Abstract
Abstract Steroid receptors of the nuclear receptor superfamily are proposed to be either: 1) located in the cytosol and moved to the cell nucleus upon activation, 2) tethered to the inside of the plasma membrane, or 3) retained in the nucleus until free steroid hormone enters and activates specific receptors. Using computational methods to analyze peptide receptor topology, we find that the “classical” nuclear receptors for progesterone (PRB/PGR), androgen (ARB/AR) and estrogen (ER1/ESR1) contain two transmembrane helices (TMH) within their ligand-binding domains (LBD).The MEMSAT-SVM algorithm indicates that ARB and ER2 (but not PRB or ER1) contain a pore-lining (channel-forming) region which may merge with other pore-lining regions to form a membrane channel. ER2 lacks a TMH, but contains a single pore-lining region. The MemBrain algorithm predicts that PRB, ARB and ER1 each contain one TMH plus a half TMH separated by 51 amino acids.ER2 contains two half helices. The TM-2 helices of ARB, ER1 and ER2 each contain 9-13 amino acid motifs reported to translocate the receptor to the plasma membrane, as well as cysteine palmitoylation sites. PoreWalker analysis of X-ray crystallographic data identifies a pore or channel within the LBDs of ARB and ER1 and predicts that 70 and 72 residues are pore-lining residues, respectively. The data suggest that (except for ER2), cytosolic receptors become anchored to the plasma membrane following synthesis. Half-helices and pore-lining regions in turn form functional ion channels and/or facilitate passive steroid uptake into the cell. In perspective, steroid-dependent insertion of “classical” receptors containing pore-lining regions into the plasma membrane may regulate permeability to ions such as Ca2+, Na+ or K+, as well as facilitate steroid translocation into the nucleus.
Key words: steroids, membrane receptors, cytosol receptors, transmembrane helix, channels
Introduction
Steroid receptors have been identified in the plasma membrane, in the cytosol and in the nucleus of target cells (reviewed in [Olefsky, 2001; Aranda and Pascual, 2001; He et al, 2010, Hammes and Levin, 2011; Levin, 2011; Stanisic et al., 2010]). It has been generally accepted that steroid hormones can circulate bound tightly to sex hormone binding globulin, bound loosely to albumin, unbound or free. The free and albumin-bound forms are thought to be available to diffuse through the membrane lipids into the cell cytoplasm, interact with their “classical” receptors, and modulate eukaryotic gene expression. Contrary to this “free hormone hypothesis”, it has been proposed that endocytosis may act as a pathway for cellular uptake of androgens and estrogens [Hammes et al., 2005, Willnow and Nykjaer, 2010]. Other evidence suggests that crosstalk occurs between extranuclear and intranuclear steroid signaling, and that this crosstalk regulates critical cellular processes [Levin, 2011]. For example, classical estrogen, progesterone, and androgen receptors (ER1 (approved symbol ESR1), PRB (approved symbol PGR) and ARB (approved symbol AR)) are post-translationally modified by lipids [Mundy, 1995]. Levin [2011] suggests that these modifications may induce tethering of the receptor-steroid complex to the plasma membrane and that membrane-localized estrogen receptor α is required for normal organ development and function. Our earlier computational analysis of the progesterone “cytosol” receptor topology indicates that PRB may insert into the plasma membrane via a transmembrane (TM) helix that is also a pore-lining region [Morrill et al., 2013].
Cell membranes consist mainly of cholesterol and aliphatic chains of phospholipids, which create a region that favors non-polar amino acids and rejects polar amino acids. Insertion of TM helices into the plasma membrane initiates complex molecular interactions among proteins, lipids and water, creating channels and altering membrane topology and function. Protein and lipid molecules are not static, but undergo continuous turnover, as well as dynamic changes [Brunori et al. 2012, Dawidowitz, 1987]. The range of motions is highly variable among different proteins, ranging from hydrogen bond formation to very large scale folding/unfolding events that may include changes in transmembrane helices [Shen et al., 1997]. Using a database of 160 3D structures, Hilldebrand et al. [2006] have defined the protein transmembrane (TM) helix as a membrane-spanning 17.3±3.1 (SD, N = 160) amino acid sequence with a hydrogen-bonded helical configuration, including α-, 310- and π-helices. The α-helix is very common, while the 310 helix is found at the ends of the α-helix. π-helices are rare. As noted by Bernsel et al. [2008], with the availability of an increasing number of 3D structures of TM proteins, it becomes clear that the helices show a significant variation in their length, slope and straightness. The lengths of the helices vary from less than 16 residues up to 40 residues. About 5% of the TM helices in the known structures are very short (<15 residues) and may only partially span the membrane. These helices are known as “half TM helices”. TM helices shorter than 10 residues are exclusively found in membrane channels [Hildebrand et al., 2006]. As reported here, several protein structure algorithms predict that the ligand binding region of the androgen, estrogen and progesterone cytosolic receptors each contain one or more transmembrane helices and/or pore-lining regions.
Materials and methods
Protein sequence and structure sources:
The amino acid sequences for the steroid binding proteins were downloaded from the ExPASy Proteomic Server of the Swiss Institute of Bioinformatics (http://www.expasy.org; http://www.uniprot.org). All receptors represent the so-called canonical sequence. Crystallographic structures of the ligand binding domains were obtained from both the RCSB PDB (www.rcsb.org/pdb/explore.do?structureld=) and the European Bioinformatics Institute (www.ebi.ac.uk/pdbsum/).
Secondary structure predictions:
Transmembrane (TM) helices were predicted using: 1) MEMSAT-SVM [Nugent et al., 2011] (http://www.bioinf.cs.ucl.ac.uk/psipred/), 2) TOPCONS consensus algorithm [Bernsel et al., 2009] (http://topcons.cbr.su.se) and 3) MemBrain algorithm [Shen and Chou, 2008] (http://www.csbio.sjtu.edu.cn/bioinf/MemBrain) and 4) Phobius, an algorithm for transmembrane topology and signal peptides [Kall et al., 2004] www.ebi.ac.uk/Tools/pfa/phobius/ . Pore-lining regions in transmembrane protein sequences were predicted using the method of Nugent and Jones [2014].
PoreWalker 1.0:
A method for the detection and characterization of TM protein channels from their 3D Structure: PoreWalker [Pellegrini-Calace et al., 2009] involves a stepwise procedure in which the pore center and pore axis are identified and optimized using geometric criteria, and the biggest and longest cavity/channel through the protein is then predicted. Pore features, including diameter profiles, pore-lining residues, size, shape and regularity of the pore are calculated, providing a characterization of the longest channel in the structure. A server is available at http://www.ebi.ac.uk/thornton-srv/software/PoreWalker/ .
CRAC and CARC domains:
CRAC is a short linear amino acid motif that mediates binding to cholesterol [Li and Papadopoulos, 1998] and stands for Cholesterol Recognition/Interaction Amino acid Consensus sequence [Fantini and Barrentes, 2013]. In a N-terminus to C-terminus direction, the motif consists of a branched apolar Leu (L) or Val (V) residue, followed by a segment containing 1-5 of any residues, followed by a mandatory aromatic Tyr (Y) residue, a segment containing 1-5 of any residues, and finally, a basic Lys or Arg. In the one letter amino acid codes the algorithm is (L/V) – X1-5 – (Y) – X1-5- (K/R). A new cholesterol binding CARC domain similar to the CRAC domain has been identified (reviewed in [Fantini and Barrantes, 2013]). The CARC domain is comparable to the CRAC domain, but exhibits the opposite orientation along the polypeptide chain (“inverted CRAC”), i.e. (K/R) –X1-5 – (Y/F) – X1-5 – (L/V). CARC is distinct from CRAC in that the central amino acid can be either Y or F.
MPRAP:
An accessibility predictor for α-helical transmembrane proteins was used to predict surface accessibility both inside and outside the cell membrane [Illergard et al., 2010]. A web-server MPRAP is available at http://mprap.cbr.su.se/.
Protein domain prediction:
The computer-assisted protein domain boundary prediction server DomPred [Bryson et al., 2007] uses the results from two different categories of method (DPS and DomSSEA), and each is individually benchmarked against one of the latest domain prediction benchmarks. The DomPred server is available at: http://bioinf.cs.ucl.ac.uk/software.html.
TMKink: A method to predict transmembrane helix kinks.
