Abstract
Background
Usefulness of next-generation sequencing (NGS) in assessing bacteria associated with oral squamous cell carcinoma (OSCC) has been undermined by inability to classify reads to the species level.
Objective
The purpose of this study was to develop a robust algorithm for species-level classification of NGS reads from oral samples and to pilot test it for profiling bacteria within OSCC tissues.
Methods
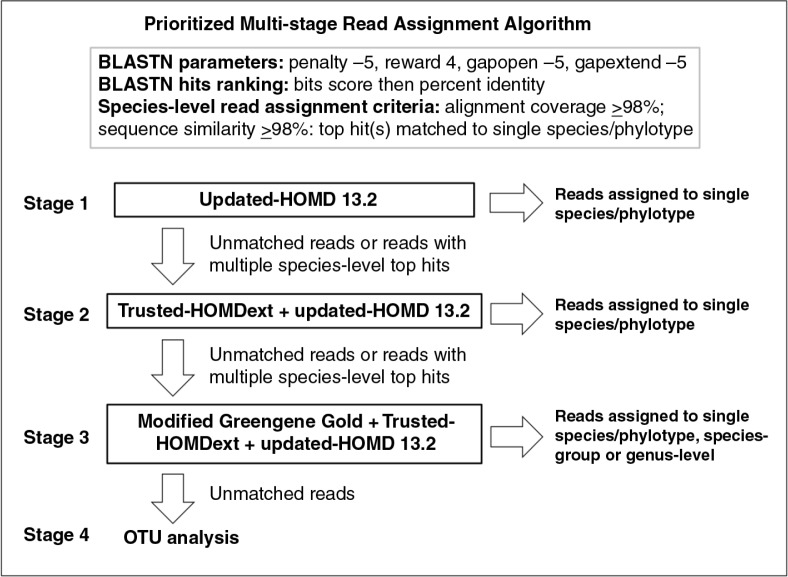
Bacterial 16S V1-V3 libraries were prepared from three OSCC DNA samples and sequenced using 454's FLX chemistry. High-quality, well-aligned, and non-chimeric reads ≥350 bp were classified using a novel, multi-stage algorithm that involves matching reads to reference sequences in revised versions of the Human Oral Microbiome Database (HOMD), HOMD extended (HOMDEXT), and Greengene Gold (GGG) at alignment coverage and percentage identity ≥98%, followed by assignment to species level based on top hit reference sequences. Priority was given to hits in HOMD, then HOMDEXT and finally GGG. Unmatched reads were subject to operational taxonomic unit analysis.
Results
Nearly, 92.8% of the reads were matched to updated-HOMD 13.2, 1.83% to trusted-HOMDEXT, and 1.36% to modified-GGG. Of all matched reads, 99.6% were classified to species level. A total of 228 species-level taxa were identified, representing 11 phyla; the most abundant were Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Actinobacteria. Thirty-five species-level taxa were detected in all samples. On average, Prevotella oris, Neisseria flava, Neisseria flavescens/subflava, Fusobacterium nucleatum ss polymorphum, Aggregatibacter segnis, Streptococcus mitis, and Fusobacterium periodontium were the most abundant. Bacteroides fragilis, a species rarely isolated from the oral cavity, was detected in two samples.
Conclusion
This multi-stage algorithm maximizes the fraction of reads classified to the species level while ensuring reliable classification by giving priority to the human, oral reference set. Applying the algorithm to OSCC samples revealed high diversity. In addition to oral taxa, a number of human, non-oral taxa were also identified, some of which are rarely detected in the oral cavity.
Keywords: bacteria, cancer, next-generation sequencing, OSCC, pyrosequencing, taxonomy
There is recently an increasing interest in the potential role of bacteria in the development of oral cancer (1). Such a trend is driven by the existing evidence on association between certain bacterial species and some types of cancer. The etiological role of Helicobacter pylori in gastric adenocarcinomas and lymphomas is a classic example (2). Other examples include the association of Chlamydia trachomatis with cervical cancer (3), Salmonella typhi with gallbladder cancer (4), and Bacteroides fragilis and Fusobacteria with colon cancer (5, 6). Mechanisms by which bacteria are thought to contribute to the development of cancer include induction of chronic inflammation, interference with eukaryotic cell cycle, or/and production of carcinogenic substances (7). Actually, the oral microbiota has been demonstrated to produce carcinogenic levels of acetaldehyde (8).
