxide (DMSO), an organosulfur compound in which hydrophobic peptides are easily dissolved. However, this organic solvent can be cytotoxic by disrupting the cell membranes or inducing apoptosis.11 The use of pools containing high numbers of peptides can lead to substantial concentration of DMSO in the culture medium, which raises concerns with regard to the detection of intact T-cell responses. Thus, we first analyzed the impact of different DMSO concentrations on CD8+ and CD4+ T-cell proliferation (Fig. S1A) and cytokine production (Fig. 1A and Fig. S1B). As reported elsewhere,12 the increase of DMSO concentration had inhibitory effects. Nevertheless we found no significant impairment of T-cell responses up to 1% DMSO (vol/vol). On this basis, a final DMSO concentration below 1% in culture is a key condition limiting the concentration and the maximum number of peptides that can be pooled to perform functional assays (i.e., <1μM for >100 peptides). This limit was respected in all subsequent experiments reported here.
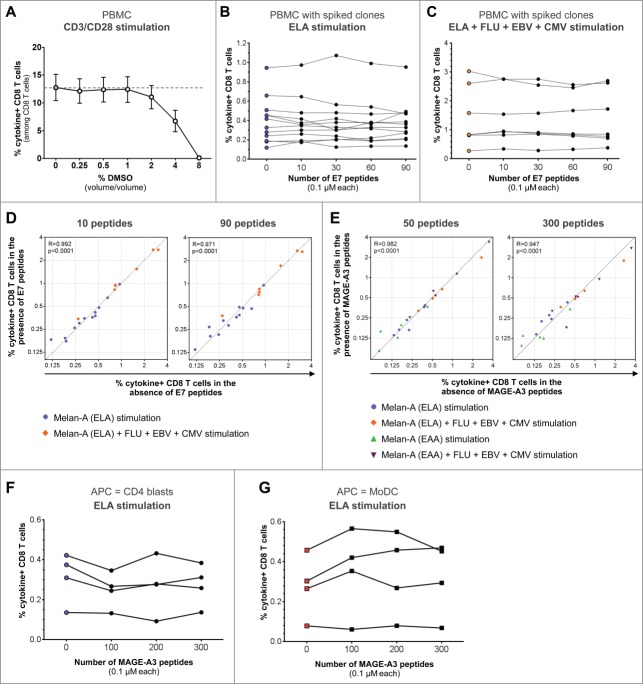
Figure 1.
Effect of large irrelevant peptide pools on the detection of spiked tumor-specific T-cell clones and endogenous viral responses. Cytokine+ CD8+ T cells refer to IFNγ- and/or TNF-α-producing cells. (A) Effect of indicated DMSO concentrations following stimulation of healthy donors PBMCs (n = 9) using plate-bound anti-CD3 and soluble anti-CD28 antibodies on cytokine+ CD8+ T cells. In panels (B-G), Melan-A-specific CD8+ T-cell clones were spiked into either PBMCs (B–E) or isolated CD8+ T cells (F–G) from healthy donors. CD8+ T cell responses were measured following stimulation with (B) ELA or (C) ELA combined with indicated viral epitopes (Table 1) in the absence or presence of pools containing 10 to 90 irrelevant peptides spanning the HPV-E7 protein. (D) Responses to ELA stimulation (circles) and to ELA plus viral epitopes combined stimulation (diamonds) were plotted in the absence versus presence of 10 (left panel) or 90 (right panel) irrelevant HPV-E7 peptide pools. Dashed line depicts the identity curve (y = x). Spearman's rank correlation coefficients “R” and corresponding p values are indicated on each panel. (E) Similar analyses were performed using 50 to 300 irrelevant peptides spanning the MAGE-A3 tumor protein. Plots additionally include data obtained when stimulation was performed using the EAA low-affinity wild-type Melan-A peptide (triangles). (F–G) Melan-A-specific CD8+ T-cell clones were spiked into isolated CD8+ T cells from healthy donors (1:200) co-cultured with APCs, CD4 blasts (F) or MoDCs (G). Cytokine+ T cells were measured following stimulation with the ELA peptide in the absence or presence of 100 to 300 irrelevant peptides from the MAGE-A3 tumor protein.