Meruelo et al. [2011] have identified distinct residue preferences in kinked versus non-kinked helices and have exploited these differences and residue conservation to predict kinked helices using a neural network algorithm. The kink predictor, TMKink, is available at http://tmkinkpredictor.mbi.ucla.edu/.
LIGPLOT v.4.5.3:
A program for automatically plotting protein-ligand interactions [Wallace et al., 1995]. www.ebi.ac.uk/thornton-srv/software/LIGPLOT/ .
Results and Discussion
Characterization of putative transmembrane helical regions within PRB, ARB, ER1 (ERα) and ER2 (ERβ)
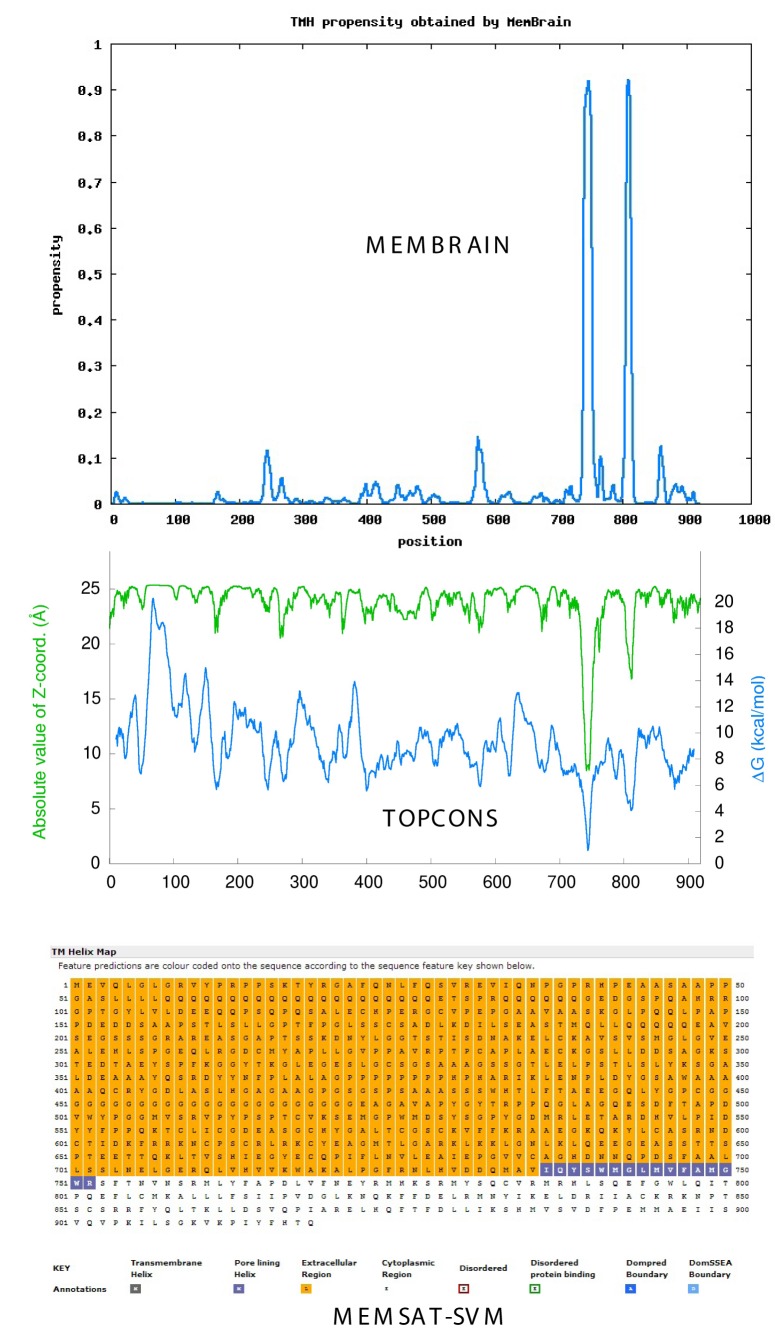
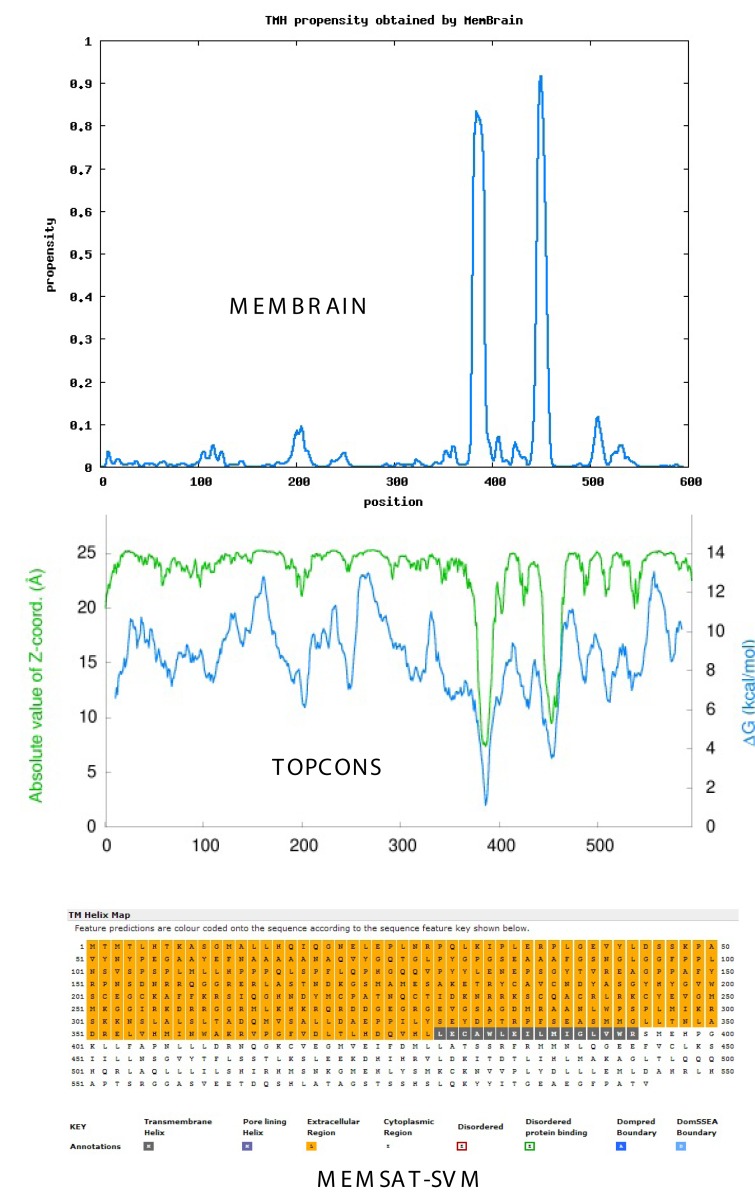
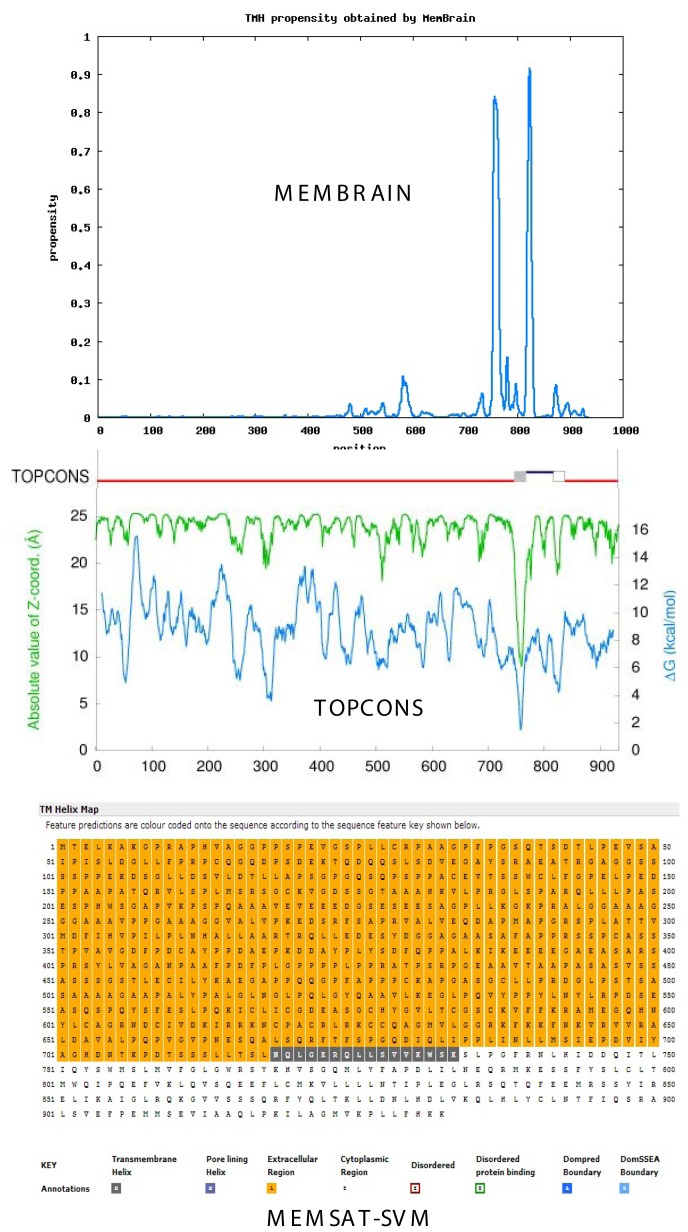
Figure 1 compares the propensity of TM helix formation in ARB using three current algorithms: the MemBrain method (top), TOPCONS consensus method (middle) and MEMSAT-SVM method (bottom). All methods predict one or more TM helices or pore-lining regions in the C-terminal region. Similar analyses of ER1and PRB are shown in Figure 2 and Figure 3, respectively. As shown for ARB, ER1 and PRB, both the TOPCONS [Bernsel et al., 2009] and MemBrain [Shen and Chou, 2008] methods predict a TM helix pair in the C-terminal region of each “classical” receptor. Analysis of ER2 demonstrates a single TM helix in the C-terminal region (not shown).
