Abstract
The effects of the direct interaction between hybridization and speciation—two major contrasting evolutionary processes—are poorly understood. We present here the evolutionary history of the Galápagos marine iguana (Amblyrhynchus cristatus) and reveal a case of incipient within-island speciation, which is paralleled by between-island hybridization. In-depth genome-wide analyses suggest that Amblyrhynchus diverged from its sister group, the Galápagos land iguanas, around 4.5 million years ago (Ma), but divergence among extant populations is exceedingly young (less than 50 000 years). Despite Amblyrhynchus appearing as a single long-branch species phylogenetically, we find strong population structure between islands, and one case of incipient speciation of sister lineages within the same island—ostensibly initiated by volcanic events. Hybridization between both lineages is exceedingly rare, yet frequent hybridization with migrants from nearby islands is evident. The contemporary snapshot provided by highly variable markers indicates that speciation events may have occurred throughout the evolutionary history of marine iguanas, though these events are not visible in the deeper phylogenetic trees. We hypothesize that the observed interplay of speciation and hybridization might be a mechanism by which local adaptations, generated by incipient speciation, can be absorbed into a common gene pool, thereby enhancing the evolutionary potential of the species as a whole.
Keywords: restriction site-associated DNA (RAD) sequencing, single-nucleotide polymorphisms, El Niño, volcanism, introgressive hybridization, morphometrics
1. Introduction
Processes of population differentiation on island systems provided the cornerstone for the development of Darwin's and Wallace's evolutionary theory [1]. If located far from the mainland, islands are rarely colonized de novo, and typically host only a limited number of clades which have often diversified across a system of spatially proximate but independent islands. Therefore, these systems can be seen as evolutionary laboratories, and provide a more simplified framework to study evolutionary processes than mainland settings. Prime examples of island-based evolutionary research include well-known adaptive radiations [2–4], Mayr's classical work on allopatric species formation [5] and compelling accounts of within-island speciation [6–8]. However, island systems do not only provide useful settings to study diversification processes leading to speciation—they also reveal insights into the processes which counteract speciation, such as hybridization. It is now broadly accepted that hybridization can be as complex a process as speciation and is a major force influencing the evolution of species [9,10].
The Galápagos, a remote oceanic archipelago of pure volcanic origin 1000 km west of the South American mainland, provides an ideal setting to study processes of evolutionary diversification and hybridization. It consists of 20 islands ranging from 1.6 to 4600 km2 in size, plus numerous smaller islets, with ages of emergence dating back 0.06–4.0 Myr [11]. This archipelago has been colonized by many taxa, progressively from older to younger islands, with subsequent speciation, but also hybridization and gene flow between islands [12]. The various effects of hybridization have been documented in Darwin's finches, which represent one of the best-studied adaptive radiations worldwide [13]. While initial speciation following colonization of the archipelago occurred on distinct islands, secondary contact between finch species has resulted in diverse outcomes [14]. On Daphne Island, it was the introgressive hybridization between Geospiza fortis and G. scandens—rather than conspecific gene flow with immigrants from other islands—that increased the genetic and morphological variation of resident populations and enhanced their evolutionary potential, enabling the species to more rapidly react to environmental changes [14]. In other cases, hybridization resulted in despeciation of sister species [15], or the complete disappearance of a species, as was the case for the large tree finch on Floreana island [16]. Further examples are available from the well-studied Galápagos giant tortoises, where lineage fusion through introgressive hybridization was recently revealed on the largest island of the archipelago, Isabela. Here, two morphologically and genetically distinct evolutionary lineages colonized the island at different times, coexisted as distinct entities for a period, and then merged into one lineage [17]. Thus, it seems that hybridization as an evolutionary process continues to offer new avenues for evolutionary biologists to explore. In this work, we investigate the evolutionary history of Galápagos marine iguanas (Amblyrhynchus cristatus) and reveal a remarkable situation whereby within-island speciation is paralleled by simultaneous between-island hybridization. In this system, hybridization masks incipient speciation, leading to far-reaching consequences for the interpretation of speciation events based on phylogenetic patterns.