In order to evaluate whether pools of irrelevant peptides − potentially competing for binding to MHC molecules – may impair the detection of endogenous T-cell responses to virus- or tumor-specific CD8+ T cells, PBMCs from twelve HLA-A2 positive healthy donors were cultured together with spiked Melan-A-specific CD8+ T-cell clones (1 clonal cell for 200 donor CD8+ T cells). Cells were stimulated for 6h with the Melan-A26–35 modified (A27L) peptide (ELA thereafter, Table 1) alone or in the presence of irrelevant peptide pools containing 10, 30, 60 or all 90 peptides at 0.1 μM each, spanning the AA sequence of the Human Papilloma Virus (HPV)-E7 protein. The frequency of CD8+ T cells expressing IFNγ and/or TNF-α was assessed by ICS. None of the patients showed detectable responses when stimulated with HPV-E7 peptides only. The mean (±SEM ) frequency of cytokine+ CD8+ T cells was 0.40% (±0.07%) following ELA stimulation in the absence of irrelevant peptide pools, and remained similar in the presence of 10, 30, 60 or all 90 HPV-E7 peptides (Fig. 1B). To further assess the effect of the presence of irrelevant peptide pools on the detection of spiked clones together with endogenous virus-specific CD8+ T-cell responses, we previously identified seven donors showing an ex vivo positive response targeting at least one of the following HLA-A2-restricted immunodominant viral peptides: FLU58–66, EBV280–288 and CMV495−503 (Table 1). We then measured the combined responses to the four peptides ELA, FLU, EBV and CMV in the co-cultures. The frequency of cytokine+ T cells (mean ± SEM of 1.42% ± 0.39%) remained stable in the presence of pools containing 10 to 90 peptides (Fig. 1C). Of note, similar results were obtained when analyzing only IFNγ+/TNF-α+ double positive T cells (data not shown). We also assessed the correlation between the antigen-specific responses measured in the absence of irrelevant peptide pools and the same responses measured in the presence of 10 to 90 HVP-E7 peptides (Fig. 1D). Spearman correlation tests showed that these responses were strongly correlated (R > 0.97 and p < 0.0001 for all peptide pools). We then tested the effect of irrelevant peptides spanning the AA sequence of a longer protein, MAGE-A3, a cancer-testis antigen expressed in several tumors including melanoma, lung and bladder cancers, and frequently investigated in cancer vaccine trials.13 Interestingly, antigen-specific responses measured in the presence of 50, 100, 150, 200, 250 and up to 300 MAGE-A3 peptides were strongly correlated to the responses measured in the absence of MAGE-A3 peptide pools (R > 0.94 and p < 0.0001 for all peptide pools) (Fig. 1E).
Table 1.
Peptide sequences and in silico-predicted characteristics of binding to HLA-A*02:01
| Binding Scores |
||||||
|---|---|---|---|---|---|---|
| Abbr. | Protein– amino acid position | Origin | Sequence | Predicted affinity (nM) * | Bimas** | IEDB*** |
| EAA | Melan-A (wt) – 26–35 | Human melanoma | EAAGIGILTV | 5164 | 0,2 | 13,00 |
| ELA | Melan-A (A27L) − 26–35 | ELAGIGILTV | 137 | 12,0 | 1,95 | |
| FLU | Matrix Protein M1 − 58–66 | Influenza virus | GILGFVFTL | 12 | 550,9 | 0,80 |
| EBV | BMLF1 − 280–288 | Epstein–Barr virus | GLCTLVAML | 112 | 49,1 | 2,40 |
| CMV | pp65 − 495−503 | Cytomegalovirus | NLVPMVATV | 29 | 159,9 | 1,00 |
affinity was calculated by the NetMHC 3.4 server (CBS, Technical University Denmark)
higher score means higher binding capacity (BIMAS HLA Peptide Binding Predictions, NIH, www-bimas.cit.nih.gov/molbio/hla_bind)
lower score means higher binding capacity (Immune Epitope Database, http://tools.immuneepitope.org/mhci)
Detection of low-affinity responses
As the “ELA” peptide shows high affinity for the MHC-I molecule (Table 1), we also tested whether detection of specific CD8+ T-cell responses targeting a low-affinity epitope may be altered by the presence of a high number of irrelevant peptides. Thus, we used − in the same settings − the wild-type Melan-A26–35 peptide (EAA thereafter, Table 1) that exhibits much lower affinity (Table 1) and immunogenicity14,15 as compared to its ELA optimized peptide analog (A27L mutation). Despite lower avidity, measured EAA-specific cytokine+ T cells also remained similar in the presence of pools containing up to 300 peptides (Fig. 1E).
