Abstract
Mechanical stimulation is crucial to bone growth and triggers osteogenic differentiation through a process involving Rho and protein kinase A. We previously cloned a gene (AKAP13, aka BRX) encoding a protein kinase A-anchoring protein in the N-terminus, a guanine nucleotide-exchange factor for RhoA in the mid-section, coupled to a carboxyl region that binds to estrogen and glucocorticoid nuclear receptors. Because of the critical role of Rho, estrogen, and glucocorticoids in bone remodeling, we examined the multifunctional role of Akap13. Akap13 was expressed in bone, and mice haploinsufficient for Akap13 (Akap13+/−) displayed reduced bone mineral density, reduced bone volume/total volume, and trabecular number, and increased trabecular spacing; resembling the changes observed in osteoporotic bone. Consistent with the osteoporotic phenotype, Colony forming unit-fibroblast numbers were diminished in Akap13+/− mice, as were osteoblast numbers and extracellular matrix production when compared to control littermates. Transcripts of Runx2, an essential transcription factor for the osteogenic lineage, and alkaline phosphatase (Alp), an indicator of osteogenic commitment, were both reduced in femora of Akap13+/− mice. Knockdown of Akap13 reduced levels of Runx2 and Alp transcripts in immortalized bone marrow stem cells. These findings suggest that Akap13 haploinsufficient mice have a deficiency in early osteogenesis with a corresponding reduction in osteoblast number, but no impairment of mature osteoblast activity.
Keywords: OSTEOPOROSIS, BRX, RHO-GEF, OSTEOGENESIS, RUNX2
Introduction
Osteoblasts play an essential role in bone formation.(1) In the postnatal organism, osteoblasts develop from bone marrow stromal cells (BMSCs) to become preosteoblasts, which in turn, become functional osteoblasts.(2) Osteoblast differentiation is regulated by various hormones and cytokines, such as bone morphogenetic proteins (BMPs), fibroblast growth factors (FGFs), transforming growth factor (TGF)-β, parathyroid hormone (PTH), and insulin-like growth factors (IGFs), and multiple transcription factors such as Runt-related transcription factor 2 (Runx2), Osterix (Osx), activating transcription factor 4 (ATF4), β-catenin, and Msx2.(3–5) In addition to local biochemical factors, mechanical stimulation is critical for initiation of osteogenic differentiation through a process involving multiple second messengers,(6,7) including Rho.(8,9)
Previously, we(10) and others(11–13) cloned a gene encoding protein A kinase-anchoring protein 13 (AKAP13). The 5.3-kb transcript that we isolated from a human breast cancer called BRX,(10) is encoded by the gene now known as AKAP13. AKAP13 contains a guanine nucleotide-exchange factor (GEF) that activates Rho GTPases.(10,12–15)Rho GTPases require GEFs, which catalyze the exchange of GTP for guanosine diphosphate (GDP) to activate the GTPase.(16,17) AKAP13 is a GEF for RhoA(12,13) and forms trimeric complexes with RhoA and protein kinase A (PKA(12)). Full-length AKAP13 transcripts encode a large modular protein with a molecular mass of 312 kDa.(12) The Lbc oncogene,(11) is a smaller AKAP13 transcript variant derived from two chromosomes, 15 and 7.(13)
We found that proteins encoded by AKAP13 bound to nuclear hormone receptors through a C-terminal nuclear receptor–interacting domain (NRID) and enhanced gene activation by estrogen receptor (ERα),(10) ERβ,(18) and the glucocorticoid receptor.(19) Of note, AKAP13 also contains a N-terminal protein kinase A anchoring domain that binds to the regulatory subunit of PKA. In addition, AKAP13 is associated with the actin cytoskeleton,(20,21) binds to α catulin in a yeast two hybrid screen (both as bait and as prey), and nucleates actin filaments to affect cell shape.(21,22) Gene targeting revealed an essential role of the gene in murine development as the Akap13−/− (null) mutation was embryonic lethal.(20)
AKAP13 proteins also coordinate signals originating from the cell membrane, including lysophosphatidic acid (LPA) receptors, the osmoreceptor(23) and serotonin receptor.(22) Application of LPA or selective expression of Gα12 and Gn13 enhanced cellular AKAP13 activation.(24) AKAP13 proteins have been shown to interact with Gα12,(12) Gα13,(25) Gα14,(26) GYq, and possibly Gs15.(19) Collectively, these functions of AKAP13, and the products of smaller transcripts such as BRX,(10) suggest thatAKAP13molecules act as integrators or docking platforms for multiple signal transduction pathways,(24,27) including Rho and mechanical signaling. Consistent with a possible role of AKAPs in bone remodeling, a genomewide analysis study revealed that AKAP11 was associated with lumbar spine bone mineral density.(28) However, despite the importance of Rho signaling for bone formation and development,(29) and the well-known effects of both estrogens and glucocorticoids on bone remodeling,(30) the possible involvement of AKAP13 in bone metabolism has not been described.
Because of our interest in AKAP13 as a molecule capable of integrating mechanical and hormonal signaling, possibly in bone, we tested the hypothesis that AKAP13 might play a role in bone homeostasis. To assess the physiological importance of AKAP13 in bone, we examined Akap13 haploinsufficient (+/−) mice. Micro-CT (µCT) analyses revealed that bone mineral density was significantly reduced in Akap13+/− mice, compared to wild-type (WT) mice, resembling the changes observed in osteoporotic bones. Haploinsufficient mice had reductions in osteoprogenitors, osteoblasts, and extracellular matrix. Experiments using BMSCs, and MC3T3-E1 cells, and knockdown of Akap13 suggest that Akap13 is required for proper osteogenesis at the earliest stages of osteoblast development.
Materials and Methods
Generation of Akap13+/− mice
Akap13+/− mice were generated and genotyped as described.(20) The study was conducted under an institutional protocol in accordance with NIH Animal Care and Use guidelines.
