Abstract
Rationale: The lung microbiome is spatially heterogeneous in advanced airway diseases, but whether it varies spatially in health is unknown. We postulated that the primary determinant of lung microbiome constitution in health is the balance of immigration and elimination of communities from the upper respiratory tract (URT; “adapted island model of lung biogeography”), rather than differences in regional bacterial growth conditions.
Objectives: To determine if the lung microbiome is spatially varied in healthy adults.
Methods: Bronchoscopy was performed on 15 healthy subjects. Specimens were sequentially collected in the lingula and right middle lobe (by bronchoalveolar lavage [BAL]), then in the right upper lobe, left upper lobe, and supraglottic space (by protected-specimen brush). Bacterial 16S ribosmal RNA–encoding genes were sequenced using MiSeq (Illumina, San Diego, CA).
Measurements and Main Results: There were no significant differences between specimens collected by BAL and protected-specimen brush. Spatially separated intrapulmonary sites, when compared with each other, did not contain consistently distinct microbiota. On average, intrasubject variation was significantly less than intersubject variation (P = 0.00003). By multiple ecologic parameters (community richness, community composition, intersubject variability, and similarity to source community), right upper lobe microbiota more closely resembled those of the URT than did microbiota from more distal sites. As predicted by the adapted island model, community richness decreased with increasing distance from the source community of the URT (P < 0.05).
Conclusions: In healthy lungs, spatial variation in microbiota within an individual is significantly less than variation across individuals. The lung microbiome in health is more influenced by microbial immigration and elimination (the adapted island model) than by the effects of local growth conditions on bacterial reproduction rates, which are more determinant in advanced lung diseases. BAL of a single lung segment is an acceptable method of sampling the healthy lung microbiome.
Clinical trial registered with www.clinicaltrials.gov (NCT02392182).
Keywords: lung, microbiome, bronchoscopy, 16S ribosomal DNA
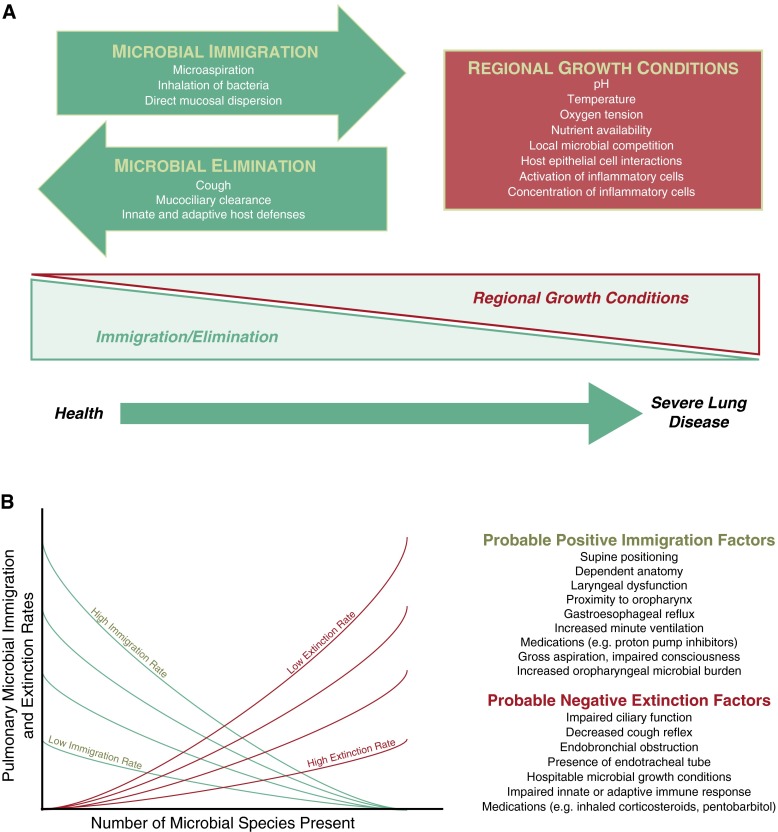
Defining the impact of the lung microbiome in health and in specific lung diseases on host immunity has great potential to advance pulmonary disease prevention and management. The constitution of the lung microbiome is determined by three factors: microbial immigration, microbial elimination, and the relative reproduction rates of its members (1, 2) (Figure 1A). Active investigation and controversy (3–6) persist regarding the relative proportion of bacteria in healthy lungs that are resident, reproducing community members (subject to local environmental growth conditions) versus transient community members (with abundance determined solely by rates of immigration and elimination). The lung bacterial microbiome shares greater community membership with communities of the mouth (4) than communities detected in air (7, 8) or other body sites (9, 10). This similarity suggests that, rather than inhalation or hematogenous spread, the upper respiratory tract (URT) is the primary source of microbial immigration to the lungs, via microaspiration (11, 12) and direct mucosal dispersion. We have proposed an adapted island model of lung biogeography (1), which postulates that, in health, more distal lung bacterial communities should have decreasing community richness and reduced similarity to their URT source community (Figure 1B).
Figure 1.
Ecological modeling of the respiratory microbiome. (A) The constitution of the respiratory microbiome is determined by three factors: microbial immigration, microbial elimination and the relative reproduction rates of its members. In health, community membership is primarily determined by immigration and elimination; in advanced lung disease, membership is primarily determined by regional growth conditions. Adapted with permission from Dickson and colleagues (2). (B) The adapted island model of lung biogeography. Community richness in health for a given site in the respiratory tract is a function of immigration and elimination factors. Adapted with permission from Dickson and colleagues (1).