APC-mediated stimulation
Instead of stimulating total PBMCs, another method to assess T-cell responses is the use of isolated CD8+ T cells in the presence of autologous antigen-presenting cells (APC), such as monocyte-derived dendritic cells (MoDC)16,17 or CD4+ T-cell blasts.18 Also in these settings, the presence of up to 300 irrelevant peptides did not impair the detection of ELA-specific CD8+ T cells (Fig. 1F–G).
Detection of tumor-specific CD4+ T-cell responses
CD4+ T cells play a central role in the development of effective adaptive immune responses and thus also need to be monitored in vaccine trials or for epitope discovery. In contrast to CD8+ T cells, CD4+ T cells recognize epitopes presented by MHC class II molecules and libraries of longer (20-mer) overlapping peptides are generally used to screen for antigen-specific responses.19 We thus measured the response of Melan-A/MART-125–36-specific CD4+ T-cell clones20 spiked into PBMCs of HLA-DQ6+ donors in the presence of 20-mers overlapping peptide pools (1 µM each). As shown in Fig. S1C, the presence of pools containing up to 120 long peptides did not alter the detection of tumor-specific cytokine+ CD4+ T cells.
Functional avidities
Although the peptide concentration used in the previous experiments is what is consensually used in most studies, this most likely represents an excess. It thus cannot be excluded that, when single epitopes are mixed into pools, the effective dose to which cells are exposed might still be affected. To address this point, we measured the threshold of responsiveness (i.e., the ability of T cells to respond to low peptide dose) in the presence of irrelevant peptides. In particular, FLU- and EBV-reactive T cells were tested by performing limiting peptide dilutions (10 to 10−4 μM) in the presence or absence of 300 MAGE-A3 peptides (0.1 μM each). Peptide concentration required to induce half maximal responses (EC50) was determined by IFNγ-ELISpot. A representative example of the responsiveness of peptide specific CD8+ T cells after stimulation with decreasing concentrations of FLU peptide is shown in Fig. 2A (left panel). Cumulative data of TCR avidity (expressed as EC50) of FLU and EBV-specific CD8+ T cells from 3 individuals is also reported (Fig. 2A, right panel). Strikingly, following antigen-specific stimulation and assessment of total IFNγ-secreting T cells, we didn't observe any significant loss in T-cell avidity if the specific stimulation occurred in the presence of 300 competing peptides.
Figure 2.