µCT and 3D analysis
Femora from Akap13+/− mice, or controls, were fixed in 4% formaldehyde in phosphate-buffered saline (PBS), washed with PBS and stored in 70% ethanol for µCT analyses.(31) Bone morphology of femora of 10-week-old and 20-week-old Akap13 haploinsufficient or WT mice were analyzed by µCT scanning using a SKYSCAN1172 (SKYSCAN, Kontich, Belgium) at 17.1-µm resolution. Quantitative data and visual models from scanned datasets of bones were obtained by using a CT analyzer (SKYSCAN). Regions of interest (ROIs) were selected with reference to a growth plate reference slice. The trabecular regions were defined as positions along the long axis of the femur relative to the growth plate reference. The trabecular region commenced 0.215 mm (50 image slices) from the growth plate level in the direction of the metaphysis, and extended from this position for a further 1.72 mm (450 image slices). The volume of interest (VOI) was automatically morphed by the Skyscan CT analyzer to edited ROI shapes between edited levels. 3D images were reconstructed by the volume rendering method and morphometric parameters, the trabecular bone mineral density (BMD), trabecular bone volume per tissue volume (BV/TV), bone surface per bone volume (BS/BV), trabecular thickness (Tb.Th), trabecular number (Tb.N), trabecular spacing (Tb.Sp), cortical area (Ct.Ar), cortical thickness (Ct. Th), maximum and minimum moments of inertia (I max and I min, respectively) were measured (CT analyzer program) in the distal femurs.(31)
RNA extraction and real-time RT-PCR
Femurs were dissected from 15-week-old mice, marrow cavities flushed, and the bones were stored at −80°C until processed. Samples were crushed with a mortar and pestle under liquid nitrogen, powder-like samples were placed in TRIzol (Invitrogen, Carlsbad, CA, USA), homogenized, and RNA was extracted according to the manufacturer’s instructions. Additionally, total cellular RNA was isolated from MC3T3-E1 cells(32) or BMSCs(33) using the RNeasy kit (QIAGEN, Valencia, CA, USA).RNA was reverse transcribed by using SuperScript III First-Strand Synthesis System (Invitrogen). cDNAs were generated from RNA extracted from the proliferative zone and the hypertrophic zone of growth plate and bones of rats as described.(34) Rats were used to facilitate accurate dissection of the two different zones, and to confirm Akap13 expression in bone in another species. Quantitative PCR was performed using an ABI PRISM 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with duplicates of the 25-µL reaction mixture of QuantiTect SYBR Green PCR Kits (QIAGEN) in MicroAmp optical 96-well, sealed reaction plates. Real-time RT-PCR was conducted using primers for mouse Akap13, Runx2, Alp, Osterix, Osteocalcin, Collagen type 1, and 18s, as noted in Supplementary Table 1.
Cell culture
Femurs were aseptically removed, cleaned of soft tissues, and the marrow cavities of bones were flushed into nutrient media (α-minimum essential medium [α-MEM], 2 mM glutamine [Invitrogen], penicillin [100 U/mL], streptomycin sulfate [100 µg/mL; Invitrogen], and 20% lot-selected, non–heat inactivated fetal bovine serum) using a nutrient media-filled syringe attached to a 25G needle. Single-cell suspensions of bone marrow were plated into a 75-cm2 flask per mouse in nutrient medium. Adherent cells (BMSCs) were passaged after treatment with collagenase type IV (Invitrogen), 0.05% trypsin-0.53 mM EDTA (Invitrogen). Cultures were maintained for up to 4 weeks with four passages and media changes every 3 days. Osteogenic differentiation was performed in nutrient medium supplemented with 1 × 10−4 M L-ascorbic acid 2-phosphate, 1 × 10−8 M dexamethasone (Sigma-Aldrich, St. Louis, MO, USA), and 5 mM β-glycerophosphate (Sigma-Aldrich) as described.(33) MC3T3-E1 cells (ATCC, Manassas, VA, USA) were maintained in α-MEM (Invitrogen) with 10% fetal bovine serum (ATCC) and 1% penicillin-streptomycin (Invitrogen) at 37°C, 5% CO2. Media was replaced every 3 days, and cells were subcultured weekly for use within 20 passages. Osteogenic differentiation of MC3T3-E1 cells was induced by differentiation medium composed of α-MEM containing 10% fetal bovine serum, 1% penicillin-streptomycin (Invitrogen), 0.2 mM L-ascorbic acid 2-phosphate (Sigma-Aldrich), and 10 mM β-glycerophosphate, as described.(32)
Alizarin red staining
Cells were rinsed with calcium and PBS, fixed with 4% paraformaldehyde for 10 min, rinsed with 95% ethanol, and air-dried. Cells were rinsed briefly with water, stained for 2 min with 1% alizarin red S solution (pH 4.2) (Sigma-Aldrich) at room temperature, and washed four times with water before analysis.
siRNA transfections
For this study, a spontaneously immortalized line of BMSCs was used (see Supplementary Fig. 1),(35) and grown using the same culture conditions as described in the cell culture section above. Transfection of Akap13-siRNA or control siRNA was performed using Lipofectamine-2000, according to the manufacturer’s instructions. For 96-well plates, 5pmol siRNA and 0.25 µL Lipofectamine with 25 µL OptiMEM (Invitrogen), and for six-well plates, 100 pmol siRNA and 5 µL Lipofectamine with 25 µL OptiMEM, each were incubated and combined. The siRNA and Lipofectamine mixture was added to the antibiotic-free growth medium followed by RNA extractions after 24 or 48 hours. Controls consisted of scrambled siRNAs.
Osteoblast quantification
Tibias of 6-week-old mice were dissected. Regions of the endosteal surface of tibias were captured by CellSens Digital imaging software (Olympus, Tokyo, Japan). Osteoblast nuclei on the endosteal surface were counted using OsteoMeasure software (OsteoMetrics, Decatur, GA, USA). Additionally, femurs from 20-week-old mice were dissected and analyzed by histomorphometry as described.(36)
Calcein and tetracycline double labeling
Intraperitoneal (i.p.) administration of 25mg/kg Calcein (Sigma) in 2% NaHCO3 buffer was given to 6-week-old mice. Ten days later, 25mg/kg tetracycline (Sigma) in 0.9% saline was given i.p. to the same mice. Mice were euthanized on day 12 and tibias were dissected, freed of muscle tissue, and fixed in 70% ethanol for several days protected from light, followed by methyl meth acrylate embedding. Embedded sections were cut and images of labeled bones were captured using a fluorescent microscope and analyzed. Inter-labeling areas were measured using the OsteoMeasure program (OsteoMetrics) and standard histomorphometry.
Tartrate-resistant acid phosphatase staining
Femurs from 20-week-old mice or calvariae of 7-week-old mice were dissected, then rinsed in ice-cold PBS. Specimens were fixed in ice-cold 70% ethanol. Calvariae were washed briefly with ice-cold PBS, fixed in acetone citrate for 30 s, washed briefly in PBS and transferred to a prewarmed garnet/tartrate solution of the Acid Phosphatase kit (Sigma-Aldrich). Stained femurs were analyzed using standard histomorphometry.(36) Calvariae were stained at 37°C, protected from light, and assessed every 5 min. Stained specimens were stored in 70% ethanol and analyzed.
Colony-forming efficiency assay
Bone marrow cells from shafts of femurs and tibias of 18-week-old mice were flushed as described for extraction of BMSCs.(37) A single-cell suspension of bone marrow was plated at low (clonal) density in a plastic culture flask (1 × 106 cells/25 cm2) with media as described in the cell culture section above and cultured for 12 days at 37°C, 5% CO2 without changing media. Next, flasks were rinsed once with Hanks’ Balanced Salt Solution (GIBCO-Life Technologies, Grand Island, NY), fixed with 100%methanol for 30 min, and stained with an aqueous solution of saturated methyl violet for 30 min. Flasks were rinsed three times with distilled water and dried overnight. Dried, stained colonies (>50 cells) were counted using a dissecting microscope, as described.(38)
Measurement of ALP mRNA in osteoprogenitors and mature osteoblasts
Osteoprogenitor cells (BMSCs) and mature osteoblasts were harvested from bone marrow and calvariae, respectively, from AKAP13 haploinsufficient or WT mice.(37) We performed excision of all non-bone surfaces; ie, sutures and cartilage. Next, the bone was digested five times 15 min in 2 mL of 4 mg/mL collagenase/dispase in PBS with gentle shaking. Cells in the first three digestions were discarded. Resultant cells were collected and depleted for CD45+ and CD 11b+ cells. A magnetic sorting system (MACS, Miltenyi Biotec Inc., Auburn, CA, USA) was used to remove hematopoietic cells from single-cell suspensions passed through a Falcon 70-µm cell strainer using antibodies to CD11b and CD45 according to manufacturer’s instructions. Effluent cells were assayed using real-time RT-PCR and primers for murine Alp transcripts as described in the RNA extraction section above.