Within the lungs of healthy individuals, substantial regional variation exists among physiological parameters that have strong in vitro effects on microbial growth rates (1, 13), including oxygen tension (13), temperature (14), pH (13), and relative deposition of inhaled particles (15). Thus, if the effects of regional growth conditions on bacterial reproduction strongly impact lung microbiome composition, then significant and consistent differences in community membership should be present at spatially separated sites (1) (e.g., the apices should, on average, harbor a distinct microbiome from the lung bases). Studies comparing the microbiota of spatially separated sites within the lungs in chronic obstructive pulmonary disease (16) and cystic fibrosis (17, 18) demonstrate considerable spatial heterogeneity in bacterial community composition within the lungs (and even lobes) of individual patients (19). However, whether comparable spatial heterogeneity in microbiota is present in the lungs of healthy subjects is unknown.
This study tested whether spatial variation exists within the lung microbiota of healthy subjects. We hypothesized that intrasubject variation, though present, would be less than variation between subjects. We asked whether consistent and distinct microbiomes could be detected across subjects at individual anatomic sites (e.g., a specific “left upper lobe [LUL] microbiome”). Furthermore, we tested the adapted island model of lung biogeography by comparing the richness and composition of a proximal lung site (right upper lobe [RUL]) to one more distal from the URT (right middle lobe [RML]), as well as to two sites (lingula and LUL) less directly exposed to microaspiration and mucosal dispersal. Bacterial communities at all intrapulmonary sites were compared with that of subjects’ supraglottic space, representing an integrated sample of the “source community” of the URT (1).
Methods
Ethics Statement
We conducted all clinical investigations according to the principles of the Declaration of Helsinki. The study protocol was approved by the institutional review board of the Veterans Affairs Ann Arbor Healthcare System. All participants understood the purpose of the study and provided written informed consent before any research procedure. All underwent a complete history and physical examination by a pulmonologist, complete pulmonary function testing, including body plethysmography and diffusing capacity, chest imaging, prospective collection of medication history, and complete blood count with differential, coagulation studies, and chemistry panel.
Participants
The 15 participants were a subset of volunteers recruited in the Lung HIV Microbiome Project from the southeast Michigan community, primarily by means of the University of Michigan clinical trials website. Inclusion criteria were men and women aged 18–80 years. Exclusion criteria were known history of pulmonary disease, reported fever, cough, or upper respiratory symptoms in the previous 4 weeks, or use of antibiotics or immunosuppressive medications in the past 3 or 6 months, respectively. All subjects (age range, 23–77 yr) were human immunodeficiency virus negative and had no known history of respiratory disease (Table 1). We analyzed samples independently of smoking status, which does not induce significant differences in the lung microbiome of otherwise healthy individuals (4).
Table 1.
Study subjects (n = 15)
| Characteristics | Value |
|---|---|
| Age, yr (mean ± SD) | 45 ± 15 |
| Female sex, n (%) | 11 (73) |
| Smoking: never/former/current | 9/3/3 |
| FEV1 % predicted (mean ± SD) | 100 ± 12 |
| FVC % predicted (mean ± SD) | 96 ± 11 |
Sample Acquisition and Processing
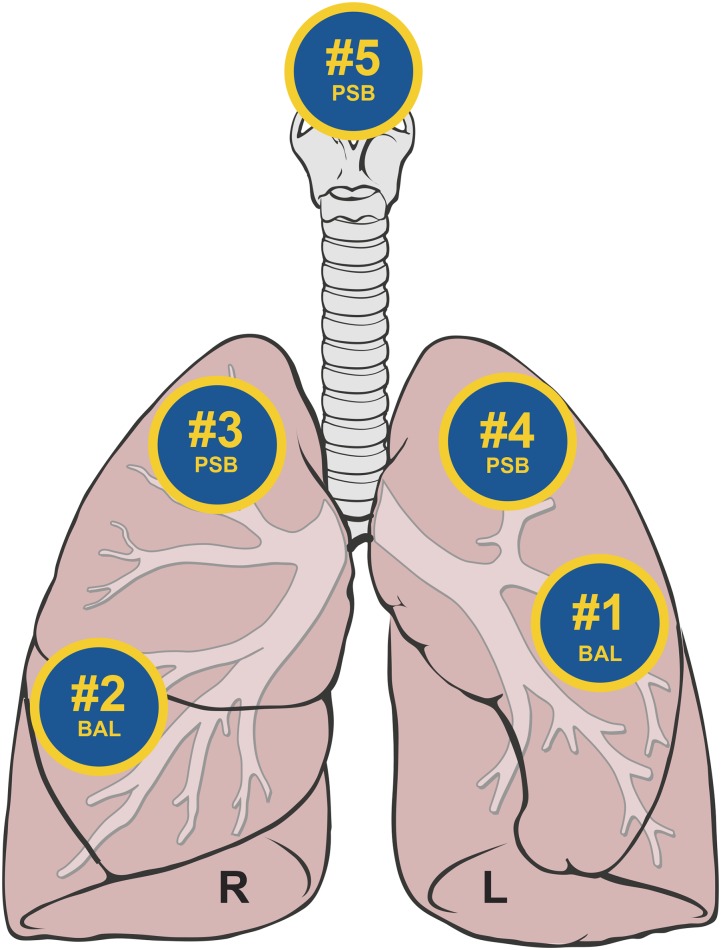
We have previously published the bronchoscopic technique used in the study (4). In previous analyses (20), we showed that aspirating saline through a bronchoscope before insertion into the subject has no statistically significant effect on the amount of bacterial DNA detected, compared with directly analyzing saline alone. Hence, we did not analyze that type of sample by MiSeq in the current study. After sedation and administration of local anesthetic to the URT (4% lidocaine to the vocal cords), the bronchoscope was inserted through the mouth and advanced quickly and without suctioning to a wedged position (Figure 2). We performed bronchoalveolar lavage (BAL) in the lingula (#1) and then in the RML (#2). Next, participants underwent an Institutional Review Board–approved modification of the previous protocol, such that we performed protected-specimen brushings (PSBs) in the RUL (#3) and LUL (#4). Next, we withdrew the bronchoscope to above the vocal cords, and performed a third PSB, sampling the posterior wall of the immediate supraglottic region (#5). We held BAL fluid and PSB specimens on ice until the time of processing. We collected reagent water controls at the time of DNA isolation and processed them in parallel with study specimens.