Bacteria associated with oral squamous cell carcinoma (OSCC) have been assessed in several studies using various methods with different types of specimens. Culture techniques have been first used to characterize bacteria on the surface of OSCC lesions (9), and later to document the presence of viable bacteria within OSCC tissues (10). Molecular techniques such as checkerboard DNA–DNA hybridization and clonal analysis of 16S rRNA have been employed to profile and compare bacterial species in tissue or saliva samples from OSCC and control subjects (11–14). While these studies identified several bacterial taxa in association with OSCC lesions or as potential markers in saliva, there seems to be no consensus among them on particular species to link to oral cancer. One possible reason, among others, for this is that cultivation and clonal analysis are limited by the number of strains/clones that can be feasibly tested, rendering reproducible detection of potentially relevant taxa, particularly low abundant ones, unlikely.
The advent of high-throughput, next-generation sequencing (NGS) techniques, such as pyrosequencing, has enabled analysis of microbial communities at significantly higher depth and coverage than classical Sanger sequencing (15). Indeed, two recent studies have employed NGS to assess the bacteriome associated with OSCC (16, 17). However, these studies have used either saliva or surface swab samples but not cancerous tissue for testing. In addition, both studies employed the typical analysis approach that involves clustering of reads into operational taxonomic units (OTUs), using a Bayesian classifier or BLAST to assign taxonomies to representative OTU sequences and describing/comparing microbial composition at the phylum and genus levels, without the capability to accurately classify individual reads to the species level, which is probably more relevant to addressing the link between bacteria and oral cancer (or any other disease). In fact, while OTU analysis and taxonomic classification to the genus level may be justified for less characterized microbial communities such as that of the soil, it is probably not for well-characterized ones like those associated with humans (18), for which well-curated databases of reference 16S rRNA gene sequences such as the Human Oral Microbiome Database (HOMD; www.homd.org) (19) and the Greengene databases (www.greengenes.lbl.gov) (20) are available. These databases do not seem to have been adequately exploited for improving the resolution of taxonomic assignment of microbial metagenomic 16S rRNA reads despite the increase in reads length obtained with NGS technologies.
The objective of this work therefore was to develop a robust, multi-stage, BLASTN-based search algorithm for classification of NGS reads from oral microbiological samples to the species level and to pilot test it for characterizing bacterial species/phylotypes within OSCC tissues. The algorithm takes advantage of three 16S rRNA reference sequence databases which were further curated in this study by removing potentially chimeric and redundant sequences and refining the associated taxonomy annotations.
Methods
OSCC DNA samples
Three samples were randomly selected from among 60 archived DNA extracts obtained from fresh OSCC biopsies in a previous study (21). All extracts had tested HPV-negative by q-PCR and had been stored at −80°C. The clinical features of the three cases selected retrospectively for the study are presented in Table 1.
Table 1.
Clinical characteristics of the OSCC cases included in the study
| Case no. | Site affected | Gender | Age (years) | Snuff dipping | Smoking |
|---|---|---|---|---|---|
| 1 | Floor of the mouth | Female | 54 | Yes | No |
| 2 | Gum | Male | 45 | Yes | Yes |
| 3 | Other and unspecified parts of the mouth | Female | 55 | No | No |
Amplicon library preparation and sequencing
Library preparation and sequencing were done at GATC Biotech (Konstanz, Germany). In a first PCR reaction, the V-V3 region of the 16S rRNA gene was amplified with the degenerate primers 27FYM (22) and 519R (23) using the reaction setup and cycling program described by Kistler et al. (24), with some modifications as follows: a second PCR reaction with few cycles was used to incorporate the GS FLX titanium adaptors FLX-A and FLX-B along with a 22-base spacer and 5-base barcodes to the amplicons. The final forward primer construct used in the second reaction was [FLX-A]-[22-base spacer]-[5-base tag]-27FYM, while that of the reverse was [FLX-B]-[22-base spacer]-519R. The three tagged amplicon libraries were pooled with another 16 libraries (another study) in equimolar amounts and sequenced unidirectionally (side A) on quarter plate using 454 GS FLX chemistry (Roche, Germany).
Preprocessing of sequencing data
The raw data were submitted to the Sequence Read Archive (SRA) under project accession number SRA204252. Data preprocessing were performed using the mothur software package version 1.33 (25). To minimize sequencing error rates, reads with any mismatch in the spacer–tag–primer sequence, base ambiguity or/and homopolymers>eight bases long were excluded, and remaining reads were trimmed so as to maintain a 50-nucleotide sliding window with an average quality score of ≥30 (26). Subsequently, the spacer–tag–primer sequence was trimmed off, and the reads were filtered to include only those with a minimum read length of 350 bases since a read length of 350–500 bases is required for identification (27). Those were aligned to SILVA reference alignment (28), and the ones with poor alignment were removed. The rest were stringently screened for chimeras with Uchime (29) and Chimera Slayer (30) sequentially, using both SILVA gold (30) (downloaded from www.mothur.org/wiki/Silva_reference_files) and, for the first time, updated-HOMD 13.2 (see the following sections) reference sequences combined as the reference set.