Figure 1. Androgen receptor.
Topology of TM helices and pore-lining regions of Homo sapiens classical androgen receptor (ARB, P102750), comparing the MemBrain algorithm (upper plot), the TOPCONS consensus method (middle plot) and MEMSAT-SVM method (lower plot). Blue squares in the MEMSAT-SVM projection indicate predicted pore-lining regions. See Methods.
Figure 2. Estrogen receptor.
Topology of TM helices and pore-lining regions of Homo sapiens classical estradiol receptor (ER1, P03372), comparing the MemBrain algorithm (upper plot), TOPCONS consensus method (middle plot) and MEMSAT-SVM method (lower plot). Black squares in MEMSAT-SVM method indicate predicted non-pore helix regions. See Methods.
Figure 3. Progesterone receptor.
Topology of TM helices and pore-lining regions of Homo sapiens classical progesterone receptor (PRB, P06401), comparing the MemBrain algorithm (upper plot), TOPCONS consensus method (middle plot) and MEMSAT-SVM method (lower plot). Black squares in MEMSAT-SVM method indicate predicted non-pore helix regions. See Methods.
Nugent and Jones [2012] have developed a computational method capable of identifying pore-lining regions in membrane proteins from sequence information alone, which can then be used to determine pore stoichiometry. As shown in the lower plot of Figure 1, the blue-colored amino acid squares in the MEMSAT-SVM projection (bottom) reflect a pore lining region in ARB and correspond to TM-1 predicted by both the MemBrain and TOPCONS algorithms, whereas ER1 (Figure 2) and PRB (Figure 3) exhibit classical TM helices (the black-colored amino acid squares) corresponding to TM-2. The pore-lining regions within groups of monomers or multimeric proteins may combine to form cavities that run parallel to the TM helices, forming a path along which ions or molecules can travel, with structural features of the pores determining specificity (reviewed in [Nugent and Jones, 2012]). MEMSAT-SVM analysis indicates that ER2, like ARB, contains a pore-lining region. Despite their apparent structural differences, both ER1 and ER2 are reported to bind 17-β-estradiol with high affinity and they bind to classical estrogen response elements in a similar but not identical fashion [Dechering et al., 2000].
Functional topology of the “classical” steroid receptors: localization of the transmembrane helices to the ligand binding domains
Table 1 summarizes the location of the “modulating” regions, DNA binding regions and ligand binding domains (LBD) within PRB, ARB, ER1 and ER2 (Uniprot database). The N-terminal (modulating) domain is hypervariable in both size and amino acid sequence. The ligand-independent activation function (AF-1) within this region is involved in gene transcription, but does not depend on ligand binding. The DNA binding region is centrally located and consists of two non-repetitive globular motifs [Schwabe et al. 1993]. The moderately conserved ligand-binding domain (LBD) may include a nuclear localization signal and amino acid sequences capable of binding both chaperones and dimerization interfaces [Wurtz et al., 1996]. The amino acid sequences predicted by the solvent accessibility predictor for α-helical transmembrane proteins: MPRAP [Illergard et al., 2010] and the TOPCONS [Bernsel et al., 2009], MemBrain [Shen and Chou, 2008] and Phobius [Kall et al., 2004] methods for predicting TM helices are indicated in rows 5–8. The bottom row indicates the number of residues within the predicted amino acid sequence between TM-1 and TM-2. As indicated, all putative TM helices identified in ARB, ER1 and PRB are within the ligand-binding domain and the number of amino acids within the peptide loop between TM-1 and TM-2 is either 50 or 51 amino acids. TOPCONS also predicts that both the N- and C-terminal ends are intracellular. Therefore, the 50-51 residue loops would be extracellular, which suggests that these residues may be associated with steroid uptake from the extracellular environment.
Table 1. Summary of the localization of ligand binding domains, DNA binding regions, modulating regions and predicted TM helices within classical nuclear steroid receptors (Homo sapiens).
| Sequence Features | PRB P06401 |
ARB P10275 |
ER1(α) P03372 |
ER2(β) Q92731 |
|---|---|---|---|---|
| Chain | 1-933 | 1-919 | 1-595 | 1-530 |
| Modulating Region | 1-566 | 1-558 | 1-184 | 1-148 |
| DNA Binding Region | 567-639 | 559-631 | 185-250 | 149-214 |
| Ligand Binding Domain | 681-933 | 690-919 | 311-551 | 215-530 |
| Residue Solvent Accessibility MPRAP |
748-768 818-838 |
735-755 804-824 |
376-396 445-465 |
None |
| TM Helices MemBrain |
750-767 817-831 |
738-752 803-813 |
380-393 444-459 |
332-345H1 397-407H1 |
| TM Helices TOPCONS |
748-768 818-838 |
734-754 802-822 |
376-396 445-465 |
328-348 |
| TM Helices Phobius |
750-768 | 736-754 803-810H1 |
376-384H1 446-453H1 |
397-406H1 |
| TM-1/TM-2 Loop Residues |
767-817 (50) |
752-803 (51) |
393-444 (51) |
….. |
1Half helix (see Methods).