Figure 2.
Sequence of bronchoscopic sampling. After sedation and administration of local anesthetic to the upper respiratory tract, the bronchoscope was inserted through the mouth and advanced quickly and without suction to a wedge position. Bronchoalveolar lavage was performed in the lingula (#1) and then in the right middle lobe (#2). protected-specimen brushing was then performed in the right upper lobe (#3), left upper lobe (#4), and supraglottic space (#5). Figure adapted from original by Patrick J. Lynch and C. Carl Jaffe, M.D., via Creative Commons Attribution 2.5 license 2006 (http://goo.gl/xuJRCO).
Sample Processing and Sequencing
We identified bacterial community members using sequencing of the bacterial 16S ribosomal RNA gene, a small and highly conserved locus of the bacterial genome that permits genus- and species-level identification. When compared with conventional, culture-based approaches, sequencing-based techniques reveal increased diversity of the respiratory microbiome and are not limited by microbe viability or the ability to grow in limited in vitro culture conditions (19, 21). We have previously described our methods of genomic DNA extraction and amplification (21, 22). We amplified the V4 region of the 16s rRNA gene from each sample using published primers (23) and the dual-indexing sequencing strategy developed by the laboratory of Patrick D. Schloss (24). We performed sequencing using the MiSeq platform and MiSeq Reagent Kit V2 (500 cycles; Illumina, San Diego, CA), according to the manufacturer’s instructions with modifications found in the Schloss standard operating procedure (25). We used Accuprime High Fidelity Taq instead of Accuprime Pfx SuperMix (Waltham, MA). Primary PCR cycling conditions were 95°C for 2 minutes, followed by 20 cycles of touchdown PCR (95°C 20 seconds, 60°C 20 s, and decreasing 0.3°C each cycle, 72°C 5 min), then 20 cycles of standard PCR (95°C for 20 s, 55°C for 15 s, and 72°C for 5 min), and finished with 72°C for 10 minutes.
Data Analysis
We processed sequence data using the software, mothur v.1.27.0 (Schloss Laboratory; University of Michigan, Ann Arbor, MI), according to the standard operating procedure for MiSeq sequence data (25) using a minimum sequence length of 250 base pairs (26). We generated a shared community file and a phylotyped (genus-level grouping) file using operational taxonomic units (OTUs) (“species”) binned at 97% identity. We assigned OTU numbers in the binning process and performed classification using the mothur implementation of the Ribosomal Database Project Classifier and the Ribosomal Database Project taxonomy training set (27).
Sequences detected in reagent control specimens clustered predominantly into two OTUs, comprising greater than 50% of the reagent control sequences. These two OTUs were then excluded from all subsequent sample analysis. Of the 20 most abundant OTUs detected at each anatomic site (lingula, RML, RUL, LUL, and supraglottic space), none were detected in greater than 0.5% abundance in reagent control specimens. When we analyzed specimen communities by which bronchoscope was used in their acquisition, we identified no detectable effect of bronchoscope contamination on community composition (P = 0.24).
We performed microbial ecology analysis using the vegan package 2.0–4 in R (28, 29). We performed all analyses in R and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA). We performed ordinations using principal component analysis on Hellinger-transformed normalized OTU tables. We determined significance of differences in community composition using permutational ANOVA (PERMANOVA), specifically running the adonis function in vegan with 1,000 permutations. We compared means via paired t test and paired ANOVA with Tukey’s multiple comparisons test, as appropriate.
Results
Adequacy of Sequencing
Sequences were obtained from 71 of 75 specimens (94.6%); the remaining four specimens (one each in lingula BAL, RML BAL, LUL PSB and supraglottic PSB, representing three unique subjects) had no detectable 16S band after primary PCR. Among all 71 samples, we identified 190 unique OTUs (“bacteria species”). After bioinformatic processing, the number of reads per sample was 11,964 (±18,337; mean [±SD]). Sequences are available online at the National Institutes of Health Sequence Read Archive (study accession no. SRP056031; National Institutes of Health, Bethesda, MD).
Lung Bacterial Communities Differed Significantly from Supraglottic Communities
We first tested whether method of sampling (BAL vs. PSB) influenced the microbiota detected. Intrapulmonary specimens acquired via BAL did not differ from specimens acquired via PSB in total reads, OTU richness, community diversity (Shannon index), or community composition (P > 0.05 for all). Accordingly, we included specimens acquired by both methods in all subsequent analyses.