Optimization of reference databases
For taxonomic assignment of reads (see the following sections), three sets of 16S rRNA gene reference sequences were used: HOMD version 13.2 (downloaded from www.homd.org/index.php?name=seqDownload&file&type=R), HOMD extended version 1.1 (downloaded from www.homd.org/index.php?name=seqDownload&file&type=R), and Greengene Gold (GGG; downloaded from www.greengenes.lbl.gov/Download/Sequence_Data/Fasta_data_files/). Due to concerns about the reliability of some reference sequences in HOMD 13.2 and HOMD extended, both sets were double-checked for and cleared off potential chimeric sequences with Uchime and Chimera Slayer, which resulted in removal of 27 and 172 sequences from HOMD 13.2 and HOMD extended, respectively. In addition, sequences in HOMD extended with better representatives in HOMD 13.2 (match at ≥98%) were removed (Floyd Dewhrist, personal communication), resulting in a final set of trusted 495 sequences (trusted-HOMDEXT). Finally, full 16S rRNA sequences of 21 novel oral taxa recently described by Camanocha and Dewhirst (31) were added to HOMD 13.2 (referred to hereafter as updated-HOMD 13.2).
GGG was also modified to only include aligned and non-redundant sequences (3,940 out of 5,441). To obtain full taxonomy annotations for these, sequences were first classified with the Wang method (32) using the 2013 greengene reference taxonomy (~202,000 taxa); the resultant classifications were then combined with the binary names (Genus species/strain no.) provided with the GGG set, which resulted in obtaining full taxonomy annotations without conflict for the majority of the sequences. For a subset, additional search in Greengene and NCBI was necessary to arrive at the right taxonomy; many of the strains initially unnamed in the GGG set have been recently named and reclassified, so taxonomy was updated accordingly.
The fasta and taxonomy files for the three sets (updated-HOMD 13.2, trusted-HOMDEXT, and modified-GGG), as well as lists of potential chimeras in HOMD and a detailed description of how they were identified can be obtained from ftp://www.homd.org/publication_data/20150120/.
Taxonomy assignment algorithm
The high-quality, non-chimeric reads were classified using a prioritized, multi-stage, BLASTN-based search against updated-HOMD 13.2, trusted-HOMDEXT, and modified-GGG as shown in Fig. 1. The ‘blastn’ command in the Nucleotide-Nucleotide BLAST 2.2.30+ was used with the following parameters: max. target seqs., 1000; penalty, -5; reward, +4; opengap, -5; gapextend, -5; outfmt, 6. For each read, hits to the reference sequences with both alignment coverage (BLASTN alignment length/read length) and identity (matches/alignment length) ≥98% were collected and ordered first by bit score then by percentage identity. Reads with a single best hit or multiple best hits (with equal bit score and identity) representing the same species/phylotype were assigned to the unique species-level taxonomy. Unmatched reads (i.e. reads with no hits at ≥98% alignment coverage and identity) and reads with best hits to multiple species were forwarded to the next stage of BLASTN search that included an additional set of reference sequences.
Fig. 1.
Prioritized, multi-stage, BLASTN search algorithm used for taxonomic assignment of the reads. Refer to the text for a description. Updated-HOMD 13.2, Human Oral Microbiome Database version 13.2 updated by removal of potential chimeric sequences and addition of new taxa; trusted-HOMDEXT, HOMD extended after clearing chimeric and redundant sequences; modified-GGG: Greengene Gold collection after removing unaligned and redundant sequences.
In the third stage, reads that hit multiple species were classified into either the ‘species-group’ level (for consistent species combinations, for example, Neisseria flavescens/subflava) or genus level (for inconsistent species combinations). Reads returning no hits from searching against all three reference sets were subjected to OTU analysis (stage 4). Reads were clustered to OTUs at 98% identity using average neighborhood. OTUs with ≤3 sequences were removed (rare OTUs), while the sequence with the smallest maximum distance to other sequences in each of the remaining OTUs was selected as a representative. The representative sequences were then BLASTN searched using the same coverage and identity criteria described above against NCBI's bacterial 16S rRNA sequences and nucleotide collection, and if any returned a hit, the corresponding OTU was assigned the species-level taxonomy of the returned hit. Finally, unmatched OTUs were labeled as potentially novel taxa, and classified to a higher rank (genus or family) using the Wang method and SILVA sequences as the reference.