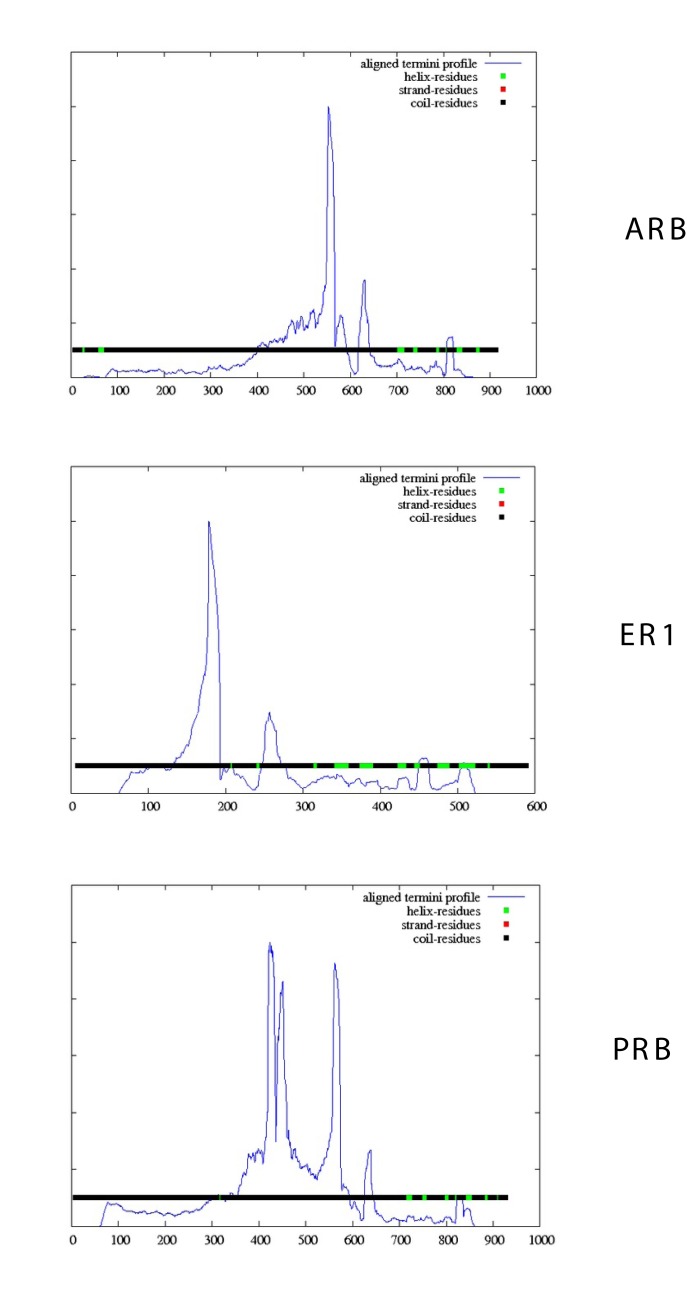
A protein domain is a conserved part of the tertiary structure that can evolve, function, and exist independently of the rest of the protein sequence (reviewed in [Alden et al., 2010]). Figure 4 compares the aligned termini profiles of the complete ARB, ER1 and PRB sequences using the DomPred server [Bryson et al., 2007]. This server provides results from two completely different categories of methods (DS and DomSSEA). A comparison of sequence features (Table 1) and domain distributions (Figure 4) indicates that each receptor exhibits a different domain profile with major peaks in the DNA binding regions and minimal peaks in the ligand binding domains. The regions containing the putative TM helices (Table 1) lack identifiable protein domains.
Figure 4. DomPred analysis of the Homo sapiens ARB (P10275), ER1 (P03372) and PRB (P06401) ligand binding domains.
Aligned termini profile (blue line) indicates the density of the end points of PSI-BLAST alignments which were generated between the query sequence and all the PSI-BLAST hits which were found, given a PSI-BLAST run against a database with all the fragments removed. See Methods.
Sequence analysis of the transmembrane helices
Table 2 compares the amino acid sequences within the individual TM helices of PRB, ARB, ER1 and ER2, as predicted by the MemBrain method. The TM-1 sequences within PRB (column 1) and ARB (column 3) each contain an amino acid sequence in common (indicated in blue): QYSWM_LMVFA{G}M{L}GW. Pedram et al. [2007] have identified a highly conserved 9-13 amino acid motif in the ligand binding domains of human/mouse ER1, ER2, PRA, PRB and ARB that is believed to be involved in steroid receptor translocation to the plasma membrane. As reported [Pedram et al., 2007], ER1 contains the motif 445FYCLKSIIINS453, ER2 the motif 397YLCVKAMIILNS408, ARB the motif 805FLCMKAIIIFS813 and PRB the motif 818FLCMKVIIIN826. As indicated in Table 2, the ARB, PRB, ER1 and ER2 translocation motifs (highlighted in red) are contained within TM-2 and account for 67-100% of the TMH residues. This indicates that the TM-2 may be essential for translocation to the plasma membrane.
Table 2. Comparison of the amino acid sequences in the transmembrane helices using the MemBrain algorithm [Shen and Chou, 2008].
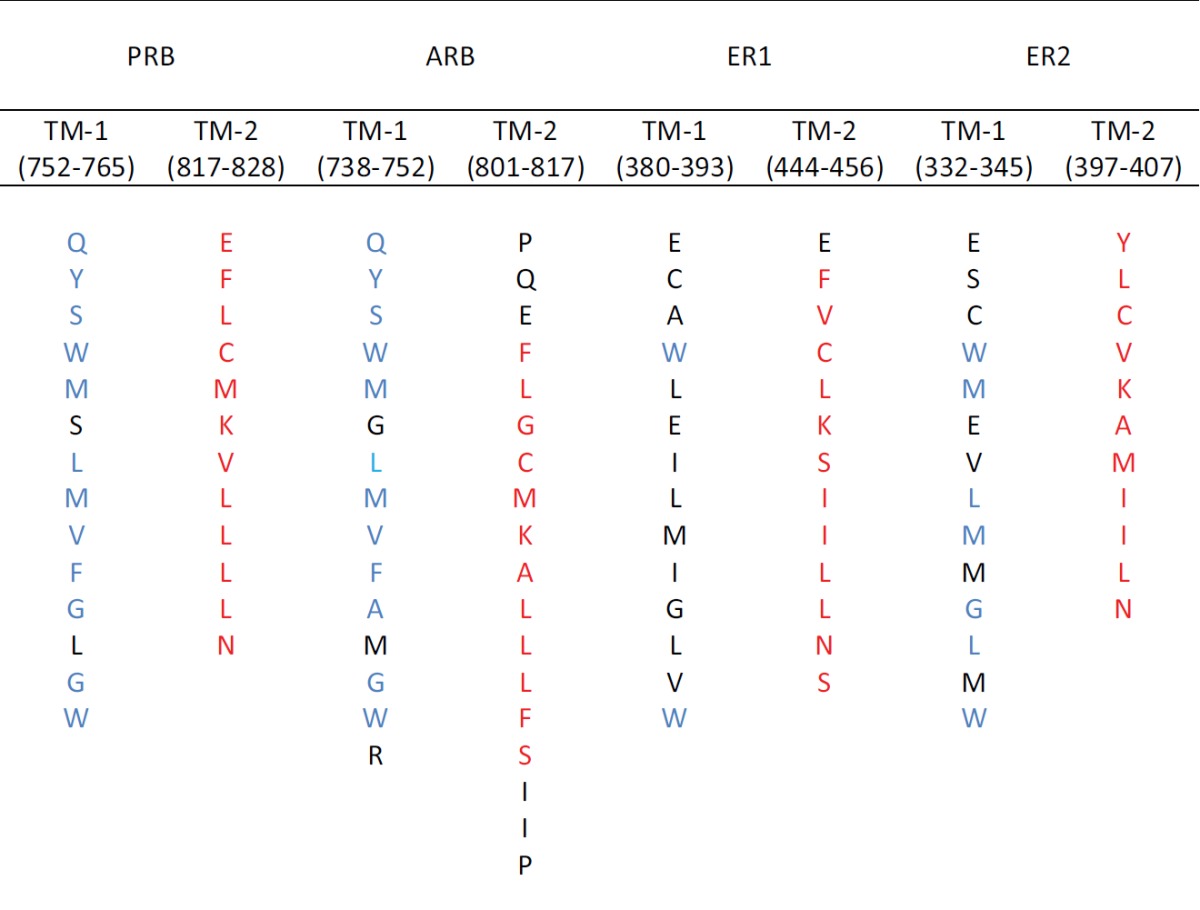 |
Red highlights indicate receptor localization motifs [Pedram et al., 2007]; blue highlights indicate sequences in common for PRB and ARB
Pedram et al. [2007] also found that C447 in cytoplasmic ER1 (but not nuclear ER1) is palmitoylated and they have identified DHHC-7 (Q9NXF8) and DHHC-21 (Q8IVQ6) as conserved palmitoylacyltransferase proteins involved in steroid palmitoylation [Pedram et al., 2012]. Cytoplasmic ER1 colocalizes with palmitoylating enzymes (DHHC-7 and DHHC-21) in the Golgi apparatus, where most probably palmitoylation occurs. It is common that palmitoylated proteins translocate into cholesterol-rich domains [Salaun et al., 2010]; there is, however, no strict consensus sequence for palmitoylation. As noted by Salaun et al. [2010], palmitoylated cysteines share certain common characteristics: 1) the surrounding amino acids tend to be basic or hydrophobic, and 2) they are frequently located in cytoplasmic regions flanking or within transmembrane helices. As shown in Table 2, the cysteines within the translocation motifs described by Pedram et al. [2007] for ARB, PRB, ER1 and ER2 exhibit these characteristics.