In situ hybridization
cDNA from 1-week-old mouse growth plates were amplified using Phusion DNA polymerase (Thermo Scientific, Waltham, MA) with primers that contained either a T7 promoter (for sense probes) or an SP6 promoter (for probes; see Supplementary Table 2). Single-stranded digoxigenin-labeled riboprobes for in situ hybridization were transcribed using DIG RNA Labeling Kit (Roche Diagnostics, Nutley, NJ) following the manufacturer’s protocol. Riboprobes were purified by Micro Bio-Spin Columns P-30 Tris RNase free (Bio-Rad, Hercules, CA), followed by alkaline hydrolysis for 30 min as described.(39) Paraffin-embedded sections of epiphyseal cartilage from1-week-old or 11-week-old mice were hybridized to digoxigenin-labeled riboprobes as described.(40) For detection, tissue sections were incubated with anti-digoxigenin alkaline phosphatase Fab fragments (Roche) for 2 hours at room temperature and treated with 4-nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Sigma) in the dark until a colorimetric change was detected. Sections were visualized using a ScanScope CS digital scanner (Aperio Technologies, Inc., Vista, CA) under bright field microscopy.
Statistical analyses
Data are expressed as mean ± SE for all values. Results were evaluated using Student’s t test or one-way analysis of variance followed by Tukey-Kramer’s post hoc test.
Results
Akap13+/− mice have lower bone mineral density than WT mice
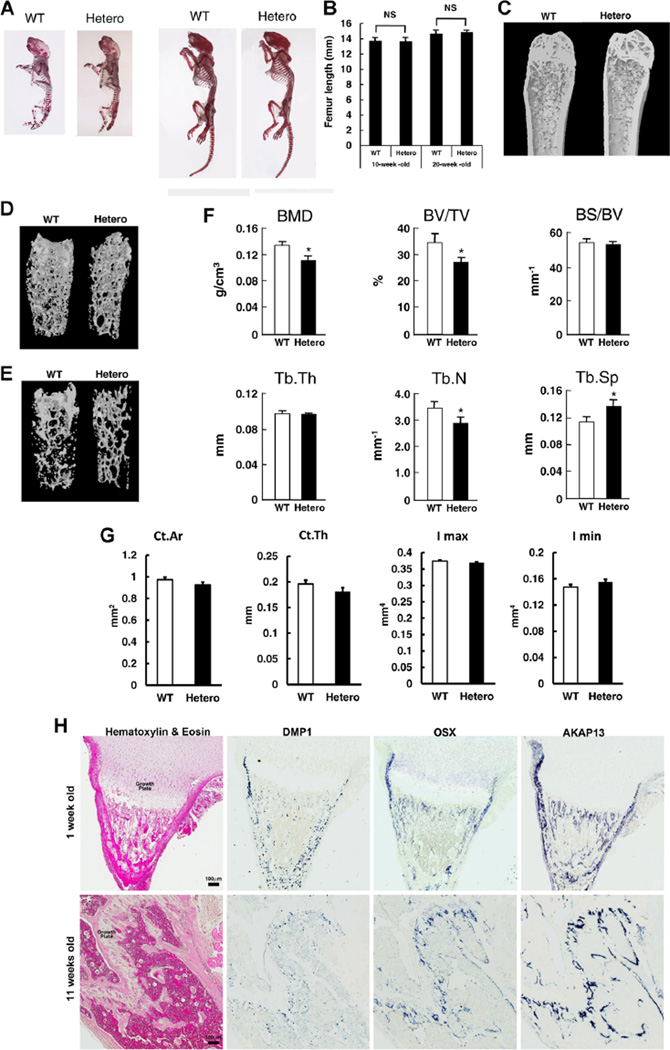
To determine whether Akap13 was involved in bone formation or maintenance, we first surveyed bone development. No skeletal anomalies or dysmorphic features were present (Fig. 1A), and growth before and after birth was not impaired as shown by analysis of femoral length of 10-week-old and 20-week old mice (Fig. 1B). However, X-ray studies suggested that Akap13+/− mice had a reduced bone content when compared with WT littermates (data not shown). We therefore quantified the in vivo bone structure of the femora of Akap13+/− and WT mice with µCT. µCT analysis of femora from 20-week-old female mice revealed decreased density, especially in trabecular bone (Fig. 1C–E). Quantitative analysis of the distal femora revealed that the BMD, BV/TV, and Tb.N were lower, and correspondingly, Tb.Sp was greater in the 20-week-old Akap13+/− mice compared to controls (Fig. 1F). Notably, cortical bone thickness was also reduced, but differences were not significant (Fig. 1G). Ten-week-old Akap13+/− mice showed similar trends, but most parameters were not significantly different at that age (Supplementary Fig. 2).
Fig. 1.
Bone phenotype of Akap13+/− mice and expression of Akap13 in bone. (A) Skeletal structure of WT and Akap13+/− mice (Hetero). Left: newborn mice. Right: 10-week-old mice. Skeletal staining by alizarin red. (B) Bone length in 10-week-old and 20-week-old Akap13+/− mice compared with WT mice. Bars represent the mean ± SE (n = 5). (C) Femurs from 20-week-old WT and Akap13+/− mice revealed decreased bone mineral density. (D, E) 3D rendering images of µCT of femora from 20-week-old WT or Akap13+/− mice showed decreased trabecular bone in the Akap13+/− mice. (F) Quantitative comparison of femoral microstructure parameters of 20-week-old WT and Akap13+/− mice. Global threshold method was used with a fixed range of grayscale values for solid pixels (white) and pixels outside this range were set as space pixels (black). Voxels range from 0 (air; HV-1000) to 255 (HV16444) with water 14.618 on the grayscale voxel. The threshold was a lower of 24 and upper of 255 voxels, the number of layers was 101, which was used for all specimens. *p < 0.05 (n = 5 mice/group). (G) Cortical parameters measured by µCT for the distal femur of 20-week-old female WT and Akap13+/− mice. (H) In situ hybridization for Akap13 transcripts. Formalin fixed, decalcified sections from proximal tibias of 1-week-old (upper panels) or 11-week-old (lower panels) mice were hybridized to DIG-labeled riboprobes for Akap13, Osterix (Osx), or Dentin matrix phosphoprotein 1 (Dmp1), producing a purple color visualized with a ScanScope CS digital scanner under bright field microscopy. Left: Hematoxylin and eosin–stained sections. Right: In situ hybridization (not counterstained). Akap13 localization in the tibia resembled that of Osterix, a specific marker for preosteoblasts and osteoblasts. BMD = trabecular bone mineral density; BV/TV = trabecular bone volume per tissue volume; BS/BV = bone surface per bone volume; Tb.Th = trabecular thickness; Tb.N = trabecular number; Tb.S = trabecular spacing; Ct.Ar = cortical area; Ct.Th = cortical thickness; Hetero = Akap13+/− mice.