Marine iguanas rank as one of the most remarkable organisms of the Galápagos. Unique among lizards worldwide, they alone have adapted to the marine environment by feeding exclusively on algae in the tidal and subtidal zones, though reproduction is purely terrestrial [18,19]. Being strong swimmers, these large and highly mobile animals have colonized all major and minor islands of the archipelago [20,21]. Amblyrhynchus is a monospecific ancient lineage forming an archipelago-endemic clade with the three species of Galápagos land iguanas (genus Conolophus) [22]. Although only a single species of Amblyrhynchus is recognized, high levels of genetic distinctiveness characterize most of its current island populations [20]. Previous work on a limited number of specimens [20] even indicated the existence of two genetically distinct Amblyrhynchus populations in the northeast (Punta Pitt—PP) and southwest (Lobería—LO; figure 3c) areas of San Cristóbal island, but further information on their distribution and evolutionary history was lacking. San Cristobal is thought to be one of the oldest of the current Galápagos Islands, having emerged between 2.4 and 4.0 Ma [11], and measuring only 550 km2 in surface area, with 140 km of shoreline.
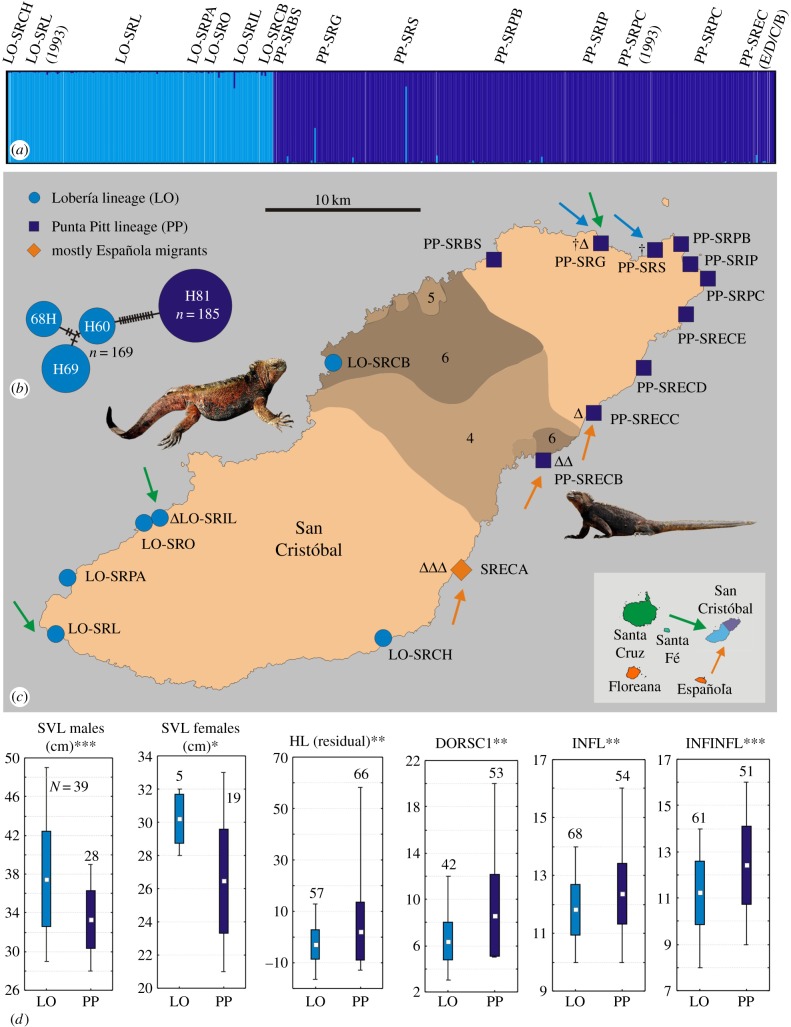
Figure 3.