As shown in Table 3, the palmitoylating enzymes DHHC-7 and -21 contain 4 transmembrane helices, as well as one pore-lining region (TM–2). Both contain a zinc finger overlapping TM-3. The TM helices varied from 14 to 29 residues in length; the RCSB PDB (see Methods) uses an algorithm that defines all TM helices as containing 20 residues. It should be noted that about half of all transmembrane helices contain bends and other deviations often referred to as “kinks” (reviewed in [Meruelo et al., 2011]). Distortions in helix geometry such as kinks may facilitate conformational changes required for protein function by providing sites of flexibility and can be important for positioning key residues precisely in the protein structure [Meruelo et al., 2011]. Based on DHHC-7 and -21 knockdown studies, these proteins are required for endogenous ER1, PRB and ARB palmitoylation, membrane trafficking, and signal transduction in cancer cells [Aicart-Ramos et al., 2011]. Since Table 3 indicates that both DHHC-7 and -21 are membrane enzymes, cytosolic steroid receptors may, at some time, be localized to the plasma membrane. As Aicart-Ramos et al. have noted [2011], this post-translational modification provides an important mechanism for regulating protein subcellular localization, stability, trafficking, aggregation, translocation to lipid rafts, and interaction with effectors.
Table 3. MEMSAT-SVM topology analysis of palmitoyltransferases involved in palmitoylation of ARB, ER1(α) and PRB.
| Protein Topology | DHHC7 (Q9NXF8) |
DHHC21 (Q8IVQ6) |
||
|---|---|---|---|---|
| TM Helix | Kinks | TM Helix | Kinks | |
| Sequence | 1 - 308 | 1 - 308 | ||
| TM-1 | 41 - 70 (29) | 50 - 71 | 9 - 37 (28) | 19 - 35 |
| TM-2 | 78 - 102 (24) | 84 - 92 | 48 - 67 (19) | (pore-lining) |
| TM-3 | 173 - 301 (28) | 134 - 161 (27) | 149 - 163 | |
| TM-4 | 217 - 246 (29) | 224 - 232 | 179 - 203 (24) | 185 - 198 |
| Zinc Finger | 130 - 180 | 90 - 140 | ||
TM helix length is indicated in parentheses.
Cholesterol recognition and interaction amino acid consensus (CRAC) motifs and CARC (inverse CRAC) motifs in “classical” nuclear steroid receptors
In addition to the TM helices and translocation motifs described above, functional regions such as CRAC motifs can also be identified within the classical nuclear steroid receptors. CRAC is a short linear amino acid motif that mediates binding to cholesterol and stands for Cholesterol Recognition/Interaction Amino acid Consensus sequence [Li and Papadopoulos, 1998; Fantini and Barrantes, 2013]. The algorithm is (L/V) – X1-5 – (Y) – X1-5- (K/R). A new cholesterol binding domain similar to the CRAC motif has been characterized and is termed a “CARC” motif (reviewed in [Fantini and Barrantes, 2013]). The so-called “CARC motif” is comparable to the CRAC domain, but exhibits the opposite orientation along the polypeptide chain (“inverted CRAC”), i.e. (K/R) – X1-5 – (Y/F) – X1-5 – (L/V), and is distinct from CRAC in that the central amino acid can be either Y or F. Molecular modeling studies have shown that CRAC/CARC motifs have a good fit for cholesterol. Wang et al. [2013] have proposed that specific interactions between steroids and the CRAC motifs of unconstrained G protein-coupled receptors account for non-genomic effects of steroids “to some extent”. Baier et al. [2011] have evidence that the CRAC motif is responsible for some of the structure/function properties of the nicotinic acetylcholine receptor. However, to date there is no direct evidence for steroid-CRAC motif interaction. Although cholesterol is concentrated in sphingolipid-enriched membrane microdomains such as “lipid rafts” [Brown and London, 2000], it is also present in the lipid disordered phase of the plasma membrane that contains high amounts of glycerolphospholipids such as phosphatidylcholine [Brown and London, 2000].
As shown in Figure 5, CRAC and CARC domains are associated with the ligand-binding domains in androgen (ARB), estrogen (ER1) and progesterone (PRB) receptors. TM helices are underlined, CRAC/CARC motifs are highlighted in red; residues V746, M749 and F764 in ARB have been identified as crucial in androgen binding [Wang et al., 2013] and are highlighted in blue. In ARB, CRAC/CARC domains predominate in the sequence and are widely distributed. CRAC or CARC motifs do not overlap the TM helices, but frequently occur within the loop region between the helices. This indicates that cholesterol binding is not associated with the membrane lipid bilayers, but is distributed throughout the ligand binding domain. Both DHHC-7 and -21 palmitoyltransferases in Table 3 above contain 4 TM helices, multiple caveolin binding motifs, as well as cholesterol binding (CRAC, CARC) domains, indicating a preference for lipid rafts. The presence of multiple transmembrane helices suggests that newly-synthesized “cytosolic” receptors may be initially localized to the Golgi, and that palmitoylation dynamics may play a role in receptor translocation to the plasma membrane.
Figure 5. The distribution of cholesterol binding CRAC/CARC domains within the ligand binding domains of the human ARB (top), ER1 (middle) and PRB (bottom).
Sequences are expressed using single letter codes and TM helices are underlined. The CRAC/CARC domains are highlighted in red. Amino acids highlighted in blue of ARB are those shown by PoreWalker analysis to be within the predicted channel and contain amino acids reported to bind DHT.
PoreWalker analysis of channel(s) in the androgen receptor
Typically, channel proteins contain a cavity (or pore) which spans the entire membrane protein with an opening on each side of the membrane. Pellegrini-Calace et al. [2009] have developed an improved computational approach (PoreWalker 1.0, see Methods) for the identification and characterization of channels in transmembrane proteins based on their three-dimensional structure. Given a set of 3D crystallography coordinates, this method can detect and identify the pore centers and axis using geometric criteria, and then the biggest and longest cavity/channel through the protein is identified. Pore features, including diameter profiles, pore-lining residues, size, shape and regularity of the pore are used to provide a quantitative and visual characterization of the channel.
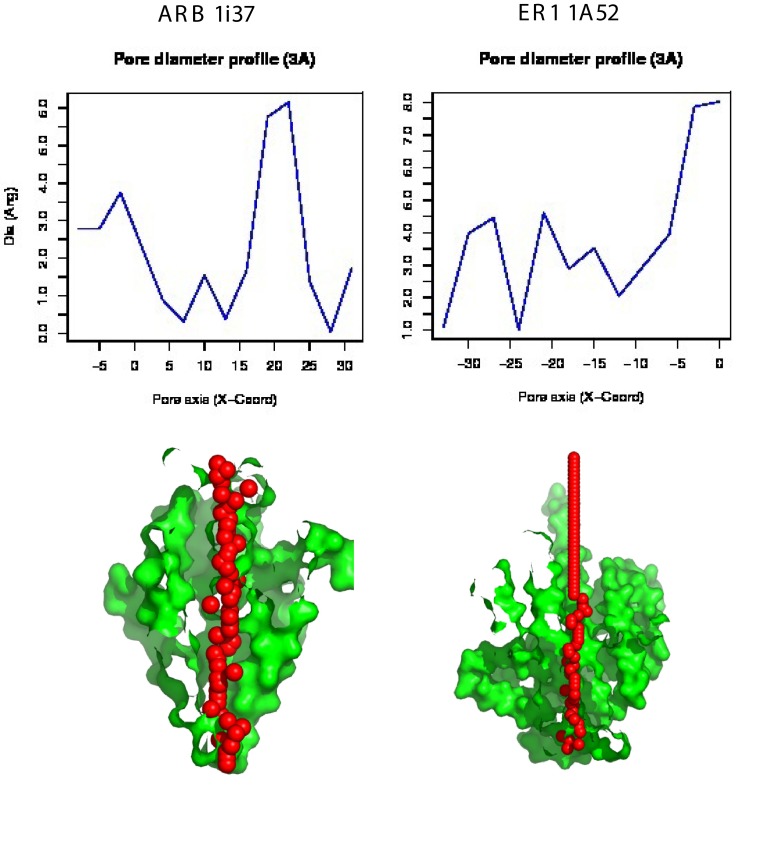
Figure 6 illustrates and compares: 1) the PoreWalker output for the ARB ligand binding domain complexed to dihydrotestosterone (2Q7I) and 2) the ER1 ligand binding domain complexed to estradiol (1A52), and identifies and characterizes the biggest and longest channel in each structure. The upper graph illustrates the pore diameter profile, whereas the lower image illustrates the features of the cavity. The protein structure is colored in green and red spheres represent pore centers at given pore heights and their diameters are proportional to the pore diameter calculated at that point. The sections were obtained by cutting the protein structure along the XZ plane. The images illustrate the respective pores in the XZ-plane section, y < 0 coordinates only. As can be seen, a channel extends across both the ligand binding domain of the human androgen receptor ARB and estrogen receptor ER1.