Next, to identify which type of bone cells expressed Akap13, we analyzed Akap13 expression in bone using in situ hybridization. Akap13 localization closely resembled that of Osterix (Fig. 1H), a specific marker for chondroprogenitor/osteoprogenitor cells, preosteoblasts, and osteoblasts.(41) Akap13 expression was not observed in growth plate chondrocytes but was present in the perichondrium. Next, Akap13 expression was quantified using RNA from the growth plate and metaphyseal bones from 3-week-old to 8-week-old rats (used in order to definitively dissect out the proliferating and hypertrophic zones and to confirm the expression in another species) (Supplementary Fig. 3). Akap13 mRNA was barely detected in the proliferative zone or hypertrophic zone, and was expressed approximately 10-fold higher in metaphyseal bone. In support of the in situ data, Akap13 protein in bone was confirmed using immunohistochemical analysis and anti-sera directed against Akap13 (Supplementary Fig. 4). The observation that Akap13 is expressed in osteoblasts and osteoblast precursors, but not to a great extent in the growth plate chondrocytes, provides a possible explanation for the reduction in bone mass but not in longitudinal bone growth of Akap13 haploinsufficient mice.
Colony formation and osteoblast number are reduced in Akap13+/− mice
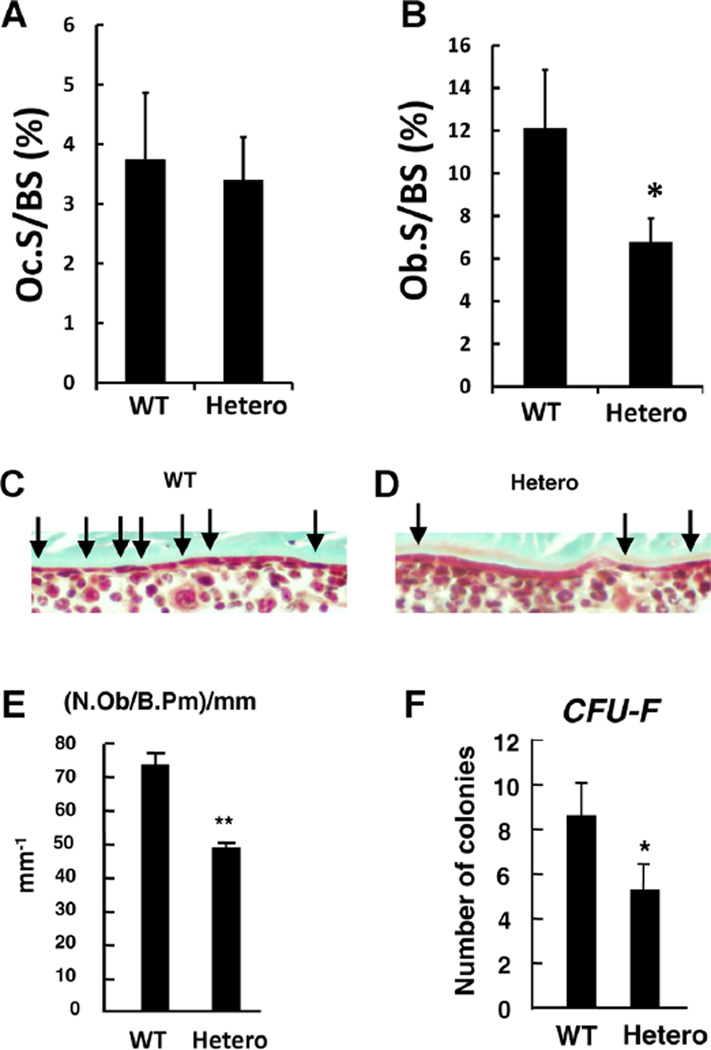
Next, we sought to determine whether the deficiency in bone mass was due to increased osteoclastic activity, or alternatively, a reduction in bone formation. BecauseAKAP13 had been shown to affect estrogen action via estrogen receptors α(10) and β,(18) we suspected that haploinsufficient mice might have increased osteoclastic activity. However, histomorphometric assessment of 20-week-old femurs using tartrate-resistant acid phosphatase (TRAP) staining revealed osteoclast surface perbone surface (OcS/BS%) to be similar in Akap13+/− mice versus controls (3.74%versus 3.42%, respectively, p = 0.41; Fig. 2A). Similarly, the number of osteoclasts per bone perimeter (N.Oc/B.Pm) were also similar, 2.0 in WT and 1.9 in haploinsufficient mice (p = 0.40). Additionally, analysis of calvariae from 7-week-old mice did not suggest increased osteoclastic activity (Supplementary Fig. 5).
Fig. 2.
Reduction in osteoblast number, but not osteoclast number in Akap13+/− mice. (A) Femurs from WT or Akap13+/− mice at 20 weeks of age were assessed for OcS/BS% using TRAP staining. Differences were not significant (p = 0.40; n = 6/group). (B) ObS/BS% was assessed using Goldner’s trichrome staining of trabecula from femurs from 20-week-old WT or Akap13 haploinsufficient mice (Hetero). n = 6 per group; *p < 0.05. (C, D) Quantification of osteoblast number by Goldner’s trichrome (bone is green, cells are stained red) in cortical bone. Micrographs showing spatial distribution of osteoblasts (black arrows) on the endosteum of WT (C) or Akap13+/− (Hetero) mice (D) showed a reduced number of osteoblasts in the Akap13+/− mice. (E) N.Ob/Ob.Pm data expressed quantitatively for WT or Akap13+/− mice (Hetero), error bars = SE, ** p < 0.01. (F) Colony forming efficiency (the number of CFU-Fs that give rise to colonies) was analyzed in BMSCs of WT and Akap13+/− mice (Hetero). Bars represent the mean ± SE, (n = 5; *p < 0.05). OcS/BS% = osteoclast surface per bone surface; TRAP = tartrate-resistant acid phosphatase; ObS/BS% = osteoblast surface per bone surface; Hetero = Akap13 haploinsufficient mice; N.Ob = osteoblast number; Ob.Pm = osteoblast perimeter.
To evaluate whether reduction in Akap13 led to a reduction in osteoblast number, quantitative histomorphometry on 20-week-old mice (Fig. 2B) was performed using Goldner’s trichrome staining and revealed that osteoblast surface per bone surface of trabecular bone (ObS/BS%) was reduced in haploinsufficient mice at an average of 6.7%, versus 12.1% in WT littermates (p < 0.05). Next, we quantified osteoblast number in the periosteum and noted a reduction in osteoblast number in 6-week-old Akap13+/− mice, compared to controls (Fig. 2C–E). These observations raised the possibility that osteogenesis was impaired in the Akap13+/− mice. The reduction in osteoblast number led us to question whether there was a decrease in the number of osteoprogenitors in Akap13+/− mice. To test that possibility, colony forming efficiency assays (the number of BMSC colonies initiated by individual CFU-Fs) were performed on Akap13+/− mice.(37) We found that numbers of colonies in the Akap13+/− mice were significantly decreased (p < 0.05), compared to those in WT mice (Fig. 2F).