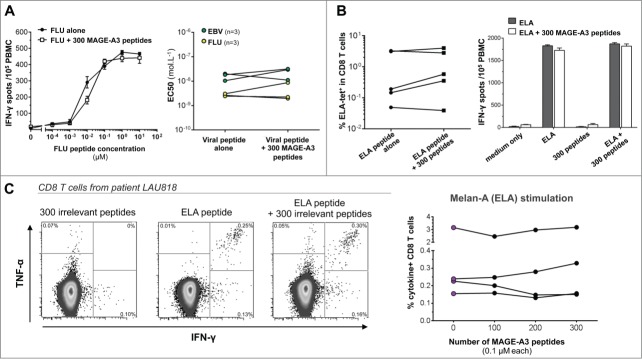
The presence of 300 irrelevant peptides does not influence the detection of viral and tumor specific T-cell responses. (A) Ex vivo TCR avidity of HLA-A2 restricted FLU58–66 and CMV495−503 specific CD8+ T-cell responses. The left panel shows representative data of the responsiveness of anti-FLU T cells after stimulation with limiting peptide dilutions (10 to 10−4 μM), in the presence or absence of 300 MAGE-A3 peptides. The right panel depicts cumulative data from six donors correlating the EC50 values of PBMC stimulated with the specific peptide alone or mixed with a pool of 300 MAGE-A3 peptides. (B) In vitro Melan-A26–35-specific T-cell proliferation and tetramer staining. PBMC from healthy donors were cultured for one week in the presence of either ELA peptide alone or ELA plus the MAGE-A3 peptide pool. At day 7, cells were collected and stained with the Melan-A26–35 tetramer (left panel) or boosted in different conditions to check for their specificity by IFNγ-ELISpot assay (right panel). Black bars show mean ± SEM response of cells stimulated for one week with ELA alone, white bars show mean ± SEM response of cells stimulated with ELA together with the pool. (C) Sensitive ex vivo detection of Melan-A26–35-specific T cells in vaccinated melanoma tumor patients. PBMCs were stimulated overnight with increasing numbers of MAGE-A3 peptides (up to 300) and then tested for cytokine production. The left panels show representative ICS analysis of PBMCs from one out of four patients tested. The right graph depicts cumulative data showing the percentages of cytokine-producing CD8+ T cells in the indicated conditions.
Proliferation of naïve precursors and recall of memory T cells
In contrast to viral antigens, the unambiguous ex vivo detection of tumor Ag-specific T cells in the peripheral blood has proven to be difficult. Therefore, a short-term in vitro stimulation assay is often performed in order to amplify T cell responses; one or multiple Ag-driven expansions of memory T cells lead to percentages of specific T cells high enough for further testing. Moreover, it has been reported that the proliferation of viral-specific CD8+ T cells, more than their functionality, could be impaired by the presence of competing peptides.12 In order to evaluate the influence of a pool of 300 irrelevant peptides on the proliferation and expansion of tumor-specific T cells, PBMCs from five healthy donors, were cultured for 1 week in the presence of ELA peptide, alone or mixed together with the MAGE-A3 peptide pool. Taking advantage of the presence of detectable ELA-specific CD8+ T cells in HLA-A2 healthy individuals,21,22 this setting allows sensitive detection of possible peptide pool inhibitory effects on low-frequency antigen-specific T-cell responses. At day 7, we compared the percentages of ELA-specific T cells by tetramer staining expanded with ELA peptide alone or in the presence of the pool. We found no significant impact of the peptide pool with regard to the expansion levels of tumor-specific T cells in all tested donors (Fig. 2B, left panel). To further investigate and confirm the specificity of expanded T lymphocytes, cultured cells were tested in an IFNγ-ELISpot assay upon re-stimulation with medium alone, ELA peptide, MAGE-A3 peptide pool or ELA peptide together with MAGE-A3 peptide pool. Relative spot numbers (Fig. 2B, right panel) showed that unrelated antigens included in the pool did not cross-react with ELA-specific T cells, not interfering with their priming and expansion. Moreover, the peptide pool did not functionally inhibit the reactivity of ELA-specific T cells.
Ex vivo detection of endogenous tumor-specific CD8+ T-cell responses in vaccinated melanoma patients
In order to further corroborate the possibility of exploiting large peptide pools in the context of immunological monitoring of cancer vaccine studies, we analyzed the effect of MAGE-A3 peptide pools on effector functions of tumor-specific T cells circulating in the peripheral blood of cancer patients. PBMCs from melanoma patients previously vaccinated with the ELA peptide23 were tested for ex vivo cytokine production following 6 h incubation with ELA alone or ELA mixed with the MAGE-A3 peptide pool (Fig. 2C). Of note, MAGE-A3 antigen expression has been reported in a substantial number of metastatic melanomas.24 However, naturally acquired MAGE-A3 specific T-cell responses were rarely detected in cancer patients and only at low frequencies.25,26 Accordingly, the 300 peptide pool alone did not increase the staining background. Interestingly, stimulation with ELA peptide together with the 300 peptide pool efficiently stimulated tumor-specific T cells, without any significant loss of reactivity as compared to ELA peptide alone (Fig. 2C).