Figure 6. PoreWalker analysis of the androgen receptor ligand binding domain complex with dihydrotestosterone (left column, PDB ID: 1i37) and the estrogen receptor ligand binding domain (right column, PDB ID: 1A52).
The upper plot illustrates the pore diameter profile, whereas the lower image illustrates the features of the cavity. The protein structure is colored in green and red spheres represent pore centers at given pore heights and their diameters are proportional to the pore diameter calculated at that point. The lower visualization of a pore section shows the position of the biggest spheres (pore centers) that can be built along the channel at 1À steps; the section was obtained by cutting the protein structure along the xy-plane. See Methods.
Table 4 compares the ARB pore lining residues identified by PoreWalker with those associated with TM-1 and TM-2 identified by MemBrain (see Table 2). The ligand binding domain of ARB involves 229 residues of the C-terminal region. Of the 65 pore-lining residues, 10 were within TM-1 and 8 within TM-2 and they constitute clusters containing about 70% of the residues within each TM helix. Similar clusters of pore-lining residues were identified within TM-1 and TM-2 of both ER1 and PRB. Therefore, the helices identified by the MemBrain algorithm are largely populated by pore lining residues in all three nuclear steroid receptors. A third cluster of 21 contact points occurs between 855R and 899I in ARB and overlaps two cholesterol binding sites (CRAC/CARC motifs) at the C-terminal end of the ligand binding domain (data not shown). Many of the residues identified as pore-lining residues by PoreWalker were confirmed as accessible to water (hydrophilic sites) using a water accessibility predictor [Illergard et al., 2010] for α-helical transmembrane proteins (MPRAP).
Table 4. Comparison of transmembrane helix regions as predicted by the MemBrain method with the pore lining residues as detected by the PoreWalker algorithm.
| ARB 1i37 | ER1 1As2 | PRB 4A2J | |||
|---|---|---|---|---|---|
| MemBrain | PoreWalker | MemBrain | PoreWalker | MemBrain | PoreWalker |
| 738 | 380 | 380 | 752 | ||
| 739 | 381 | 381 | 753 | ||
| 740 | 382 | 382 | 754 | 754 | |
| 741 | 741 | 383 | 383 | 755 | 755 |
| 742 | 742 | 384 | 384 | 756 | 756 |
| 743 | 743 | 385 | 385 | 757 | 757 |
| 744 | 744 | 386 | 758 | 758 | |
| 745 | 745 | 387 | 759 | 759 | |
| 746 | 746 | 388 | 760 | 760 | |
| 747 | 747 | 389 | 389 | 761 | 761 |
| 748 | 748 | 390 | 762 | ||
| 749 | 749 | 391 | 763 | 763 | |
| 750 | 392 | 764 | 764 | ||
| 751 | 751 | 393 | 765 | 765 | |
| 752 | |||||
| 803 | 803 | 444 | 444 | 817 | |
| 804 | 804 | 445 | 818 | 818 | |
| 805 | 446 | 819 | 819 | ||
| 806 | 806 | 447 | 820 | 820 | |
| 807 | 807 | 448 | 448 | 821 | 821 |
| 808 | 808 | 449 | 822 | 822 | |
| 809 | 450 | 823 | 823 | ||
| 810 | 610 | 451 | 451 | 824 | |
| 811 | 811 | 452 | 452 | 825 | 825 |
| 812 | 812 | 453 | 826 | 826 | |
| 813 | 454 | 454 | 827 | 827 | |
| 455 | 455 | 828 | 828 | ||
| 456 | 456 | 829 | |||
| 457 | 830 | ||||
| 458 | 831 | ||||
| 459 | 459 | ||||
Secondary structure and binding sites for steroids within the ligand binding domain
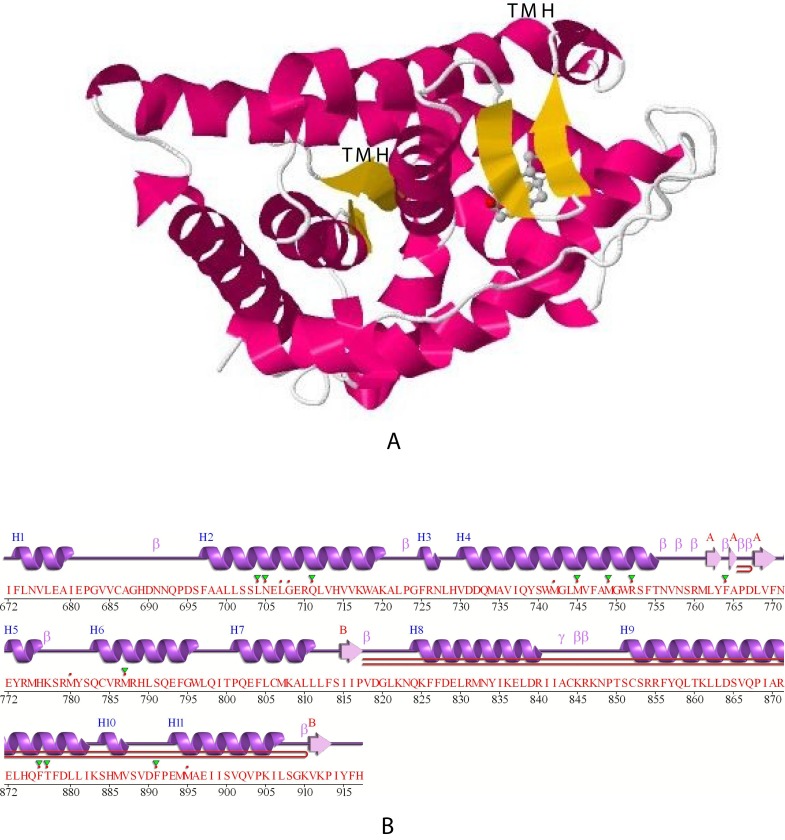
The upper part (A) of Figure 7 illustrates the crystal structure of the ligand binding domain of rat ARB (1i37, P15207). The transmembrane helices (TMH) identified in Figure 1 are indicated both above the center and in the upper right of the graphic in Figure 7 by “TMH” and correspond to residues 738Q – R752 (center TMH) and residues 803E – F813 (upper right) as predicted in Table 4. As noted in Table 4, both putative TM helices contain residues identified as pore-lining amino acids by PoreWalker (Table 4). The lower part (B) of Figure 7 illustrates the secondary structure of the 246 residues of rat ARB 1i37 (P15207). Eleven helical regions are indicated as purple coils and defined as H1, H2 etc. The TM helices predicted in the JMol projection (Figure 8) correspond to H3 – H4 and H5-β-H6 in the “classical” PDB projection in B (lower image). The region between 753S and Q802 corresponds to the putative 51 amino acid extracellular loop (described above) and contains 3 β strand structures (indicated in yellow).
Figure 7. Jmol projection and PDB-predicted secondary structure of rat ARB. The upper image (A) illustrates a JMol projection of the crystal structure of the ligand binding domain of rat ARB (1i37, P15207).
The two transmembrane helices are identified by “TMH” in the center (dark red) and upper right (dark red) and correspond to residues 738Q – R752 and 803E – F813, respectively. The lower projection (B) indicates the α-helices and the β strand structures (indicated in yellow), as predicted by PDB. See Methods.