Reduced osteoblast number was accompanied by a reduction in extracellular matrix
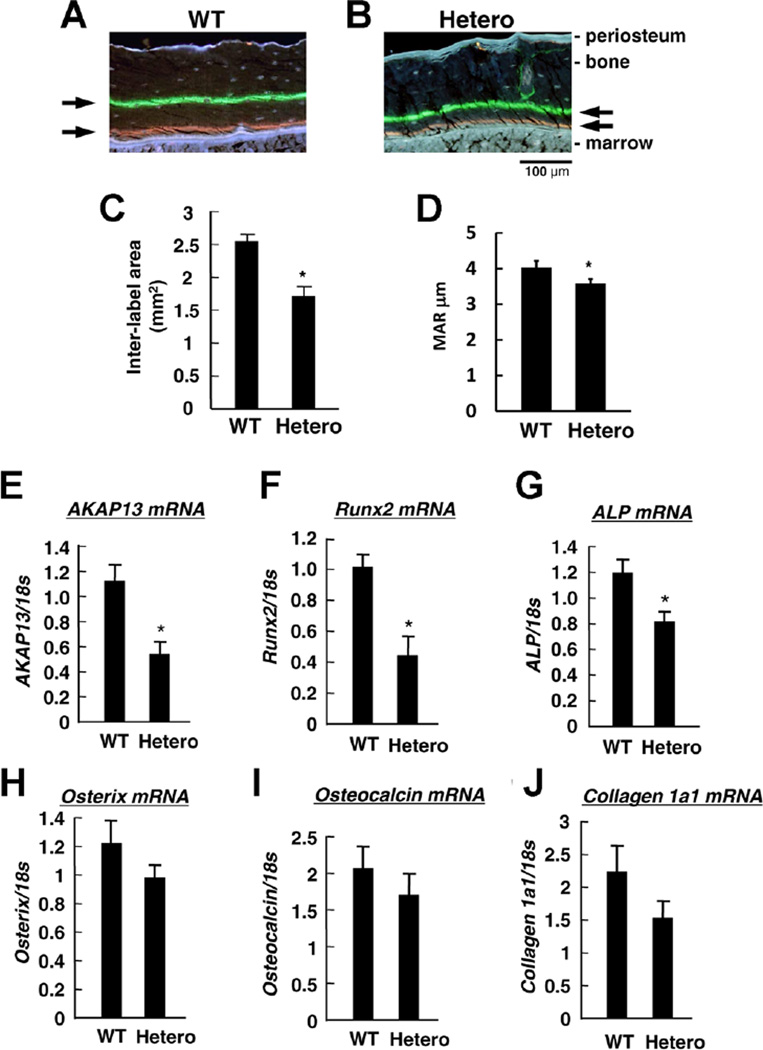
The reduction in bone mass and osteoblast number led us to quantify synthesis and deposition of bone extracellular matrix using calcein double labeling experiments on WT and Akap13+/− mice. Of note, mice haploinsufficient for Akap13 exhibited reduced matrix deposition, compared to littermate controls (Fig. 3A–D, Supplementary Fig. 6) at 6 weeks. The impairment in matrix production was supported by analysis of the trabecular compartment at 6 weeks, which suggested an increase in single-labeled surfaces, and a reduction in double-labeled surfaces (Supplementary Table 3). In the 20-week-old mice, the WT mice showed a reduction in bone formation consistent with aging, but interestingly, bone formation in the haploinsufficient mice continued. We interpreted these data to suggest a reduction in extracellular matrix stemming from the reduced osteoblast number.
Fig. 3.
Assessment of bone formation and expression of Akap13 and osteogenic markers in mouse femoral bone. (A, B) New bone formation (black arrows) as determined by calcein and tetracycline double labeling of WT (A) or Akap13+/− mice (Hetero) (B) in the mid-shaft of the tibias at 6 weeks of age. Representative micrograph. Note the reduction in space between arrows in the Hetero mice. Bar = 100 µm. (C) Quantitative measurement of matrix deposition using Osteo Measure determined by region of double staining mean value ± SE (n = 3 per group), *p < 0.05. (D) Endocortical MAR in µm per day in WT or Akap13+/− mice (Hetero) (n = 5 per group; *p = 0.04, one-tailed t test). (E) Relative mRNA expression of Akap13, (F) Runx2, (G) Alp, (H) Osterix, (I) Osteocalcin, and (J) Collagen Type 1a1, in the femoral bones from WT or Akap13+/− mice (Hetero) as shown. Data were normalized to 18s ribosomal RNA. Bars = mean ± SE, *p < 0.05 (n = 8 mice/group). Hetero = Akap13+/− mice; MAR = mineral apposition rate.
Runx2 and Alp were decreased in femora from Akap13+/− KO mice
Next, to determine whether reduction in Akap13 affected osteoblast differentiation, we examined critical osteogenic markers known to regulate bone formation in the haploinsufficient mice. As expected, quantification of transcripts in femora of 15-week-old mice using real-time PCR showed that Akap13 mRNA levels in Akap13+/− mice were 50%lower than in WT mice (Fig. 3E). Notably, Runx2 and Alp transcripts were significantly reduced in the Akap13+/− mice (Fig. 3F, G), suggesting that Akap13 may be involved in early steps of osteoblast differentiation. Runx2 is one of the earliest osteoblast differentiation factors and a critical early determinant of osteoblast differentiation(42–45); however, the upstream factors that regulate Runx2 are incompletely understood. On the other hand, real-time PCR did not reveal significant differences between haploinsufficient and WT mice in mRNA abundance of Osterix, Osteocalcin, and Collagen 1a1 mRNA (Fig. 3H–J). To further assess osteoprogenitor function in the Akap13+/− mice, we measured Alp transcript levels in osteoprogenitors and mature osteoblasts. Levels of Alp mRNA were reduced in osteoprogenitor cells of Akap13+/− mice, but not in mature osteoblasts (Supplementary Fig. 7), consistent with an effect on osteoprogenitor commitment, although other approaches might be used to further assess function of the mature osteoblasts. Taken together, these findings coupled with the colony forming efficiency assay are consistent with a role for Akap13 in early osteogenesis.