Results
Pyrosequencing information
A total of 33,810 raw reads were obtained (~11,000 reads per sample). Filtering by read quality and length as described above removed 24,178 reads (71.5%). Additional 1,881 reads with poor alignment were excluded. One-thousand chimeras were identified with Uchime and another 46 with Chimera Slayer, leaving a final of 6,705 non-chimeric reads (mean of 2,235±336 reads/sample) with an average length of 416 bases.
Taxonomic assignment of reads
Using the novel, multi-stage, assignment algorithm, about 96% of the reads were matched to the reference sequences as follows: 92.8% to updated-HOMD 13.2, 1.83% to trusted-HOMDEXT, and 1.36% to modified-GGG. The majority of these reads (91.7%) were assigned to single species/phylotypes, 7.9% consistently matched to two or a few species/phylotypes so were assigned to the ‘species-group’ level, and only 0.4% were classified to the genus level in the final stage. That is, 99.6% were classified to the species level. OTU analysis of the remaining 4% unmatched reads, generated 17 non-rare, species-level OTUs, of which 12 matched reference sequences in NCBI including seven oral clones described in recent studies (33–35) but not included in HOMD. One OTU could not be classified even to the phylum level.
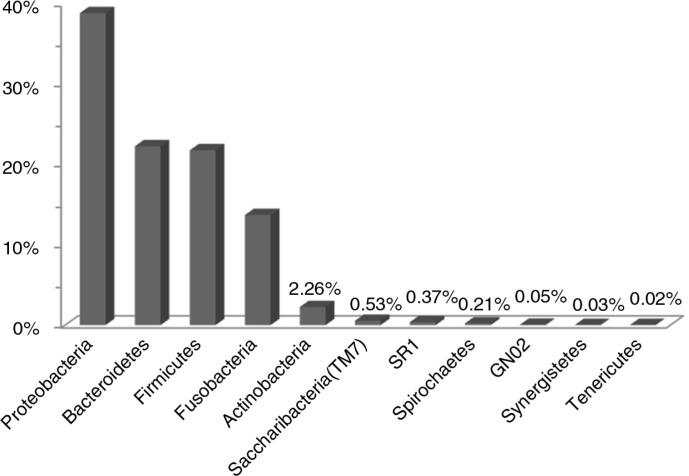
Bacteria within OSCC – phylum level
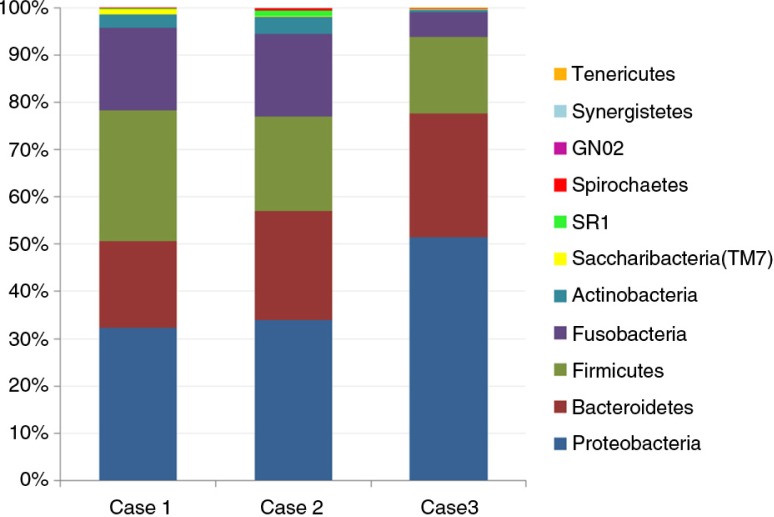
Eleven bacterial phyla were identified (Fig. 2), of which eight were present in all samples. The most abundant in order were: Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Actinobacteria, accounting for 98.8% of the reads. Phyla Tenericutes, Synergistetes, and GN02 were represented by few reads in single samples. Cases 1 and 2 had comparable phylum distribution, while case 3 had higher proportion of Proteobacteria at the expense of Fusobacteria and Actinobacteria (Fig. 3).
Fig. 2.
Relative abundance (%) of 11 phyla detected in the OSCC samples. GN02, Synergistetes, and Tenericutes were found in single samples.
Fig. 3.
Distribution of the detected phyla in each of the study OSCC samples.