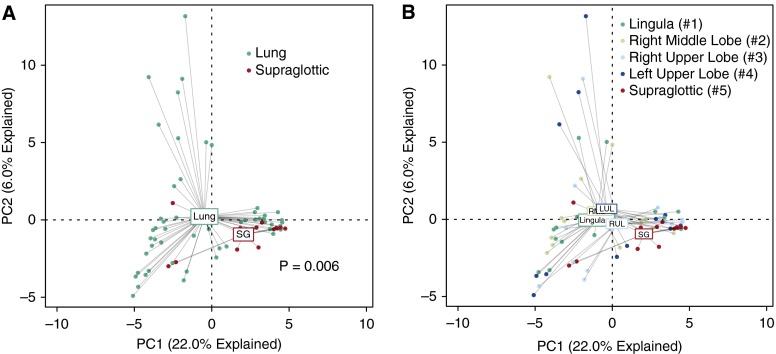
Next, to confirm that lung communities were not exclusively attributable to bronchoscopic carryover of organisms, we compared the microbiota of intrapulmonary sites with that of the URT. We first used the data visualization technique of unsupervised ordination (principal components analysis). This method distinguishes specimens based upon relative similarity of their bacterial communities, and identifies the “average” of a given specimen type by connecting individual points at a centroid. When specimens were labeled by anatomic site of origin, most supraglottic specimens clustered tightly around their centroid, whereas lung specimens exhibited broader variation in community composition (Figure 3A). Although some lung specimens were similar in community composition to supraglottic specimens, many were dissimilar. The distinct separation between the centroids of supraglottic specimens and lung specimens demonstrates a difference in the average community composition between upper and lower respiratory tracts. Permutation testing confirmed that, as a group, intrapulmonary communities differed significantly from those of the supraglottic space (P = 0.006).
Figure 3.
Ordination of supraglottic and intrapulmonary bacterial communities. (A and B) Unsupervised ordination (principal components analysis) of supraglottic (SG) versus combined intrapulmonary specimens (A) or individual intrapulmonary specimens (B), labeled by specimen site. Numbers in B refer to the order of sampling during bronchoscopy. Supraglottic communities were significantly distinct from lungs collectively (P = 0.006) and, with the exception of the right upper lobe (RUL), individually (see Table 2 for significance). LUL = left upper lobe; RML = right middle lobe.
Repeating this ordination approach with analysis of the four intrapulmonary sites disclosed that, on average, the RUL, the third intrapulmonary site sampled, most closely resembled the supraglottic site (Figure 3B). Permutation testing confirmed that the communities of the other three lung sites were statistically distinct from that of the supraglottic space (Table 2).
Table 2.
Significance of PERMANOVA (adonis) comparison of microbial communities by site
|
P Value |
|||||
|---|---|---|---|---|---|
| Intrapulmonary (All) | Lingula (#1) | Right Middle Lobe (#2) | Right Upper Lobe (#3) | Left Upper Lobe (#4) | |
| Supraglottic (#5) | 0.006 | 0.015 | 0.005 | 0.161 | 0.036 |
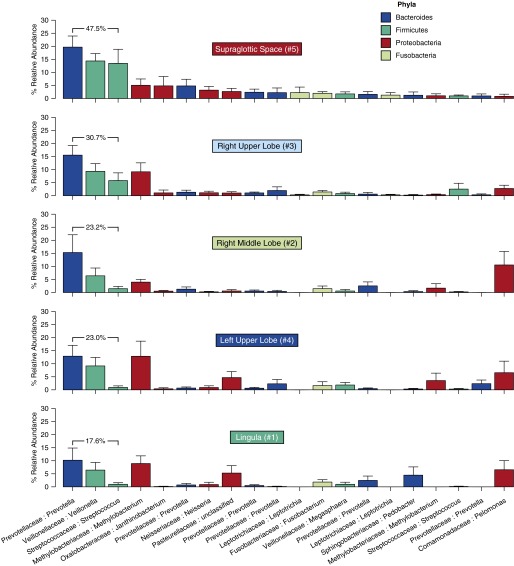
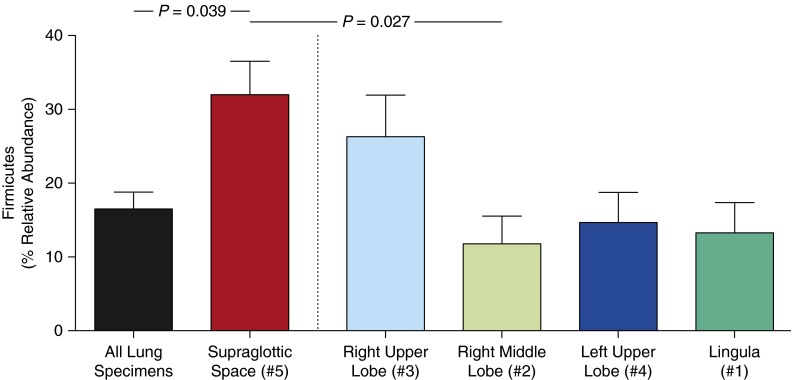
To explore further the significant differences between upper and lower respiratory tract communities, we directly compared the relative abundance of prominent URT bacteria in each site. We plotted the 20 most abundant OTUs in supraglottic specimens, ranked in descending order of mean abundance, for each specimen site (Figure 4). As suggested by ordination (Figure 3B), the RUL more closely resembled the URT than did other intrapulmonary sites, as evidenced by total relative abundance of the three most abundant OTUs (labeled). Differences between supraglottic and lung sites were attributable largely to relative abundance of the Firmicutes phylum, which was more abundant in the URT than in the lungs (P = 0.04; Figure 5). All but one subject had a greater relative abundance of Firmicutes in supraglottic specimens than in the RML (P = 0.03); the same was true for the LUL and lingula, with the exception of four and two subjects, respectively. In turn, all but two subjects had a greater relative abundance of Firmicutes in the RUL than in the more distal RML. This difference in Firmicutes was driven by a prominent Streptococcus OTU that was more abundant in supraglottic specimens (P = 0.02; Figure 4). As described previously here, community composition did not vary predictably by sampling sequence.