Role of Akap13 in osteoblast differentiation
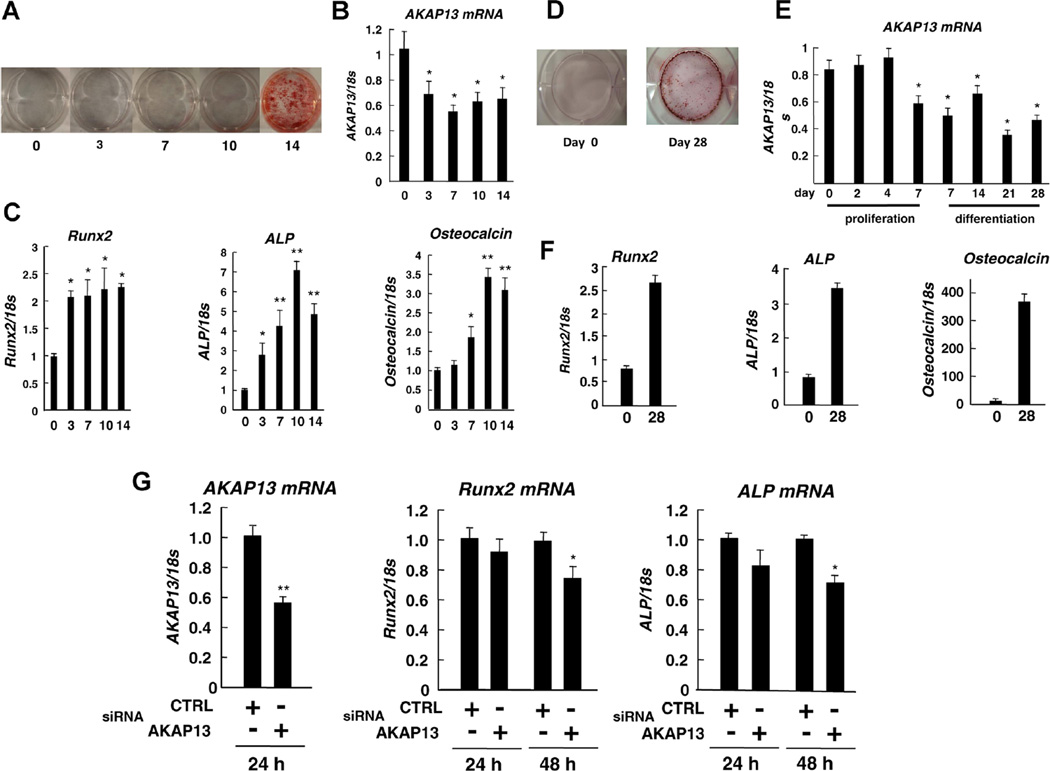
To explore the role of Akap13 in osteoblast differentiation in greater detail, we sought to examine levels of expression of Akap13 during osteogenesis in vitro. Therefore, we assessed expression of Akap13 transcripts during proliferation and differentiation stages using two in vitro models of osteoblast differentiation, BMSCs, and MC3T3-E1 cells, a murine preosteoblast cell line(32) (Fig. 4B, E, respectively). Real-time PCR was performed to analyze levels of mRNAs of Akap13, Runx2, Alp, and Osteocalcin. As expected, and consistent with prior reports (Fig. 4A, D), these osteogenic markers were increased as BMSCs and MC3T3-E1 differentiated as demonstrated by alizarin red S staining (Fig. 4C, F). Of note, in both models of osteoblast differentiation, Akap13 transcripts were initially high in the proliferating cells, and transcript levels fell during differentiation of both BMSCs and MC3T3-E1 cells (Fig. 4B, E). It was also observed that Akap13 mRNA was decreased by day 7 of proliferation, prior to initiation of differentiation. Taken together, these results also suggest a role for Akap13 in very early steps or stages of osteoblast differentiation.
Fig. 4.
Levels of Runx2 transcripts are dependent, in part, upon Akap13. Relative expression of Akap13, Runx2, Alp, and Osteocalcin during the osteogenic differentiation of BMSCs or MC3T3-E1 cells. (A) BMSCs were cultured in osteogenic induction medium and total RNA was collected at 0, 3, 7, 10, and 14 days, and cells were stained with Alizarin red. (B) mRNA expression levels of Akap13 at different days as noted. (C) Transcripts of Runx2, ALP, and osteocalcin were also quantified in cells similarly cultured. Data were normalized to 18s ribosomal RNA. Bars represent the mean ± SE (n = 3), *p < 0.05 and **p < 0.01 compared with day 0. (D) MC3T3-E1 cells were cultured in growth medium and after reaching 100% confluence on day 7, osteogenic induction media was added and alizarin red S staining was performed on days 0 and 28. (E) Akap13 mRNA was analyzed at 0, 2, 4, and 7 days during proliferation and at 7, 14, 21, and 28 days during differentiation of MC3T3-E1 cells. (F) Analysis of osteogenesis in MC3T3-E1 cells. Cells were differentiated and Runx2, Alp, or Osteocalcin transcripts were quantified of day 0 and 28 by real-time RT-PCR. Bars represent the mean ± SE (n = 3), *p < 0.05. (G) Knockdown of Akap13 in immortalized BMSCs during proliferation. SiRNA for Akap13 or Control (CTRL) was added, as noted. A significant reduction in Runx2 and Alp transcripts was observed at 48 hours, *p < 0.05 and **p < 0.01 compared to control-treated cells.
Runx2 and Alp mRNA were reduced in Akap13-siRNA–treated cells
To further investigate the effects of Akap13 on bone-regulatory genes, we knocked down Akap13 with siRNA in immortalized BMSCs (Supplementary Fig. 1)(35) during proliferation. We confirmed Akap13 mRNA was efficiently reduced, and by using real-time PCR, there were significant reductions in mRNA encoding Runx2 and Alp mRNA at 48 hours in Akap13 siRNA treated cells, compared to control (Fig. 4G). These results confirmed that reduction of Akap13 leads to a reduction of Runx2 transcripts, and suggests that Akap13 acts upstream of Runx2 to influence bone formation.
Discussion
We report that mice haploinsufficient for Akap13 have an osteoporotic phenotype, as shown by a reduction in bone mass and increased trabecular spacing. Colony-forming efficiency assays demonstrated a reduced number of osteoprogenitors, accompanied by a reduction in the number of osteoblasts, and there was a corresponding reduction in production of extracellular matrix in the Akap13 haploinsufficient mice. We noted reduction in bone mass, but differences in cortical thickness, although reduced, were not significant. Taken together, the results suggest that normal levels of Akap13 are required at an early stage of osteogenic differentiation.
Osteoporosis is an important health care issue resulting in annual costs of $17.9 billion to the US and £1.7 billion to the UK healthcare systems, respectively.(46) These figures underscore the importance of understanding factors affecting bone development and remodeling. One of the least understood processes are the events that trigger commitment of uncommitted progenitors to an osteoblast fate. Although Rho, specifically RhoA, and mechanical stimulation, were reported to play an important role in osteogenic differentiation,(7,9,29) the responsible RhoA GEF(s) have not been previously identified. Notably, gene association studies in humans have linked development of osteoporosis to at least one GEF, the anti-Rho guanine nucleotide exchange factor-3 (ARHGEF3)(47); and one GTPase-activating protein that opposes the action of GEFs, anti- Rho GAP-1 (ARHGAP1).(48) Furthermore, in European men, SNPs in AKAP11 and RANKL emerged as associated with osteoporosis.(28) Interestingly, given the requirement for AKAP13 in induction of MEF2c levels in the heart,(20) MEF2C was one of 13 BMD loci of significance (p < 5 × 10−8) identified in subjects of Northern European descent.(48) Thus, a role for GEFs, and possibly for AKAP13, in osteoporosis finds support in genome-wide association studies (GWASs).