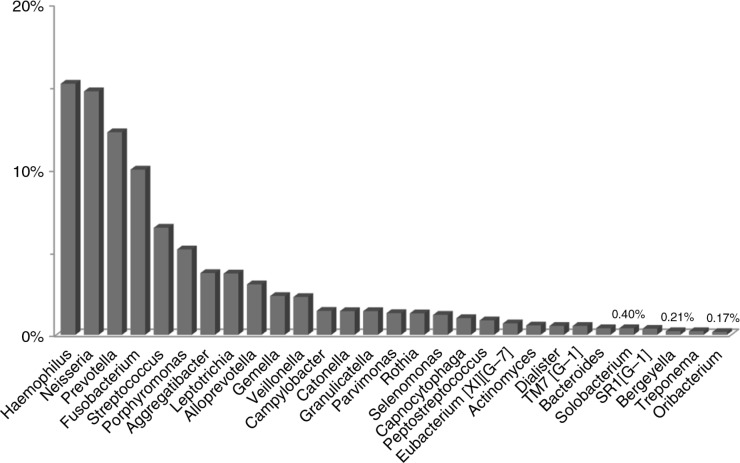
Bacteria within OSCC – genus level
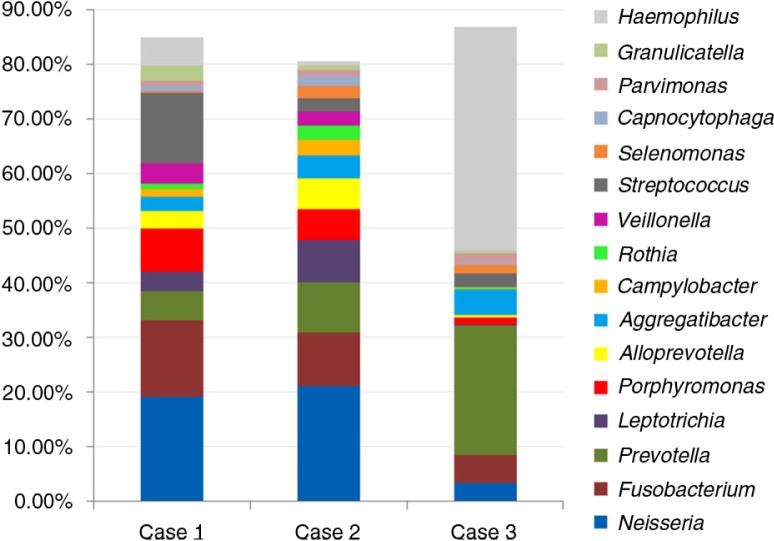
The reads were classified into 78 genera, of which 29 were detected in all samples. Overall, Haemophilus, Neisseria, Prevotella, Fusobacteria, Streptococcus, Porphyromonas, Leptotrichia, and Aggregatibacter were the most abundant in order (Fig. 4). Sixteen genera accounted for >80% of the reads. The distribution of these in each of the three samples is shown in Fig. 5. Case 3 had exceptionally very high relative abundance of genus Haemophilus, resulting in a significantly different profile than that of cases 1 and 2.
Fig. 4.
Relative abundance (%) of 29 genera detected in all OSCC samples. Abundance of Haemophilus was inflated by the presence of high level of H. influenzae in the sample from case 3 (see Fig. 5).
Fig. 5.
Distribution of 16 genera accounting for >80% of the reads in each of the OSCC sample. Profiles of cases 1 and 2 are comparable, while that of case 3 deviates significantly due to high levels of Haemophilus.
Bacteria within OSCC – species level
Excluding 102 rare OTUs, a total of 228 species-level taxa (mean of 118±18 taxa/subject) were identified in the samples as follows: 222 species/phylotypes, 2 species groups, and 4 potentially novel OTUs. The vast majority of these were human, oral taxa representing 191 sequences in updated-HOMD 13.2, 16 in trusted-HOMDEXT, 10 in modified-GGG, and 7 in the NCBI's nucleotide collection. A list of detected taxa sorted by number of positive samples and relative abundance is presented in Supplementary Table 1.
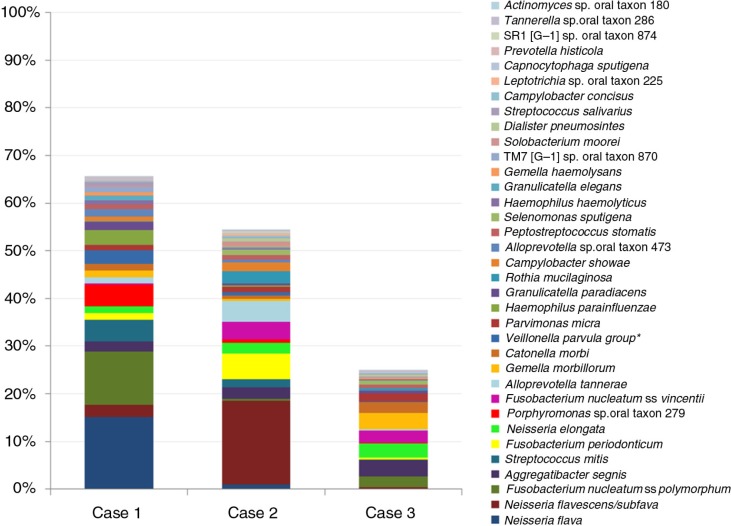
Thirty-five of the species-level taxa were detected in all the three samples (Fig. 6), while 54 were found in two of them, that is, 89 taxa were identified at least twice. The rest were identified in single samples at very low abundance, the only exception being Haemophilus influenzae, which was detected at 40.5% in the sample from case 3. Excluding the latter, the most abundant taxa, on average, were Prevotella oris, Neisseria flava, N. flavescens/subflava, Fusobacterium nucleatum ss polymorphum, Aggregatibacter segnis, Streptococcus mitis, Fusobacterium periodonticum, Neisseria elongata, Porphyromonas sp. oral taxon 279, and Alloprevotella tannerae. The distribution of these and other taxa, however, varied considerably among the three samples.