Concluding Remarks
Competition between cognate and irrelevant peptides for binding to MHC molecules at the cell surface can alter the sensitivity of T-cell stimulation. Here, we showed that efficient in vitro immune monitoring of viral but also tumor antigen specific T cells could be achieved by using a single large pool of peptides (up to 300 peptides) without affecting the detection of the specific responses and regardless of the read-out (ICS, ELISpot, proliferation) or the method of stimulation (total PBMCs, APCs). Although MAGE-A3 and several other classical tumor-associated proteins tested in cancer vaccines such as NY-ESO-1 or Melan-A are in the range of 100–300 AA in length, it remains to be determined whether larger peptide pools (i.e., >300 peptides) could be used when monitoring vaccine responses to bigger proteins (e.g., HER2/neu with 1255 AA). However, in that case, DMSO concentration (exceeding 1%) becomes the limiting factor and may require to reduce the concentration of each peptide below 0.1 μM which may not be optimal for sensitive detection of antigen-specific T cells. Altogether, the knowledge obtained from this study is important for future clinical trials or epitope mapping since it allows high-throughput monitoring of immune responses using limiting amount of biological material.
Material and Methods
Patients
HLA-A*0201–positive patients with stage III/IV metastatic melanoma were included in the Ludwig Institute for Cancer Research clinical trial LUD 00–018, approved by institutional review boards and regulatory agencies. Patients received monthly low-dose vaccinations s.c. with 100 μg Melan-A peptide (Melan-A26–35(A27L)) adjuvanted with CpG (500 μg PF-3512676/7909; provided by Pfizer/Coley Pharmaceutical Group) and IFA (300–600 μL Montanide ISA-51).23
Peptides
Specific peptides used in the study are described in Table 1. For CD8+ T-cell experiments, we used 9-mers overlapping by 8 AA and covering the whole sequence of HPV-E7 or MAGE-A3 proteins. For CD4 T cell experiments, pools were constituted of a mix of 20 AA peptides overlapping by 10 AA coming from different tumor antigens: Mesothelin, Gp100, CEA, Tyrosinase, and Her-2/neu. Peptides were synthesized at the Protein and Peptide Chemistry Facility, University of Lausanne (Switzerland) and analyzed by mass spectrometry. The homogeneity of all peptides was >90%, as indicated by analytical HPLC. HPV-E7 overlapping peptides were kindly provided by Prof. Van Der Burg SH (Leiden, Netherlands).
In vitro stimulation
Twelve and five healthy donors previously identified as positive for the HLA-A2 and HLA-DQ6 allotypes, respectively, were selected. Cryopreserved PBMCs were thawed and rested overnight in RPMI 1640 containing 10% FBS at a density of 2×106 cells/mL. PBMCs were cultured together with Melan-A-specific CD8+ or CD4+ T-cell clones15 and stimulated for 6 h with indicated tumor and/or viral peptides (0.1 μM) alone or in the presence of irrelevant peptide pools containing 10 to 300 peptides spanning the AA sequence of the HPV-E7 or MAGE-A3 proteins, for CD8+ T-cell experiments, and Mesothelin, Gp100, CEA, Tyrosinase, and Her-2/neu, for CD4 T-cell experiments. Alternatively, CD8+ T cells were isolated from healthy donors using Dynabeads human CD8+ T-cell isolation kit (Invitrogen) and cultured with autologous antigen presenting cells, i.e., monocyte-derived dendritic cells (MoDCs) or “CD4+ PHA blasts”.16,18 MoDCs were generated from adherent monocytes cultured for 5 d in RPMI 8% human serum with GM-CSF (450 U/mL) and IL-4 (300 U/mL) followed by 48h maturation with LPS (1 μg/mL). CD4 blasts were generated by isolating CD3+CD4+ T cells using Dynabeads human CD4+ T cell isolation kits (Invitrogen), followed by stimulation with PHA (1 μg/mL) in IL-2 (150 U/mL, Proleukin) supplemented RPMI 8% human serum. CD4+ blasts were used after at least 10 d of culture. For long-term in vitro stimulation, PBMCs from 5 HLA-A2+ donors were stimulated in 24 well-plates (4 × 106 cells/well) for one week with 0.1 μM Melan-A26–35 (A27L) in the presence or absence of MAGE-A3 peptide pool (0.1 μM each peptide). After 48 h of incubation at 37°C, 100 U/mL IL-2 were added to the culture. On day 7, expanded T cells were collected and tested.