AKAP13 is unique, because it is the only GEF that couples a protein A-kinase regulatory protein-binding domain with carboxyl regions capable of integrating multiple cellular signals. In support of a possible role of the protein A kinase region of Akap13 in activation of bone marrow-derived stromal cells, a pyrazole-pyridine compound (CW008) that activates cAMP/PKA/CREB signaling has been shown to stimulate osteoblast differentiation and osteogenesis in ovariectomized mice.(49) Conversely, we observed that Akap13+/− mice had reductions in Runx2 transcripts, and that knockdown of Akap13 in uncommitted BMSCs and osteogenic MC3T3E1 cells led to significant reductions in Runx2 transcripts. Runx2 is a master regulator and a transcription factor in bone development.(50) Reduction of Runx2 in the Akap13 haploinsufficient mice supports a role for Akap13 in early osteoblast differentiation.
The Rho-type small GTP-binding proteins (Rho GTPases) are known to play an important role in regulating cytoskeletal formation, cell proliferation, and cell differentiation, and several lines of evidence indicate the initial events for commitment to osteogenesis require intact RhoA signaling.(7–9) Notably, the role of RhoA in osteogenesis and osteoblast differentiation is complex, and is not as simple as an “on” or “off” switch. Levels of Akap13 are high in preosteoblasts, but become reduced during differentiation. At later stages of osteoblast differentiation, Rho has been shown to inhibit bone accumulation, as shown by mice that conditionally expressed an osteoblast-specific RhoA dominant negative mutant, which had a high bone mass and Type 1 collagen transcripts.(51) Interestingly, our data (Supplementary Table 3) support this finding, because dynamic labeling experiments at 20 weeks indicated resident osteoblasts haploinsufficient for AKAP13 exceeded matrix production of osteoblasts from the WT mice, which showed reductions in bone formation due to aging. RhoA and Rho-associated protein kinase (ROCK) inhibit the phosphorylation of insulin receptor substrate 1 (IRS-1), which is the crucial step in IGF-1 signaling favoring osteoblast differentiation. Moreover, it was previously reported that inhibition of ROCK increases BMP2 and OSTEOCALCIN in human osteoblasts.(52) It should be emphasized that extracellular cues and substrate can greatly affect levels of active RhoA, which underscores the importance of observations in an in vivo murine model of bone formation and development.(53,54)
In conclusion, Akap13 haploinsufficient mice exhibited an osteoporotic phenotype characterized by a reduction in BMD, reduction in production of extracellular matrix, and reduced numbers of osteoprogenitors and mature osteoblasts. The root cause of the reduction in bone mass appears to be impairment in differentiation of osteoprogenitors resulting in deficient osteoblast numbers.
Supplementary Material
Acknowledgments
We acknowledge critical assistance by Kimberlyn Maravet Baig-Ward and Dr. Vardit Kram. We greatly appreciated assistance of Jim Herrick and Donna Jewison at the Mayo Bone Histomorphometry Laboratory, contributions by Dr. Xiu-Ye Xing and support by Dr. Alan H. DeCherney. Dr. Sergei Kuznetsov provided the immortalized bone marrow stem cells.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
Authors’ roles: Study design: HK, JHS, KH, MFY, PGR, PD, IT, TK. Study conduct: HK, KH, XCG, JB, JCL, CQ, TC, JHS. Data collection: HK, KH, JCL, CQ, TC, XCG. Data analysis: HK, JHS, KH, MFY, PGR, CQ, JB, IT. Data interpretation: HK, KH, PGR, MFY, JHS, TK. Drafting manuscript: HK, JS. JS, PGR. HK and JHS take responsibility for the integrity of the data analysis. All authors approved the final version of the manuscript.
References
- 1.Olsen BR, Reginato AM, Wang W. Bone development. Ann Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 2.Cohen MM., Jr The new bone biology: pathologic, molecular, and clinical correlates. Am J Med Gen A. 2006;140(23):2646–2706. doi: 10.1002/ajmg.a.31368. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi A, Komori T, Suda T. Regulation of osteoblast differentiation mediated by bone morphogenetic proteins, hedgehogs, and Cbfa1. Endocrine Rev. 2000;21(4):393–411. doi: 10.1210/edrv.21.4.0403. [DOI] [PubMed] [Google Scholar]
- 4.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99(5):1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 5.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13(7):791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 6.Rangaswami H, Schwappacher R, Marathe N, et al. Cyclic GMP and protein kinase G control a Src-containing mechanosome in osteoblasts. Sci Signal. 2010;3(153):ra91. doi: 10.1126/scisignal.2001423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnsdorf EJ, Tummala P, Kwon RY, et al. Mechanically induced osteogenic differentiation-the role of RhoA, ROCKII and cytoskeletal dynamics. J Cell Sci. 2009;122:546–553. doi: 10.1242/jcs.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welsh CF, Roovers K, Villanueva J, et al. Timing of cyclin D1 expression within G1 phase is controlled by Rho. Nat Cell Biol. 2001;3(11):950–957. doi: 10.1038/ncb1101-950. [DOI] [PubMed] [Google Scholar]
- 9.McBeath R, McBeath R, Pirone DM, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 10.Rubino D, Driggers P, Arbit D, et al. Characterization of Brx, a novel Dbl family member that modulates estrogen receptor action. Oncogene. 1998;16(19):2513–2526. doi: 10.1038/sj.onc.1201783. [DOI] [PubMed] [Google Scholar]
- 11.Toksoz D, Williams DA. Novel human oncogene lbc detected by transfection with distinct homology regions to signal transduction products. Oncogene. 1994;9(2):621–628. [PubMed] [Google Scholar]
- 12.Diviani D, Soderling J, Scott JD. AKAP-Lbc anchors protein kinase A and nucleates Galpha 12-selective Rho-mediated stress fiber formation. J Biol Chem. 2001;276(47):44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 13.Sterpetti P, Hack AA, Bashar MP, et al. Activation of the Lbc Rho exchange factor proto-oncogene by truncation of an extended C terminus that regulates transformation and targeting. Mol Cell Biol. 1999;19(2):1334–1345. doi: 10.1128/mcb.19.2.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossman KL, Der CJ, Sondek J. GEF means go: turning on Rho GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y. Dbl family guanine nucleotide exchange factors. Trends Biochem Sci. 2001;26(12):724–732. doi: 10.1016/s0968-0004(01)01973-9. [DOI] [PubMed] [Google Scholar]
- 16.Ishizaki T, Ishizaki T, Maekawa M, et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15(8):1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16(13):1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 18.Driggers PH, Segars JH, Rubino DM. The proto-oncoprotein Brx activates estrogen receptor beta by a p38 mitogen-activated protein kinase pathway. J Biol Chem. 2001;276(50):46792–46797. doi: 10.1074/jbc.M106927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kino T, Souvatzoglou E, Charmandari E, et al. Rho family Guanine nucleotide exchange factor Brx couples extracellular signals to the glucocorticoid signaling system. J Biol Chem. 2006;281(14):9118–9126. doi: 10.1074/jbc.M509339200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayers CM, Wadell J, McLean K, et al. The Rho guanine nucleotide exchange factor AKAP13 (BRX) is essential for cardiac development in mice. J Biol Chem. 2010;285(16):12344–12354. doi: 10.1074/jbc.M110.106856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park B, Nguyen NT, Dutt P, et al. Association of Lbc Rho guanine nucleotide exchange factor with alpha-catenin-related protein, alpha-catulin/CTNNAL1, supports serum response factor activation. J Biol Chem. 2002;277(47):45361–45370. doi: 10.1074/jbc.M202447200. [DOI] [PubMed] [Google Scholar]