Fig. 6.
Subject-level distribution of 35 species-level taxa identified in all the three OSCC samples. *Veillonella parvula group: V. parvula, V. dispar and V. rogosae.
A number of human, non-oral taxa were also identified including B fragilis, Leptotrichia trevisani, Fusobacterium varium, Haemophilus pittmaniae, Propionibacterium granulosum, and Wolinella succinogenes. The former was detected in two samples, while the rest was found in only one sample. In addition, two environmental species, Sphingopyxis alaskensis and Cupriavidus metallidurans were identified in case 2.
Discussion
This report describes the use of a novel prioritized, multi-stage algorithm for species-level taxonomy assignment of bacterial 16S rRNA NGS reads from oral samples in a pilot study involving three OSCC samples. To our best knowledge, this is the first attempt to profile bacteria within OSCC tissues to the species level using NGS.
Sequencing was achieved at ~11,000 reads per sample. However, filtering by length and quality parameters removed 70% of the reads which is too high compared to other oral microbiome studies (16, 24). In fact, similar processing of reads from another 16, primarily bacterial DNA samples run in parallel with the three OSCC samples resulted in removal of only 30% of the reads. It seems, therefore, that the presence of high human DNA background in OSCC samples adversely affects reads quality and length. Optimization of extracts from OSCC tissues for library preparation, for example, by enriching bacterial DNA, should be considered in future work. The high-quality reads were then subjected to exceptionally stringent chimera check by using two effective chimera detection software and combining HOMD 13.2 and SILVA gold sequences to make the reference set, an approach never reported before. Using updated-HOMD 13.2 as a reference for chimera detection may, however, be viewed as a more reliable alternative to one established method in which high abundance sequences in the dataset itself is used as a reference (26). Indeed, including HOMD 13.2 sequences resulted in detection of significantly more chimeras compared to when only SILVA gold was used (data not shown).
Matching of non-chimeric reads to reference databases was performed using sequence% identity cutoff of 98% as previously described (27, 36) but a more stringent alignment coverage cutoff (98% vs. 95% in previous studies). Priority was given to the reference sequences in updated-HOMD 13.2 based on the rational that 1) they represent the highest quality, best curated full 16S rRNA gene sequences from oral bacteria 2) reads from a sample are most likely derived from species belonging to the same environment from which the sample was taken. Indeed, the vast majority of reads in this study (~93%) matched reference sequences in updated-HOMD 13.2. Surprisingly, neither of the two previous studies (16, 17) used HOMD as a reference in their analysis, which probably explains why one of them reported classification of only 16.7% of the reads (16).
Next in priority was the extended set of the HOMD 16S rRNA sequences which contained additional collection of oral taxa with less reliable, partial sequences that have, nevertheless, proved valuable and been previously used for classification of short clone sequences (27). The purpose of including this set was to increase the assignment rate of reads to known human, oral taxa. However, an additional effort was carried out to clear it first of potential chimera as well as sequences already present in the HOMD 13.2, resulting in a more reliable reference sequence set (trusted-HOMDEXT). The third reference set exploited by the algorithm, modified-GGG, comprises reference sequences of both human oral and non-oral taxa as well as a number of well-characterized, environmental species/phylotypes, thus allowing classification of reads from non-oral taxa possibly present in the samples. Including trusted-HOMDEXT and modified-GGG increased the proportion of reads that could be successfully classified to 96%, which is very close to that reported for classification of short clone sequences, using a comparable algorithm (27). An additional search of non-rare OTU representatives against NCBI also returned top hits, mostly sequences of oral clones not included in HOMD. These shorter clone oral sequences in NCBI thus served as good attractor sequences for relevant taxa, which reflect the need to regularly update HOMD to include such sequences and, in turn, minimize the need for the slow, system-demanding NCBI search.