Functional avidity
HLA-A2 positive healthy subjects were screened for the presence of specific T-cell responses to FLU-M158–66 and EBV-BMLF1280–288 by ex vivo IFNγ-ELISpot assay. Functional avidity of FLU-responding T cells from three individuals and of EBV-specific T cells from three other individuals was then analyzed. In brief, PBMCs were thawed, rested overnight and stimulated for 18 h by incubating 105 total PBMCs per well in pre-coated 96-well ELISpot plates (Mabtech), in triplicate. Plates were developed following the manufacturer's instructions. Spot-forming units were counted using a Bioreader-6000-E automated counter (Byosis). EC50 was determined as the peptide concentration needed to achieve a half-maximal response as previously described.27
Intracellular cytokine staining / Flow cytometry
IFNγ and TNF-α cytokine secretion was investigated by ICS. The following monoclonal antibodies were used at predetermined optimal concentrations: anti-CD3−PerCP-Cy5.5 (eBioscience), anti-CD8+−AF488, anti-CD4−PE-Cy7, anti-IFNγ−BV421, anti-TNFα−AF647 (Biolegend). Surface staining was performed for 20 min. at 4°C. Cells were then stained for dead cell exclusion with aqua live/dead stain kit (Life Technologies) according to the manufacturer's instructions. FcR Blocking Reagent (Miltenyi Biotec) was used to increase staining specificity by blocking unwanted binding of antibodies. For intracellular staining of IFNγ and TNF-α, cells were fixed and permeabilized using the “FoxP3 Staining Buffer Set” (eBioscience) according to the manufacturer's recommendations. Sample acquisition was performed on the Gallios Flow-Cytometer (Beckman Coulter) and data were analyzed using the FlowJo Software (TreeStar). The background obtained with corresponding irrelevant peptide pools alone or non-stimulated cells were subtracted from values obtained in the presence of cognate peptide + irrelevant peptide pool or specific peptide alone, respectively. The proliferation of Melan-A26–35 –specific CD8+ T cells was assessed by tetramer staining using PE-labeled Melan-A26–35 tetramer (TcMetrix), FITC conjugated CD8+ and aqua live/dead stain kit.
Statistical analysis
Statistical analysis was done using GraphPad Prism (version 6). Non-parametric methods (Wilcoxon matched-pairs test and Spearman rank correlation) were used to evaluate group differences and correlations as sample sizes were too small to ascertain normal distribution of values. p values below 0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by the Swiss National Foundation Grant 32003B_146638 (LD); the Swiss Cancer League Grant KLS-02744–02–2011 (LD); The Krebsliga Fellowship KFS 3064–08–2012 (PR).