Genetic and morphological differentiation of LO and PP lineages on San Cristóbal Island. LO-SRL and PP-SRPC refer to the original Lobería and Punta Pitt localities, photos show adult LO and PP males. (a) Assignment of 454 individuals based on 18 microsatellite loci, after exclusion of inter-island hybrids and migrants. Abbreviations show sampling locations and 1993 marks specimens sampled in that year. (b) Haplotype network of control region sequences (mtDNA) for LO and PP specimens. (c) Map of sampling localities; arrows indicate migrants/hybrids from Santa Cruz (green), Española (orange) and Lobería (blue); dagger symbols denote locations of within-island hybrids between PP and LO; triangles denote locations of inter-island hybrids. Population SRECA contains Española migrants/hybrids only. Shaded areas mark lava groups 4–6 aged less than 0.1 Ma [40]. (c) Mean, standard deviation and range of morphological variables differing between LO and PP. ***p < 0.001, **p < 0.01, *p < 0.05; sample sizes above each plot, details and abbreviations in Results and electronic supplementary material.
To investigate the contradictory pattern of shallow phylogenetic divergence [21] versus strong genetic population structure [20] in marine iguanas, we first reconstruct a temporal framework for their evolution. We estimate the age of the Conolophus–Amblyrhynchus split from an iguanine time-tree based on protein-coding nuclear genes and multiple temporal calibrations across squamates [23]. As these genes are not variable within Amblyrhynchus, we additionally use mitochondrial DNA (mtDNA), as well as nuclear DNA (nucDNA) from genome-wide restriction site-associated DNA sequencing (RADSeq), to infer the age of marine iguana lineages. On the population level, we derive genetic clusters from an archipelago-wide analysis of microsatellite loci and reconstruct phylogenetic relationships between these clusters based on nucDNA. Finally, we search for significant morphometric differences between the two units on San Cristóbal and investigate the geological past of this island. We find PP and LO to be reproductively isolated and morphologically differentiated sister lineages. As such, the San Cristóbal system represents an exceptional case of within-island divergence of a large and mobile lizard, probably initiated by recurrent volcanism. Hybridization of PP and LO with populations from other islands offers further insights into how marine iguanas—an apparently monospecific lineage—integrate processes of diversification and local adaptation into a common evolutionary gene pool. This mechanism may enhance the evolutionary potential of the species, and enable them to withstand severe climatic oscillations and successfully occupy diverse habitats along the entire Galápagos archipelago.
2. Material and methods
(a). Sampling
During 2011–2014, the majority of the coastline of San Cristóbal was surveyed for marine iguana colonies. Blood samples were obtained from 460 specimens at 17 sites spaced at maximum distances of roughly 10 km, except in the southeast where no iguanas were located along a major part of the coast. An additional 53 samples were obtained from previous fieldwork in 1993.
(b). Molecular genetic analyses
Seven different molecular datasets (A–H) were assembled for phylogenetic and population genetic analysis. For detailed laboratory and phylogenetic analysis protocols, see the electronic supplementary material, in which tables S1 and S2 provide an overview of samples used in each dataset.
(i). Squamate time-tree based on nuclear gene sequences
To identify the closest relative of the Galápagos iguanas and date their origin on the archipelago, we sequenced the RAG1, BDNF, R35 and NKTR genes for six focal species, and combined them with 72 squamates [23] in a concatenated alignment of 3000 bp. Phylogenetic analysis was conducted by partitioned Bayesian inference (BI) with MrBayes v. 3.2 [24]. Divergence times were estimated using Beast v. 1.7.2 [25] with 18 time-constraints across squamates [23].
(ii). Time tree of Galápagos iguanas based on mitochondrial DNA
A representative selection of the three main haplotype lineages within Amblyrhynchus [20], including PP and LO, and all species of Conolophus, were sequenced for 5557 bp from seven mitochondrial genes. Analyses were performed as for dataset A, with a secondary time constraint for the Amblyrhynchus–Conolophus split based on the estimate for this split obtained from analysis A.
(iii). Mitochondrial differentiation and phylogeography of marine iguanas
Complete mitochondrial control region sequences of 1181 bp in length were sequenced from 310 marine iguanas from San Cristóbal and 34 previously unused samples from various small islands (electronic supplementary material, table S2). These were added to existing data [20] to give a total of 1491 sequences to reconstruct a haplotype network (electronic supplementary material, figure S2) and perform mitochondrial-based assignment of San Cristóbal samples.