Figure 4.
Relative abundance of upper respiratory tract microbiota in lung specimens. The 20 most abundant operational taxonomic units (OTUs) in supraglottic specimens are ranked in descending order of mean relative abundance and shown across specimen sites. Specimen sites are labeled by anatomic site and sequence of sampling during bronchoscopy. OTUs are labeled by family:genus. Bar plots are colored by phylum (see Phyla legend); error bars represent SEM. The sum of the three most abundant upper respiratory tract OTUs (Prevotella sp., Veillonella sp., Streptococcus sp.) is shown.
Figure 5.
Relative abundance of Firmicutes phylum in spatially separated sites in the respiratory tract. Supraglottic specimens contained a significantly greater relative abundance of Firmicutes-classified operational taxonomic units than did lung specimens collectively (P = 0.04) or than right middle lobe specimens (P = 0.03; data are mean ± SEM).
We attempted to validate previous studies suggesting that select bacteria (specifically, Tropheryma whipplei [4, 30]) are disproportionately abundant in the lung relative to the URT, implying the influence of regional growth conditions on community membership. We identified a Tropheryma OTU in lung specimens of 4 of 14 subjects (28.5%). In two subjects, it was detected at all four intrapulmonary sites; in the other two, it was detected in a single site. This Tropheryma OTU was completely absent from all 14 supraglottic specimens and from reagent controls. Thus, our findings support the concept that the healthy lung microbiome harbors minority members distinct from that of the URT.
Lung Bacterial Community Richness Decreased with Distance from the URT
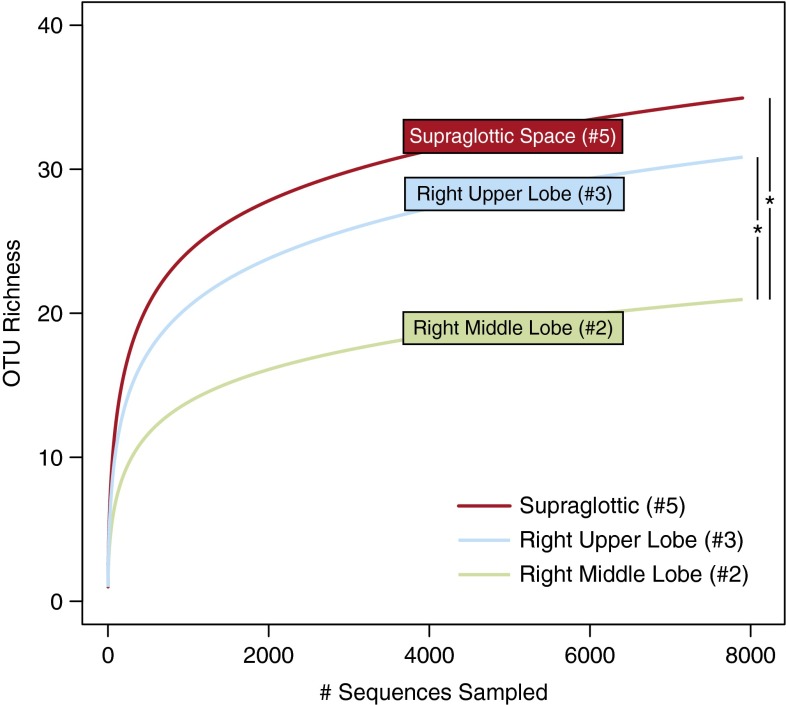
If the URT is the primary source of microbial immigration to the lungs, as postulated by the adapted island model of lung biogeography (1) (Figure 1B), community richness (i.e., the number of unique OTUs [“species”] at a given site) should decrease at greater distances from the larynx. We tested this hypothesis using rarefaction analysis (which assesses species richness independent of sampling depth) of communities at various sites along the entire respiratory tract. Community richness was greatest in supraglottic communities, followed by the most proximal lung site (RUL), then the RML (P ≤ 0.05) (Figure 6). LUL and lingula, both closer to the URT in linear distance than the RML, but protected from microaspiration by the acute-angle takeoff of the left mainstem bronchus, were of intermediate richness compared with the RUL and RML (P > 0.05 for all direct comparisons). Importantly, richness did not decrease with each subsequent sample during the bronchoscopy, nor did richness depend on whether the sample was collected by BAL or PSB.
Figure 6.
Operational taxonomic unit (OTU) richness decreases with increased distance from the source community. Rarefaction analysis demonstrated that OTU richness was greater in the supraglottic space than in intrapulmonary sites. Richness was greater in the right upper lobe (RUL) than in the right middle lobe (RML), as predicted by the adapted island model of lung biogeography. OTU richness of the lingula (#1) and left upper lobe (#4) were intermediate between RUL and RML (see main text). Numbers refer to order of bronchoscopic sampling. *P ≤ 0.05.