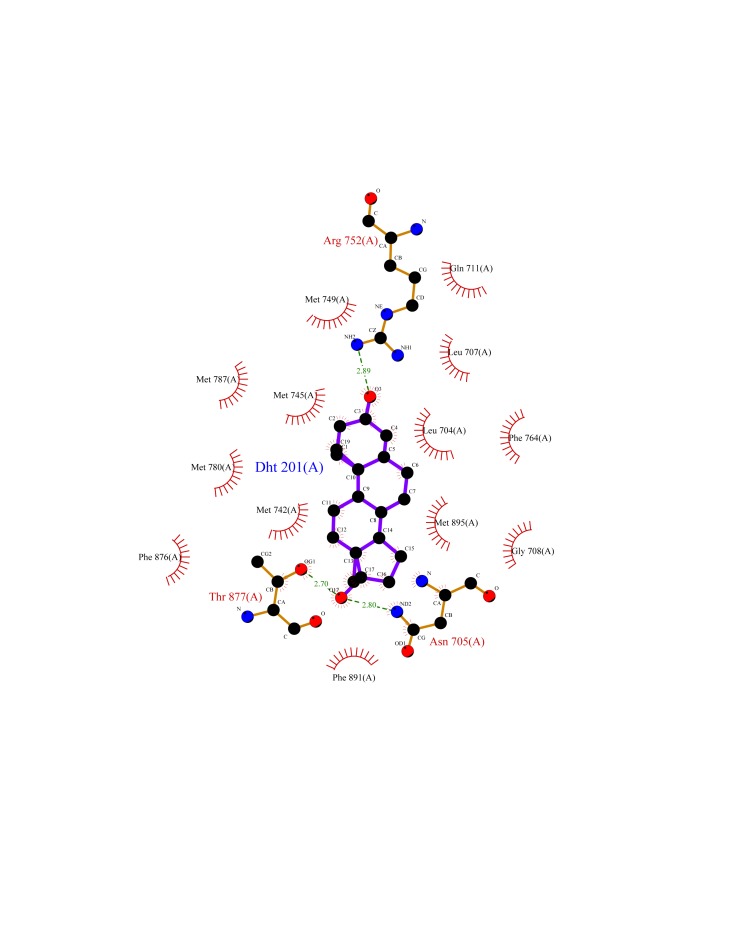
Figure 8. A schematic 2-D representation of a protein-ligand complex of dihydrotestosterone (DHT) and rat ARB (1i37, P15207) using LIGPLOT [Wallace et al., 1995] and the standard PDB file input.
The interactions shown are those mediated by hydrogen bonds and by hydrophobic contacts. Hydrogen bonds are indicated by dashed lines between the atoms involved, while hydrophobic contacts are represented by an arc with spokes radiating towards the ligand atoms they contact. The contacted atoms are shown with spokes radiating back.
Wang et al. [2013] have used Docking and comparative molecular similarity index analysis (CoMSIA) to study the mechanism for binding of steroids and non-steroidal chemicals to androgen receptors. The obtained docking conformations and predictive CoMSIA models identified the primary interaction site and key residues involved in the binding process. The major factors that influence the binding of steroids and non-steroidal chemicals were electrostatic and hydrophobic interactions. Their results indicate that androgen receptor residues V746, M749 and F764 are crucial for steroid binding, in addition to previously reported E711, R752 and T877 [Wang et al., 2013]. Figure 8 illustrates the dihydrotestosterone (DHT) – 1i37 interactions using a Jmol projection. Analysis of the positions of the crucial residues demonstrates that V746, M749 and R752 are within the TM-1 pore-lining region and F764 is within the external loop between TM-1 and TM-2. PoreWalker of ARB further indicates that the DHT binding site is within a cholesterol-rich pore-lining TM helix region (TM-1). This suggests that, at least for androgens, the plasma membrane ligand “docking” site coincides with the pore-lining region (TM-1) associated with the PoreWalker predicted channel.
Conclusions
Our results suggest that the movement and localization of “nuclear steroid receptors” is somewhat more complex than generally recognized. Extranuclear ARB, ER1 and PRB proteins are currently thought to be largely cytoplasmic, with smaller amounts “tethered” to the inside of the plasma membrane by palmitoylation and therefore localized outside the nucleus [Hammes and Levin, 2011; Levin 20115]. However, measuring precisely how much receptor is dissolved in cytosol of intact cells is difficult, since some proteins appear to be weakly associated with membranes and are released into solution upon cell lysis (see [Clegg, 1984; Feig and Sugila, 2013]). Attachment of lipid moieties such as myristic and/or palmitic acid can also play an important role in altering the properties and localization of proteins (reviewed in [Aicart-Ramos et al., 2011]).
As predicted here, the ligand binding domains of ARB, ER1 and PRB each contain two transmembrane helices (Table 1) with multiple cholesterol binding (CRAC/CARC) motifs (Figure 4), possibly associated with membrane lipid rafts [Fantini and Barrantes, 2013; Aicart-Ramos et al., 2011; Brown and London 2000]. As indicated in Table 2, a cysteine-containing 11-13 residue sequence within the TM helices of ARB, ER1 and PRB is localized to the plasma membrane and forms a helix-loop-helix structure with the 50-51 residue loops extending into the extracellular fluid (Table 1). Palmitoylation of the cysteine in predicted TM-2 by membrane DHHC-7 and /or -21 palmitoylacyltransferase (Table 3), believed to occur in the Golgi [Salaun et al., 2010], may facilitate transfer of the TM helices to the plasma membrane, accounting for the reported “tethering”. As indicated in Figure 7, the 51 amino acid extracellular loop between TM-1 and TM-2 of ARB contains three β-strands and multiple cholesterol binding motifs, and facilitates steroid uptake. It should also be noted that numerous cellular and physiological studies have implicated both ER subtypes (ER1 and ER2) and PRB as critical regulators of gap junctional intercellular communication in reproductive tissue (reviewed in [Firestone and Kapadia, 2012]), further contributing to steroid regulation of cell function.
The N-attachment (scaffolding) domain of caveolin is thought to be an essential element in the interaction between caveolin and the proteins involved in signal transduction (e.g. [Lajoie and Nabi, 2010]). Acconcia et al. [2005] found that palmitoylation increases the physical association of ER1 with caveolin-1 and that the palmitoylation of 447C is essential for promoting caveolin interactions and membrane translocation [Aicart-Ramos et al., 2011]. This observation was extended by Pedram et al. [2007] to ER2, PRB and ARB. As seen in Table 2, the cysteine palmitoylation site is within MemBrain predicted TM-2 (residues 445F-V465) of the ER1 receptor, TM-2 (residues 802-822) of ARB, and TM-2 (residues 818-838) of PRB, and coincides with the predicted plasma membrane-spanning regions that interact with caveolin-rich lipid rafts. Both DHHC-7 and -21 are membrane enzymes (Table 3), and palmitoylation within the TM-2 sequence may direct the steroid receptor to a site on the plasma membrane which is, in turn, concentrated within caveolin-rich lipid rafts. Therefore, cytosolic steroid receptors must be localized to the cell membrane at some point in their life cycle for palmitoylation to occur. Internalization of ligands and receptors by the lipid raft domains occurs via a process defined as raft-dependent endocytosis [Lajiie and Nabi, 2010], which accounts for the uptake of both ligands and steroids into the cell. Our findings here and elsewhere [Morrill et al., 2013] indicate that circulating steroids act at the cell surface to initiate the internalization of a steroid-receptor complex, and/or formation of channels from the pore-lining regions [Morrill et al., 2013] that serve to regulate movement of specific ions (e.g. Ca2+, Na+, H+, and Cl-) essential for cell function. As noted previously [Morrill et al., 2013], multiple steroid-response systems may be required for ion regulation, as well as to facilitate accumulation of a cholesterol-steroid-receptor complex [Willnow and Nykjaer, 2010, Epand, 2006] into the cytoplasm.
Acknowledgements
This research was supported in part by National Institutes of Health research grants HD-10463 and GM-071324.
Abbreviations:
- ARB/AR
androgen receptor
- PRB/PGR
progesterone receptor
- ER1/ESR1
estrogen receptor α
- ER2/ESR2
estrogen receptor β
- LBD
ligand-binding domains
- TMH
transmembrane helices
Footnotes
There are no competing interests.