References
- 1.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol 2003; 3:952-61; PMID:14647477; http://dx.doi.org/ 10.1038/nri1250 [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Brameshuber M, Zeng X, Xie J, Li QJ, Chien YH, Valitutti S, Davis MM. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 2013; 39:846-57; PMID:24120362; http://dx.doi.org/ 10.1016/j.immuni.2013.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science 1996; 274:94-6; PMID:8810254; http://dx.doi.org/ 10.1126/science.274.5284.94 [DOI] [PubMed] [Google Scholar]
- 4.Appay V, Rowland-Jones SL. The assessment of antigen-specific CD8+ T cells through the combination of MHC class I tetramer and intracellular staining. J Immunol Methods 2002; 268:9-19; PMID:12213338; http://dx.doi.org/ 10.1016/S0022-1759(02)00195-3 [DOI] [PubMed] [Google Scholar]
- 5.Kern F, Surel IP, Brock C, Freistedt B, Radtke H, Scheffold A, Blasczyk R, Reinke P, Schneider-Mergener J, Radbruch A, et al.. T-cell epitope mapping by flow cytometry. Nat Med 1998; 4:975-8; PMID:9701254; http://dx.doi.org/ 10.1038/nm0898-975 [DOI] [PubMed] [Google Scholar]
- 6.Miyahira Y, Murata K, Rodriguez D, Rodriguez JR, Esteban M, Rodrigues MM, Zavala F. Quantification of antigen specific CD8+ T cells using an ELISPOT assay. J Immunol Methods 1995; 181:45-54; PMID:7537312; http://dx.doi.org/ 10.1016/0022-1759(94)00327-S [DOI] [PubMed] [Google Scholar]
- 7.Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, Persaud N, Trigona W, Fu TM, Sinclair E et al.. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods 2001; 255:27-40; PMID:11470284; http://dx.doi.org/ 10.1016/S0022-1759(01)00416-1 [DOI] [PubMed] [Google Scholar]
- 8.Tobery TW, Wang S, Wang XM, Neeper MP, Jansen KU, McClements WL, Caulfield MJ. A simple and efficient method for the monitoring of antigen-specific T cell responses using peptide pool arrays in a modified ELISpot assay. J Immunol Methods 2001; 254:59-66; PMID:11406153; http://dx.doi.org/ 10.1016/S0022-1759(01)00397-0 [DOI] [PubMed] [Google Scholar]
- 9.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, et al.. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol 2003; 77:2081-92; PMID:12525643; http://dx.doi.org/ 10.1128/JVI.77.3.2081-2092.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Precopio ML, Butterfield TR, Casazza JP, Little SJ, Richman DD, Koup RA, Roederer M. Optimizing peptide matrices for identifying T-cell antigens. Cytometry A 2008; 73:1071-8; PMID:18781655; http://dx.doi.org/ 10.1002/cyto.a.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galvao J, Davis B, Tilley M, Normando E, Duchen MR, Cordeiro MF. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB JK 2014; 28:1317-30; PMID:24327606; http://dx.doi.org/ 10.1096/fj.13-235440 [DOI] [PubMed] [Google Scholar]
- 12.Suneetha PV, Schlaphoff V, Wang C, Stegmann KA, Fytili P, Sarin SK, Manns MP, Cornberg M, Wedemeyer H. Effect of peptide pools on effector functions of antigen-specific CD8+ T cells. J Immunol Methods 2009; 342:33-48; PMID:19135447; http://dx.doi.org/ 10.1016/j.jim.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 13.Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, Suciu S, Kruit WH, Eggermont AM, Vansteenkiste J et al.. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol 2013; 31:2388-95; PMID:23715562; http://dx.doi.org/ 10.1200/JCO.2012.44.3762 [DOI] [PubMed] [Google Scholar]
- 14.Valmori D, Fonteneau JF, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini JC, Romero P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J Immunol 1998; 160:1750-8; PMID:9469433 [PubMed] [Google Scholar]
- 15.Derre L, Ferber M, Touvrey C, Devevre E, Zoete V, Leimgruber A, Romero P, Michielin O, Levy F, Speiser DE. A novel population of human melanoma-specific CD8 T cells recognizes Melan-AMART-1 immunodominant nonapeptide but not the corresponding decapeptide. J Immunol 2007; 179:7635-45; PMID:18025209; http://dx.doi.org/ 10.4049/jimmunol.179.11.7635 [DOI] [PubMed] [Google Scholar]