The Healthy Lung Does Not Contain Anatomically Distinct Bacterial Communities
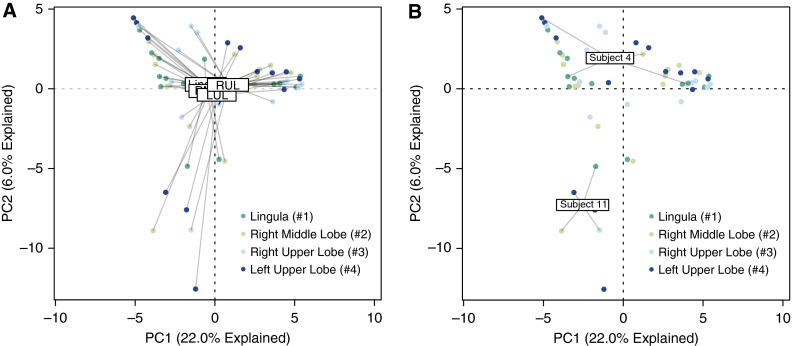
To compare the community membership of the intrapulmonary sites directly, we again visualized the data using principal components analysis. The centroids of the four intrapulmonary sites were all in close proximity, implying similarity in average community composition (Figure 7A). This finding was confirmed by permutation testing, which identified no significant difference in average community composition between any two sites (P > 0.05 for all site-to-site comparisons). No detectable difference was found in community diversity (Shannon index) when intrapulmonary sites were compared with each other (P > 0.05 for all site-to-site comparisons).
Figure 7.
Ordination of intrapulmonary bacterial communities. (A) Unsupervised ordination (principal components analysis) of intrapulmonary specimens, labeled by anatomic site. Numbers in key refer to order of sampling during bronchoscopy. Community membership was not significantly distinct across sites (P > 0.05 for all comparisons). (B) Principal components analysis labeled by representative subjects. Subject 11 exhibited similarity across intrapulmonary sites, whereas subject 4 exhibited heterogeneity in community membership. LUL = left upper lobe; RML = right middle lobe; RUL = right upper lobe.
We next compared the four intrapulmonary communities of individual subjects, but, for clarity, show only two representative participants (Figure 7B). In some (e.g., subject 11), lung sites clustered tightly in ordination space, implying similarity in community composition. In others (e.g., subject 4), considerable heterogeneity in community composition was observed across intrapulmonary sites. Relative heterogeneity of intrapulmonary communities (as measured by mean Bray-Curtis distance, a statistic that quantifies the dissimilarity in membership of distinct communities) was not associated with patient factors (demographics or lung function; P > 0.05 for all comparisons). When compared directly, the average variation in microbial community composition within individual subjects was significantly less than the variation at the same anatomical site across subjects (Bray-Curtis distance 0.80 vs. 0.86; P = 0.00003). In other words, a given subject’s RML community more closely resembles her/his own LUL community than it does other subjects’ RML communities.
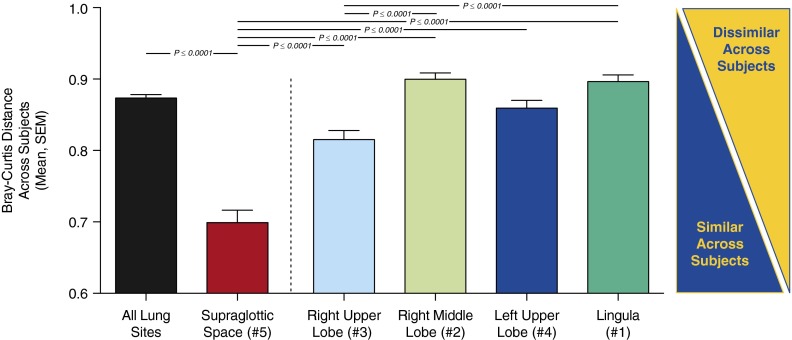
Finally, we asked if various sites in the respiratory tract differ across subjects in the relative variability of their bacterial communities. We calculated the Bray-Curtis distance between all communities obtained at the same anatomic site (e.g., the lingular community for subject 1 vs. the lingular communities of all other subjects) and compared them by anatomic site. Supraglottic communities were significantly more similar across subjects than were communities at intrapulmonary sites (Figure 8; P ≤ 0.001 for all comparisons). Of the intrapulmonary sites, communities obtained from the RUL were significantly more similar across subjects than were those of the RML or lingula (P ≤ 0.001).
Figure 8.
Comparison of intersubject similarity in microbiota by anatomic site. Microbiota detected in the supraglottic space were more similar across subjects than were microbiota detected at intrapulmonary sites (P ≤ 0.001 for all). Microbiota detected in the right upper lobe were more similar across subjects than were microbiota detected in the right middle lobe or lingula (P ≤ 0.001).
Discussion
This study systematically examined spatial variation of the respiratory tract bacterial microbiomes of 15 healthy subjects using Illumina MiSeq analysis of 16S rRNA–encoding genes. We compared microbiota of four lung lobes with each other and with the microbiota of the supraglottic space. Results show that, with the exception of the RUL, intrapulmonary sites contained a microbiome significantly distinct from that of the URT. When compared with each other, intrapulmonary sites did not contain consistently distinct microbiota. Although spatial variation in lung microbiota was observed within some individual subjects, intersubject variation at given anatomic sites significantly exceeded intrasubject variation. Microbiota from the RUL more closely resembled those of the supraglottic space in numerous important ecologic parameters (community richness, community composition, intersubject variability, and similarity to source community) than did microbiota at more distal lung sites. Our findings support the adapted island model of lung biogeography (1, 31).
These observations are important clinically, because they imply that the community composition of the healthy lung is determined principally by immigration and elimination, with minimal contribution from selective pressures on bacterial reproduction rates. One corollary is that increasing bacterial immigration, as in sizeable aspiration, permits pneumonia even in a previously healthy individual (particularly if accompanied by decreased cough, as with intoxication or otherwise altered mental status). Similarly, appreciating the nonsterile nature of the healthy lungs helps explain how viral lower respiratory infections can facilitate bacterial pneumonias in the absence of previous lung disease, simply by reducing bacterial elimination and possibly by favoring bacterial survival or proliferation in damaged epithelium.