References
- Acconcia F., Ascenzi P., Bocedi A., Spisni E., Tomasi V., Trentalance A., Visca P., Marino M. (2005) Palmitoylation-dependent estrogen receptor alpha membrane localization: regulation by 17beta-estradiol. Mol Bio Cell 16, 231-37. 10.1091/mbc.E04-07-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aicart-Ramos C., Valero R.A., Rodriguez-Crespo I. (2011) Protein palmitoylation and subcellular trafficking. Biochim Biophys Acta 1808, 2981-94. 10.1016/j.bbamem.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Alden K., Veretnik S., Bourne P.E. (2010) dConsensus: a tool for displaying domain assignments by multiple structure-based algorithms and for construction of a consensus assignment. BMC Bioinfomatics 10, 310. 10.1186/1471-2105-11-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A., Pascual A. (2001) Nuclear hormone receptors and gene expression. Physiol Rev 81, 1269-1304. [DOI] [PubMed] [Google Scholar]
- Baier, C.J., Fantini, J., Barrantes, F.J. (2011) Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci Rep 69, DOI 10.1038. [DOI] [PMC free article] [PubMed]
- Bernsel A., Viklund H., Falk J., Lindahl E., von Heijne G., Wolfson A. (2008) Prediction of membrane-protein topology from first principles. Proc Natl acad Sci USA 105, 7177-81. 10.1073/pnas.0711151105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsel A., Viklund H., Hennerdal A., Elofsson A. (2009) TOPCONS: consensus prediction of membrane topology. Nucleic Acids Res 37, 465-8. 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.A., London E. (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem 275, 17221-24. 10.1074/jbc.R000005200 [DOI] [PubMed] [Google Scholar]
- Brunori M., Gianni S., Giri R., Morrone A., Travaglini-Allocatelli C. Morphogenesis of a protein: folding pathways and the energy landscape. Biochem Soc Transact (2012) 40, 429-432. 10.1042/BST20110683 [DOI] [PubMed] [Google Scholar]
- Bryson K., Cozzetto D., Jones D.T. (2007) Computer-assisted protein domain boundry prediction using DomPred server. Curr Protein Pept Sci 8, 181-8. 10.2174/138920307780363415 [DOI] [PubMed] [Google Scholar]
- Clegg J.S. (1984) Properties and metabolism of the aqueous cytoplasm and its boundaries. Am J Physiol 246, R133-51. [DOI] [PubMed] [Google Scholar]
- Dawidowicz E.A. (1987) Dynamics of membrane lipid metabolism and turnover. Ann Rev Biochem 56, 43-57. 10.1146/annurev.bi.56.070187.000355 [DOI] [PubMed] [Google Scholar]
- Dechering K, Boersma C, Mosselman S. (2000) Estrogen receptors alpha and beta: two receptors of a kind? Curr Med Chem 7, 561-76. 10.2174/0929867003375010 [DOI] [PubMed] [Google Scholar]
- Epand R.M. (2006) Cholesterol and the interaction of proteins with membrane domains. Prog Lipid Res 45, 279-94. 10.1016/j.plipres.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Fantini J., Barrantes F.J. (2013) How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Frontiers in Physiology 4, 1-9. 10.3389/fphys.2013.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig M., Sugila Y. (2013) Reaching new levels of realism in modeling biological macromolecules in cellular environments. J Mol Graph Model 45, 144-56. 10.1016/j.jmgm.2013.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestone G.L., Kapadia B.J. (2012) Mini Review: Regulation of Gap Junctions Dynamics by Nuclear Hormone Receptors and their Ligands. Mol Endocrinol 26, 1798-1807. 10.1210/me.2012-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes A., Andreasson T.K., Spoelgen R., Raila J., Hubner J.N., Schulz H., Metzger J., Schweigert F.J., Luppa P.B., Nykjaer A., Willnow T.E. (2005) Role of endocytosis in cellular uptake of sex steroids. Cell 122, 751-62. 10.1016/j.cell.2005.06.032 [DOI] [PubMed] [Google Scholar]
- Hammes S.R, Levin E.R. (2011) Minireview: Recent advances in extranuclear steroid receptor actions. Endocrinology 152, 4489-95. 10.1210/en.2011-1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Cheng Q, Xie W. (2010) Minireview: Nuclear receptor-controlled steroid hormone synthesis and metabolism. Mol Endocrinol 24, 11-21. 10.1210/me.2009-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilldebrand P.W., Lorenzen S., Goede A., Preissner R. (2006) Analysis and prediction of helix-helix interactions in membrane channels and transporters. Proteins 64, 253-62. 10.1002/prot.20959 [DOI] [PubMed] [Google Scholar]
- Illergard K., Callegari S., Elofsson A. (2010) MPRAP: An accessibility predictor for α-helical transmembrane proteins that performs well inside and outside the membrane. BMC Bioinfomatics 11, 333-44. 10.1186/1471-2105-11-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L. (2004) A combined transmembrane topology and signal peptide prediction method. J Mol Biol, 338, 1027-36. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Lajoie P., Nabi I.R. (2010) Lipid rafts, caveolae, and their endocytosis. Internat Rev Cell Mol Biol 282, 135-63. 10.1016/S1937-6448(10)82003-9 [DOI] [PubMed] [Google Scholar]
- Levin E.R. (2011) Minireview: Extranuclear steroid receptors: Roles in modulation of cell functions. Mol. Endocrinol 25, 377-84. 10.1210/me.2010-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Papadopoulos V. (1998) Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139, 4991-7. [DOI] [PubMed] [Google Scholar]
- Meruelo A.D., I., Samish I., Bowie J.U. (2011) TMKink: a method to predict transmembrane helix kinks, Protein Sci. 20, 1256-64. 10.1002/pro.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrill G.A., Kostellow A.B., Gupta R.K. (2013) A computational analysis of non-genomic plasma membrane progestin binding proteins: Signaling through ion channel-linked cell surface receptors. Steroids 78, 1233-44. 10.1016/j.steroids.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Mundy D.L. (1995) Protein palmitoylation in membrane trafficking. Biochem Soc Trans 23, 572-76. [DOI] [PubMed] [Google Scholar]
- Nugent T., Ward S., Jones D.T. (2011) The MEMPACK alpha-helical transmembrane protein structure prediction server. Bioinformatics 27, 1438-39. 10.1093/bioinformatics/btr096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent T., Jones D.T. (2012) Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinfomatics 13, 169-78. 10.1186/1471-2105-13-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky J.M. (2001) Nuclear Receptor Minireview Series. J Biol Chem, 276, 36863-64. 10.1074/jbc.R100047200 [DOI] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Sainson R.C., Kim J.K., Hughes C.C., Levin E.R. (2007) A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem 282, 22278-88. 10.1074/jbc.M611877200 [DOI] [PubMed] [Google Scholar]
- Pedram A., Razandi M., Deschenes R.J., Levin E.R. (2012) DHHC-7 and -21 are palmitoylacyltranserases for sex seroid receptors. Mol Biol Cell 23, 188-99. 10.1091/mbc.E11-07-0638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini-Calace M., Malwald T., Thornton J.M. (2009) PoreWalker: A novel tool for the identification and characterization of channels in transmembrane proteins from their three-dimensional structure. PloS Computational Biology 5, 1-16. 10.1371/journal.pcbi.1000440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaun C., Greaves J., Chamberlain L.H. (2010) The intracellular dynamic of protein palmitoylation. J Cell Biol 191, 1229-38. 10.1083/jcb.201008160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe J.W., Chapman L., Finch J.T. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell 75, 567-78. 10.1016/0092-8674(93)90390-C [DOI] [PubMed] [Google Scholar]
- Shen L., Bassolino D., Stouch T. Transmembrane helix structure, dynamics, and interactions: Multi-nanosecond molecular dynamics simulations. Biophys J (1997) 73, 3-20 10.1016/S0006-3495(97)78042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Chou J.J. (2008) MemBrain: Improving the accuracy of predicting transmembrane helices. PLoS ONE 3 (6), e2399. 10.1371/journal.pone.0002399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisic V., Lonard D.M., O’Malley B.W. (2010) Modulation of steroid hormone receptor activity. Prog Brain Res 181, 153-76. 10.1016/S0079-6123(08)81009-6 [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski R.A., Thorton J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interaction. Protein Eng 8, 127-34. 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang C., Li Y.J., Cao J.M. (2013) Specificity out of clutter: a hypothetical role of G-protein-coupled receptors in the non-genomic effect of steroids. FEBS Lett 587, 823-5. 10.1016/j.febslet.2013.02.025 [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Shi W., Giesy J.P., Yu H., Wang Y. (2013) Docking and CoMsia studies on steroids and non-steroidal chemicals as androgen receptor ligands. Ecotoxicology Environmental Safety 2013, 143-149. 10.1016/j.ecoenv.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Willnow T.E., Nykjaer A. (2010) Cellular uptake of steroid carrier proteins-Mechanisms and implications. Mol Cell Endocrinol 316, 93-102. 10.1016/j.mce.2009.07.021 [DOI] [PubMed] [Google Scholar]
- Wurtz J-M., Bourguet W., Renaud J-P., Vivat V., Chambon P., Moras D., Gronemeyer H. (1996) A canonical structure for the ligand-binding domain of nuclear receptors. Nature Structural & Molecular Biology 3, 87-94. 10.1038/nsb0196-87 [DOI] [PubMed] [Google Scholar]