In addition to the use of multiple reference databases that are prioritized by relevance and sequence quality, a major strength of the algorithm described here is that reads are individually matched to the reference sequences, which ensures most accurate assignment for each read. This is not the case with OTU-based algorithms that assign taxonomy to representative OTU sequences since reads called into an OTU using a certain sequence identity cutoff may not belong to a single species. This is especially true for those species with sequence similarities greater than the cutoff used. For example, in the genus Streptococcus, many species have greater than 99% sequence similarity to each other even based on the full-length 16S rRNA. Thus, a cutoff of 98% identity will lump multiple species of this genus together in an OTU. One common option is to use a Bayesian classifier to obtain consensus OTU taxonomy labels, but this usually reduces the taxonomic resolution to the genus, if not higher level. Our algorithm scarifies the speed for higher accuracy by searching individual reads against the most relevant reference sequences using the BLASTN program, which accounts for both sequence percentage identity and alignment length. The slower speed however is being quickly caught up by the availability of lower cost and higher performance, multi-core CPU computing resource, such as the cloud computing platform. Thus, the read-by-read, reference-based approach should be more commonly adopted for studying microbial communities with good quality reference source.
Five bacterial phyla – namely Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Actinobacteria – have been consistently detected in tissue OSCC samples, obviously because they are the most abundant. In this study, additional six phyla were identified, with G02 reported for the first time within OSCC tissues. Firmicutes was not the most abundant phylum, while Bacteroidetes and Fusobacteria formed substantial proportions, which seems to be the characteristic of both cancerous and normal tissues of patients with OSCC (14, 17). Seventy-nine genera were identified which is double the highest number previously reported (13). Of the genera detected at high abundance in this report, Prevotella, Fusobacterium, and Leptotrichia have previously been shown to be characteristically more abundant in samples from OSCC patients compared to those from healthy controls (17). While Streptococcus was among the most abundant genera, it accounted for less than 10% of the reads on average, which is comparable to findings reported by Schmidt et al. (17) and Bebek et al. (14). In contrast, Pushalkar et al. (13), found it to represent around 50% of the sequences in both tumor and non-tumor tissues. Such a considerable variation may be explained by PCR and cloning biases, or may be due to targeting different regions of the 16S rRNA gene.
A total of 228 species-level taxa were identified in this study, which is the highest diversity reported for bacteria within OSCC tissues. Using culture methods, Hooper et al. (10) detected 80 viable species within OSCC tissues. They later, employing classical 16S rRNA gene clonal analysis, identified additional 28 species in the same samples (11), bringing diversity to 108 species, of which 38 species were identified in this study. The results described by Pushalkar et al. (13), however, are probably the most comparable to the findings in this report, since they used HOMD for identification of their sequences. Indeed, 60 out of 80 species/phylotypes identified in that study were also detected in this study. Of course, much more species/phylotypes were identified here because of the higher sequencing throughput offered by pyrosequencing compared to the classical sequencing.
The vast majority (~95%) of the species/phylotypes identified in the OSCC samples represented oral taxa. While these are probably commensals adapting to the tumor tissue environment, the possibility that a few of them may contribute to the development with OSCC or modify its clinical course cannot be excluded. The role of highly abundant species, particularly those with pathogenic potential such as P. oris, A. segnis, and Fusobacterium spp., should be explored further, probably testing them against oral epithelium in vitro. In addition to oral taxa, a number of human, non-oral taxa were also identified, some of which are rarely detected in the oral cavity, such as B. fragilis. The latter species may, in fact, be of relevance to the development of OSCC since it is linked in the literature to colon cancer. Actually, one proteomic work identified six proteins from this species in saliva of OSCC patients (37). This, therefore, warrants further investigation.
In conclusion, we describe a robust algorithm that assigns individual NGS reads to species level by searching against multiple sets of high-quality, 16S rRNA reference sequences. The assignment is based on the best hits of single reads to the reference based on both sequence identity and alignment length. For biologically sensible taxonomy assignment, the algorithm gives priority to the taxonomy information provided by the highest quality, most relevant reference set which, in the case of oral samples, is the HOMD set. Appling the algorithm to a dataset from three OSCC samples resulted in unambiguous classification of the majority of the reads to the species level. The number of bacterial species-level taxa detected is the highest reported so far for OSCC tissues, with a number of species being reported for the first time in oral samples. However, the biological significance of specific taxa in OSCC was not the focus of this report and it remains to be evaluated in large-scale, controlled studies.
Supplementary Material
Acknowledgements
The study was funded by the Substance Abuse Research Center (SARC) at Jazan University, Saudi Arabia (grant no. 1010/2010).