Our results validate and extend important observations of previous studies of the healthy human lung microbiome, as explained in the next three paragraphs. The finding that the human lung microbiome is distinct from that of the URT, its primary source community, despite considerable overlap in community membership, is consistent with previous reports (4, 5). We also confirmed the disproportionate abundance in the lung of a Tropheryma OTU, reported to be lung enriched in prior studies (4, 30), whereas most other lung community members were shared by the upper and lower respiratory tracts. Both this example and the novel converse finding of a specific Streptococcus OTU in low abundance in the lungs relative to the supraglottic sample imply that the lung environment exerts at least a minor influence on community composition (2, 4).
Regarding spatial variation within the lung microbiome of individual subjects, our data are congruent with those of Charlson and colleagues (3), who analyzed the left lower lobe and RML of six healthy subjects, and Segal and colleagues (5), who analyzed the lingula and RML of 29 healthy subjects. We extend those studies by sampling a greater number of sites per subject, which permitted detection of the disparity of RUL versus other sites, and by our use of PSB, the gold-standard sampling modality in culture-dependent bronchoscopic studies. An important methodological difference between this study and that of Segal and colleagues (5) is our isolation of bacterial DNA from unfractionated BAL rather than cell-free supernatant; we have previously shown that removing eukaryotic cells significantly influences community composition of BAL fluid (32). The congruence of our collective findings, despite this significant methodological difference, speaks to the strength of the underlying biological signal. The consistent lack of spatial variation seen in the total of 50 healthy subjects of these three studies is perhaps surprising, given the known considerable regional variation within the lungs in oxygen tension, pH, and other physiological parameters relevant to relative bacterial growth rates (1, 2, 4, 13–15). Nevertheless, relative absence of spatial variation supports the transient rather than resident nature of most lung microbiome members in healthy subjects.
Importantly, our finding of greater intersubject than intrasubject variation in microbiota at distinct sites in health contrasts with previous studies in advanced lung diseases (16–18). The disparity likely results from magnified local changes in factors favoring microbial survival and even reproduction in the diseased lung (2). Support for this possibility comes from the well described association between disease severity and identification of characteristic persistent bacterial species (“colonizers”) adapted to specific conditions of diseased airways (33–35). Further studies are needed to identify the molecular mechanisms by which colonization regional growth conditions permit such in specific chronic and acute lung diseases (1, 2, 10). Such results could lead to better personalized targeting of airways diseases, thus reducing the indiscriminate use of broad-spectrum antibiotics.
The findings that the microbiota of the RUL more closely resembled that of the URT than did other lobes in four ecologically important respects, and that the RUL was intermediate between the supraglottic space and other lobes with regard to community richness, relative abundance of the Firmicutes phylum, and intersubject variation, have practical implications for sampling the airways for culture-independent analysis. Because the RUL was always the third of five sites sampled, its ecological similarity to the URT cannot plausibly be ascribed to bronchoscopic carryover. These results extend our finding that, although the microbial communities of the mouth and nose differ significantly (9), in a previous study, insertion of the bronchoscope via the mouth versus the nose has no appreciable effect on BAL microbiome community membership (2, 22). Another theoretical source of sequence contamination is bacterial DNA present in sterilized bronchoscope lumens, and our analysis did not include bronchoscopic rinse specimens as controls, as discussed in Methods. However, we identified no appreciable effect of bronchoscope contamination when specimen communities were analyzed by which bronchoscope was used. Together with the lack of difference between BAL and PSB samples in the current study, these results lead us to conclude that bronchoscopic carryover and contamination contribute minimally to analysis of lung microbiota using bronchoscopy.
Instead, we postulate that these findings are better explained by consideration of the anatomy of the respiratory tract and with microaspiration and direct mucosal dispersion as the primary routes of microbial immigration to the lung (1, 11, 12). The left mainstem bronchus arises from the trachea at a 45–50° angle, whereas the right mainstem bronchus is a relatively straight extension of the trachea (36), and thus more subject to microaspiration. Compared with the RML, which abuts the diaphragm, the RUL is more proximal to the URT source community by the length of the bronchus intermedius (2–3 cm). Thus, our findings regarding the RUL are also consistent with predictions of the adapted island model of lung biogeography (1).
Acknowledgments
Acknowledgment
The authors thank Sean Crudgington, Nicole Falkowski, and Dayana Rojas for assistance in specimen processing and sequencing, Drs. Patrick Schloss, Thomas Schmidt, Vincent Young, and all the members of the Lung HIV Microbiome Project Steering Committee for stimulating discussions and critiques, and the staff of the Veterans Affairs Ann Arbor Healthcare System Endoscopy Suite for expert care of the participants.
Footnotes
Supported by National Institutes of Health grants T32HL00774921 (R.P.D.), U01HL098961 (J.M.B., G.B.H., and J.L.C.), R01HL114447 (G.B.H.), Biomedical Laboratory Research and Development Service Merit Review Award 1 I01 BX001389 (C.M.F.), and Department of Veterans Affairs faculty salary support (J.M.B. and J.L.C.). Additional support provided by the Host Microbiome Initiative of the University of Michigan.