- 16.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med 1994; 179:1109-18; PMID:8145033; http://dx.doi.org/ 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guermonprez P, Valladeau J, Zitvogel L, Thery C, Amigorena S. Antigen presentation and T cell stimulation by dendritic cells. Ann Rev Immunol 2002; 20:621-67; PMID:11861614; http://dx.doi.org/ 10.1146/annurev.immunol.20.100301.064828 [DOI] [PubMed] [Google Scholar]
- 18.Atanackovic D, Matsuo M, Ritter E, Mazzara G, Ritter G, Jager E, Knuth A, Old LJ, Gnjatic S. Monitoring CD4+ T cell responses against viral and tumor antigens using T cells as novel target APC. J Immunol Methods 2003; 278:57-66; PMID:12957396; http://dx.doi.org/ 10.1016/S0022-1759(03)00209-6 [DOI] [PubMed] [Google Scholar]
- 19.Klyushnenkova EN, Link J, Oberle WT, Kodak J, Rich C, Vandenbark AA, Alexander RB. Identification of HLA-DRB1*1501-restricted T-cell epitopes from prostate-specific antigen. Clin Cancer Res 2005; 11:2853-61; PMID:15837732; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-1927 [DOI] [PubMed] [Google Scholar]
- 20.Jandus C, Bioley G, Dojcinovic D, Derre L, Baitsch L, Wieckowski S, Rufer N, Kwok WW, Tiercy JM, Luescher IF et al.. Tumor antigen-specific FOXP3+ CD4 T cells identified in human metastatic melanoma: peptide vaccination results in selective expansion of Th1-like counterparts. Cancer Res 2009; 69:8085-93; PMID:19808957; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-2226 [DOI] [PubMed] [Google Scholar]
- 21.Pittet MJ, Valmori D, Dunbar PR, Speiser DE, Lienard D, Lejeune F, Fleischhauer K, Cerundolo V, Cerottini JC, Romero P. High frequencies of naive Melan-A/MART-1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J Exp Med 1999; 190:705-15; PMID:10477554; http://dx.doi.org/ 10.1084/jem.190.5.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero P, Speiser DE, Rufer N. Deciphering the unusual HLA-A2/Melan-A/MART-1-specific TCR repertoire in humans. Eur J Immunol 2014; 44:2567-70; PMID:25154881; http://dx.doi.org/ 10.1002/eji.201445004 [DOI] [PubMed] [Google Scholar]
- 23.Speiser DE, Lienard D, Rufer N, Rubio-Godoy V, Rimoldi D, Lejeune F, Krieg AM, Cerottini JC, Romero P. Rapid and strong human CD8+ T cell responses to vaccination with peptide, IFA, and CpG oligodeoxynucleotide 7909. J Clin Invest 2005; 115:739-46; PMID:15696196; http://dx.doi.org/ 10.1172/JCI23373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brasseur F, Rimoldi D, Lienard D, Lethe B, Carrel S, Arienti F, Suter L, Vanwijck R, Bourlond A, Humblet Y, et al.. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int J Cancer 1995; 63:375-80; PMID:7591235; http://dx.doi.org/ 10.1002/ijc.2910630313 [DOI] [PubMed] [Google Scholar]
- 25.Dhodapkar MV, Young JW, Chapman PB, Cox WI, Fonteneau JF, Amigorena S, Houghton AN, Steinman RM, Bhardwaj N. Paucity of functional T-cell memory to melanoma antigens in healthy donors and melanoma patients. Clin Cancer Res 2000; 6:4831-8; PMID:11156242 [PubMed] [Google Scholar]
- 26.Hanagiri T, van Baren N, Neyns B, Boon T, Coulie PG. Analysis of a rare melanoma patient with a spontaneous CTL response to a MAGE-A3 peptide presented by HLA-A1. Cancer Immunol Immunother 2006; 55:178-84; PMID:16187089; http://dx.doi.org/ 10.1007/s00262-005-0063-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vigano S, Bellutti Enders F, Miconnet I, Cellerai C, Savoye AL, Rozot V, Perreau M, Faouzi M, Ohmiti K, Cavassini M, et al.. Rapid perturbation in viremia levels drives increases in functional avidity of HIV-specific CD8 T cells. PLoS Pathog 2013; 9:e1003423; PMID:23853580; http://dx.doi.org/ 10.1371/journal.ppat.1003423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.