- 22.Bear MD, Li M, Liu Y, et al. The Lbc Rho guanine nucleotide exchange factor α-catulin axis functions in serotonin-induced vascular smooth muscle cell mitogenesis and RhoA/ROCK activation. J Biol Chem. 2010;285:32919–32926. doi: 10.1074/jbc.M109.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kino T, Takatori H, Manoli I, et al. Brx mediates the response of lymphocytes to osmotic stress through the activation of NF AT5. Sci Signal. 2009;2(57):ra5. doi: 10.1126/scisignal.2000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diviani D, Baisamy L, Appert-Collin A. AKAP-Lbc: a molecular scaffold for the integration of cyclic AMP and Rho transduction pathways. Eur J Cell Biol. 2006;85(7):603–610. doi: 10.1016/j.ejcb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Slice LW, Rozengurt E. Activation of protein kinase D by signaling through Rho and the alpha subunit of the heterotrimeric G protein G13. J Biol Chem. 2001;276(42):38619–38627. doi: 10.1074/jbc.M105530200. [DOI] [PubMed] [Google Scholar]
- 26.Dutt P, Kjoller L, Giel M, et al. Activated Galphaq family members induce Rho GTPase activation and Rho-dependent actin filament assembly. FEBS Lett. 2002;531(3):565–569. doi: 10.1016/s0014-5793(02)03625-6. [DOI] [PubMed] [Google Scholar]
- 27.Kino T, Segars JH, Chrousos GP. The guanine nucleotide exchange factor Brx: a link between osmotic stress, inflammation and organ physiology and pathophysiology. Expert Rev Endocrinol Metab. 2010;5(4):603–614. doi: 10.1586/eem.10.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roshandel D, Thomson W, Pye SR, et al. A validation of the first genome-wide association study of calcaneus ultrasound parameters in the European Male Ageing Study. BMC Med Genet. 2011 Jan 28;12:19. doi: 10.1186/1471-2350-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khatiwala CB, Kim PD, Peyton SR, et al. ECM compliance regulates osteogenesis by influencing MAPK signaling downstream of RhoA and ROCK. J Bone Miner Res. 2009;24:886–898. doi: 10.1359/JBMR.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krum SA. Direct transcriptional targets of sex steroid hormones in bone. J Cell Biochem. 2011;112:401–408. doi: 10.1002/jcb.22970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouxsein ML, Boyd SK, Christiansen BA, et al. Guidelines for assessment of bone microstructure in rodents using micro–computed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 32.Gopalakrishnan R, Suttamanatwong S, Carlson AE, et al. Role of matrix Gla protein in parathyroid hormone inhibition of osteoblast mineralization. Cells Tissues Organs. 2005;181:166–175. doi: 10.1159/000091378. [DOI] [PubMed] [Google Scholar]
- 33.Ren J, Jin P, Sabatino M, et al. Global transcriptome analysis of human bone marrow stromal cells (BMSC) reveals proliferative, mobile and interactive cells that produce abundant extracellular matrix proteins, some of which may affect BMSC potency. Cytotherapy. 2011;13:661–664. doi: 10.3109/14653249.2010.548379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazarus JE, Hegde A, Andrade AC, et al. Fibroblast growth factor expression in the postnatal growth plate. Bone. 2007;40(3):577–586. doi: 10.1016/j.bone.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Krebsbach PH, Kuznetsov SA, Satomura K, et al. Bone formation in vivo: comparison of osteogenesis by transplanted mouse and human marrow stromal fibroblasts. Transplantation. 1997;63(8):1059–1069. doi: 10.1097/00007890-199704270-00003. [DOI] [PubMed] [Google Scholar]
- 36.Parfitt AM, Drezner MK, Glorieux FH, et al. Bone histomorphometry: standardization of nomenclature, symbols, and units: report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987 Dec;2(6):595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 37.Chen XD, Shi S, Xu T, et al. Age-related osteoporosis in biglycan-deficient mice is related to defects in bone marrow stromal cells. J Bone Miner Res. 2002;17(2):331–340. doi: 10.1359/jbmr.2002.17.2.331. [DOI] [PubMed] [Google Scholar]
- 38.Krebsbach PH, Kuznetsov SA, Bianco P, et al. Bone marrow stromal cells: characterization and clinical application. Crit Rev Oral Biol Med. 1999;10(2):165–181. doi: 10.1177/10454411990100020401. [DOI] [PubMed] [Google Scholar]
- 39.Cox KH, DeLeon DV, Angerer LM, et al. Detection of mRNAs in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101(2):485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 40.Bandyopadhyay A, Kubilus JK, Crochiere ML, et al. Identification of unique molecular subdomains in the perichondrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev Biol. 2008 Sep 1;321(1):162–174. doi: 10.1016/j.ydbio.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaback LA, Soung do Y, Naik A, et al. Osterix/Sp7 regulates mesenchymal stem cell mediated endochondral ossification. J Cell Physiol. 2008;214(1):173–182. doi: 10.1002/jcp.21176. [DOI] [PubMed] [Google Scholar]
- 42.Banerjee C, McCabe LR, Choi JY, et al. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J Cell Biochem. 1997;66(1):1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 43.Ducy P, Zhang R, Geoffroy V, et al. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 44.Harada H, Tagashira S, Fujiwara M, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J Biol Chem. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 45.Satomura K, Krebsbach P, Bianco P, et al. Osteogenic imprinting upstream of marrow stromal cell differentiation. J Cell Biochem. 2000;78(3):391–403. [PubMed] [Google Scholar]
- 46.Holroyd C, Cooper C, Dennison E. Epidemiology of osteoporosis. Best Pract Res Clin Endocrinol Metab. 2008;22(5):671–685. doi: 10.1016/j.beem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Xu XH, Dong SS, Guo Y, et al. Molecular genetic studies of gene identification for osteoporosis: the 2009 update. Endocr Rev. 2010;31:447–505. doi: 10.1210/er.2009-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivadeneira F, Styrkársdottir U, Estrada K, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim JM, Choi JS, Kim YH, et al. An activator of the cAMP/PKA/CREB pathway promotes osteogenesis from human mesenchymal stem cells. J Cell Physiol. 2013;228(3):617–626. doi: 10.1002/jcp.24171. [DOI] [PubMed] [Google Scholar]
- 50.Lee KS, Kim HJ, Li QL, et al. Runx2 is a common target of transforming growth factor beta 1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Negishi-Koga T, Shinohara M, Komatsu N, et al. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat Med. 2011;17(11):1473–1480. doi: 10.1038/nm.2489. [DOI] [PubMed] [Google Scholar]
- 52.Ohnaka K, Shimoda S, Nawata H, et al. Pitavastatin enhanced BMP-2 and osteocalcin expression by inhibition of Rho-associated kinase in human osteoblasts. Biochem Biophys Res Comm. 2001 Sep 21;287(2):337–342. doi: 10.1006/bbrc.2001.5597. [DOI] [PubMed] [Google Scholar]
- 53.Ducy P. Cbfa1: a molecular switch in osteoblast biology. Dev Dyn. 2000;219(4):461–471. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1074>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 54.Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008 Dec 1;121(Pt 23):3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.