Author Contributions: conception and design—R.P.D., J.R.E.-D., G.B.H., and J.L.C.; acquisition of data—C.M.F., L.M., and J.L.C.; analysis and interpretation of data—R.P.D., J.R.E.-D., J.M.B., G.B.H., and J.L.C.; drafting or revising of manuscript—R.P.D., J.R.E.-D., C.M.F., J.M.B., G.B.H., and J.L.C.; final approval of the manuscript—R.P.D., J.R.E.-D., C.M.F., L.M., J.M.B., G.B.H., and J.L.C.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, Collman RG. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Chen H, Berger KI, Goldring RM, Rom WN, Blaser MJ, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [Published erratum appears in Microbiome 2:21.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Twigg HL, III, Morris A, Ghedin E, Curtis JL, Huffnagle GB, Crothers K, Campbell TB, Flores SC, Fontenot AP, Beck JM, et al. Use of bronchoalveolar lavage to assess the respiratory microbiome: signal in the noise. Lancet Respir Med. 2013;1:354–356. doi: 10.1016/S2213-2600(13)70117-6. [DOI] [PubMed] [Google Scholar]
- 7.Bertolini V, Gandolfi I, Ambrosini R, Bestetti G, Innocente E, Rampazzo G, Franzetti A. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of northern Italy. Appl Microbiol Biotechnol. 2013;97:6561–6570. doi: 10.1007/s00253-012-4450-0. [DOI] [PubMed] [Google Scholar]
- 8.Bowers RM, Sullivan AP, Costello EK, Collett JL, Jr, Knight R, Fierer N. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol. 2011;77:6350–6356. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, Beck JM, Curtis JL, Huffnagle GB. Analysis of the upper respiratory tract microbiota as the source of the lung and gastric microbiotas in healthy individuals. MBio. 6(2):e00037-15. doi:10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed]
- 11.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. 1997;111:1266–1272. doi: 10.1378/chest.111.5.1266. [DOI] [PubMed] [Google Scholar]
- 12.Huxley EJ, Viroslav J, Gray WR, Pierce AK. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med. 1978;64:564–568. doi: 10.1016/0002-9343(78)90574-0. [DOI] [PubMed] [Google Scholar]
- 13.West JB. Regional differences in the lung. Chest. 1978;74:426–437. doi: 10.1378/chest.74.4.426. [DOI] [PubMed] [Google Scholar]
- 14.Ingenito EP, Solway J, McFadden ER, Jr, Pichurko B, Bowman HF, Michaels D, Drazen JM. Indirect assessment of mucosal surface temperatures in the airways: theory and tests. J Appl Physiol. 1987;63:2075–2083. doi: 10.1152/jappl.1987.63.5.2075. [DOI] [PubMed] [Google Scholar]
- 15.Hatch TF. Distribution and deposition of inhaled particles in respiratory tract. Bacteriol Rev. 1961;25:237–240. doi: 10.1128/br.25.3.237-240.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, Conrad D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012;6:471–474. doi: 10.1038/ismej.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML, Fligner CL, Singh PK. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci USA. 2012;109:13769–13774. doi: 10.1073/pnas.1107435109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. Cell-associated bacteria in the human lung microbiome. Microbiome. 2014;2:28. doi: 10.1186/2049-2618-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. Analysis of culture-dependent vs. culture-independent techniques for the identification of bacteria in clinically-obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52:3605–3613. doi: 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson RP, Erb Downward JR, Freeman CM, Walker N, Scales BS, Beck JM, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. Changes in the lung microbiome following lung transplantation include the emergence of two distinct pseudomonas species with distinct clinical associations. PLoS One. 2014;9:e97214. doi: 10.1371/journal.pone.0097214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16s rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD.Miseq SOP—mothur. 2015 [accessed 2013 Oct 27]. Available from: http://www.mothur.org/wiki/miseq_sop
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Center for Microbial EcologyRdp release 11, update 3. 2015 [accessed 2013 Oct 27]. Available from: https://rdp.cme.msu.edu/
- 28.R Core Team. Vienna: R Foundation for Statistical Computing; 2013. R: a language and environment for statistical computing. [Google Scholar]
- 29.Oksanen JF, Blanchet G, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: community ecology package. R package version 2.0-4. 2012.
- 30.Lozupone C, Cota-Gomez A, Palmer BE, Linderman DJ, Charlson ES, Sodergren E, Mitreva M, Abubucker S, Martin J, Yao G, et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am J Respir Crit Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whiteson KL, Bailey B, Bergkessel M, Conrad D, Delhaes L, Felts B, Harris JK, Hunter R, Lim YW, Maughan H, et al. The upper respiratory tract as a microbial source for pulmonary infections in cystic fibrosis: parallels from island biogeography. Am J Respir Crit Care Med. 2014;189:1309–1315. doi: 10.1164/rccm.201312-2129PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, Schmidt TM. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 2015;6:pii:e02284–14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cystic Fibrosis FoundationPatient registry report. 2012 [accessed 2014 Apr 25]. Available from: http://www.cff.org/uploadedfiles/research/clinicalresearch/patientregistryreport/2012-cff-patient-registry.pdf
- 34.Evans SA, Turner SM, Bosch BJ, Hardy CC, Woodhead MA. Lung function in bronchiectasis: the influence of Pseudomonas aeruginosa. Eur Respir J. 1996;9:1601–1604. doi: 10.1183/09031936.96.09081601. [DOI] [PubMed] [Google Scholar]
- 35.Zalacain R, Sobradillo V, Amilibia J, Barron J, Achotegui V, Pijoan JI, Llorente JL. Predisposing factors to bacterial colonization in chronic obstructive pulmonary disease. Eur Respir J. 1999;13:343–348. doi: 10.1034/j.1399-3003.1999.13b21.x. [DOI] [PubMed] [Google Scholar]
- 36.Kubota Y, Toyoda Y, Nagata N, Kubota H, Sawada S, Murakawa M, Fujimori M. Tracheo-bronchial angles in infants and children. Anesthesiology. 1986;64:374–376. doi: 10.1097/00000542-198603000-00015. [DOI] [PubMed] [Google Scholar]