Conflict of interest and funding
The authors declare that they have no competing interests.
References
- 1.Meurman JH. Oral microbiota and cancer. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5195. 5195, doi: http://dx.doi.org/10.3402/jom.v2i0.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter S, Beglinger C. Helicobacter pylori and gastric cancer: the causal relationship. Digestion. 2007;75:25–35. doi: 10.1159/000101564. [DOI] [PubMed] [Google Scholar]
- 3.Markowska J, Fischer N, Markowski M, Nalewaj J. The role of Chlamydia trachomatis infection in the development of cervical neoplasia and carcinoma. Med Wieku Rozwoj. 2005;9:83–6. [PubMed] [Google Scholar]
- 4.Nagaraja V, Eslick GD. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther. 2014;39:745–50. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 5.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–6. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 6.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lax AJ. Opinion: bacterial toxins and cancer – a case to answer? Nat Rev Microbiol. 2005;3:343–9. doi: 10.1038/nrmicro1130. [DOI] [PubMed] [Google Scholar]
- 8.Marttila E, Uittamo J, Rusanen P, Lindqvist C, Salaspuro M, Rautemaa R. Acetaldehyde production and microbial colonization in oral squamous cell carcinoma and oral lichenoid disease. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;116:61–8. doi: 10.1016/j.oooo.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Nagy KN, Sonkodi I, Szoke I, Nagy E, Newman HN. The microflora associated with human oral carcinomas. Oral Oncol. 1998;34:304–8. [PubMed] [Google Scholar]
- 10.Hooper SJ, Crean SJ, Lewis MA, Spratt DA, Wade WG, Wilson MJ. Viable bacteria present within oral squamous cell carcinoma tissue. J Clin Microbiol. 2006;44:1719–25. doi: 10.1128/JCM.44.5.1719-1725.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper SJ, Crean SJ, Fardy MJ, Lewis MA, Spratt DA, Wade WG, et al. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J Med Microbiol. 2007;56:1651–9. doi: 10.1099/jmm.0.46918-0. [DOI] [PubMed] [Google Scholar]
- 12.Mager DL, Haffajee AD, Devlin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: a descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. J Transl Med. 2005;3:27. doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pushalkar S, Ji X, Li Y, Estilo C, Yegnanarayana R, Singh B, et al. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. doi: 10.1186/1471-2180-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bebek G, Bennett KL, Funchain P, Campbell R, Seth R, Scharpf J, et al. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum Mol Genet. 2012;21:1557–65. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siqueira JF, Jr., Fouad AF, Rocas IN. Pyrosequencing as a tool for better understanding of human microbiomes. J Oral Microbiol. 2012;4 doi: 10.3402/jom.v4i0.10743. 10743, doi: http://dx.doi.org/10.3402/jom.v4i0.10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushalkar S, Mane SP, Ji X, Li Y, Evans C, Crasta OR, et al. Microbial diversity in saliva of oral squamous cell carcinoma. FEMS Immunol Med Microbiol. 2011;61:269–77. doi: 10.1111/j.1574-695X.2010.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz EL, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9:e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fettweis JM, Serrano MG, Sheth NU, Mayer CM, Glascock AL, Brooks JP, et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2013;13(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasher AT, Al-Hebshi NN, Al-Moayad EE, Suleiman AM. Viral infection and oral habits as risk factors for oral squamous cell carcinoma in Yemen: a case–control study. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;118:566–72. doi: 10.1016/j.oooo.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–70. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–9. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PLoS One. 2013;8:e71227. doi: 10.1371/journal.pone.0071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49:1464–74. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camanocha A, Dewhirst FE. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol. 2014;6 doi: 10.3402/jom.v6.25468. 25468, doi: http://dx.doi.org/10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–74. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolivar I, Whiteson K, Stadelmann B, Baratti-Mayer D, Gizard Y, Mombelli A, et al. Bacterial diversity in oral samples of children in Niger with acute noma, acute necrotizing gingivitis, and healthy controls. PLoS Negl Trop Dis. 2012;6:e1556. doi: 10.1371/journal.pntd.0001556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeshita T, Nakano Y, Kumagai T, Yasui M, Kamio N, Shibata Y, et al. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J. 2009;3:65–78. doi: 10.1038/ismej.2008.91. [DOI] [PubMed] [Google Scholar]
- 36.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie H, Onsongo G, Popko J, de Jong EP, Cao J, Carlis JV, et al. Proteomics analysis of cells in whole saliva from oral cancer patients via value-added three-dimensional peptide fractionation and tandem mass spectrometry. Mol Cell Proteomics. 2008;7:486–98. doi: 10.1074/mcp.M700146-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.