(iv). Archipelago-wide microsatellite loci genotyping of marine iguanas
Alleles of newly genotyped samples were scored with Genemarker (v. 1.95; Applied Biosystems) and added to an existing dataset of 12 microsatellite loci [20], resulting in an available pool of almost 1500 genotyped samples from all major islands. To avoid overrepresentation of populations [26], the dataset was standardized by random pruning to approximately 50 samples per island, excluding islands with fewer than 20 samples. For San Cristóbal, 124 samples were used, with 50 each for LO and PP, plus 24 specimens from the previously unsampled East coast. Altogether 614 individuals from 11 islands (electronic supplementary material, table S2) were included for model-based Bayesian clustering analysis [27] to infer archipelago-wide population structure.
(v). Microsatellite loci genotyping of San Cristóbal marine iguanas
The 513 available samples from San Cristóbal were scored at 18 microsatellite loci [28] and any resampled animals or individuals with more than 6% missing data were removed (n = 39). In order to identify occasional migrants from other islands, whose original population are not represented in the dataset, we used assignment tests [29] in Geneclass [30]. Prior to population structure analysis, 20 individuals who did not assign with a probability of more than 80% to either of the San Cristóbal populations, or had a mitochondrial haplotype associated with another island, were removed. We inferred the fine-scale structure of populations on San Cristobal from the remaining 454 samples. Demographic history was assessed using Bottleneck [31] and MsVar v. 1.3 [32] with various priors (electronic supplementary material, tables S5 and S6). Evolution under gene flow versus drift was tested with 2mod [33].
(vi). Genome-wide nuclear DNA analysis by restriction site associated DNA sequencing
Samples of eight outgroups and 33 marine iguana individuals spanning all major islands, including four LO and five PP samples (electronic supplementary material, table S2), were subjected to RADSeq by Floragenex [34]. Genomic DNA was digested with SbfI, and libraries were sequenced in two lanes of an Illumina HiSeq 2000 platform using single-end 90 bp chemistry. A mean of 4 981 040 ± 1 195 064 reads per sample was obtained and processed with PyRad v. 2.15 [35]. Base calls with a Phred quality score of less than 33 were coded as N. Sorted reads passing the filter (4 556 898 ± 1 086 834 reads per sample) were aligned into within-sample clusters using a similarity threshold of 0.95 (average 50.7 ± 199.6 reads per cluster). We retained clusters with more than 20 reads coverage, less than twice the standard deviation of coverage depth, less than five undetermined sites, less than 5 heterozygous sites and with two or fewer alleles. Heterozygous sites were coded using ambiguity codes and consensus sequences aligned across samples using the same similarity threshold. Loci with identical polymorphic sites in more than three samples were excluded as potential paralogues. A sparse alignment containing loci represented in more than five samples was retained (electronic supplementary material, figure S5). We conducted maximum-likelihood phylogenetic inference with a GTR + Γ model in RAxML v. 8 [36] with 100 rapid bootstrap replicates and calculated a time-calibrated phylogeny in Beast v. 2.0 [37], using calibrations as in dataset B.
(vii). Timetree based on restriction site-associated DNA sequencing data
We dated splits within Amblyrhynchus with a four-taxon subset of dataset F, containing one land iguana outgroup and three marine iguana specimens, representing one of the deepest splits within marine iguanas, and the PP/LO split. After exclusion of missing/ambiguous sites, the final matrix contained 1 793 845 sites, which we analysed in Beast v. 2.0 under a coalescent tree prior (constant growth), time-calibrating the root according to analysis of dataset A at 4.6 Ma (normal prior, standard deviation 0.3) and with a GTR substitution model.
(viii). Single-nucleotide polymorphism-based species tree analysis of marine iguanas
To obtain a more detailed picture of marine iguana diversification, we repeated the PyRad procedure for a subset of data including only the 33 ingroup samples. Settings were identical, except for the last step in which we retained a strict matrix with 6893 loci, including 579 304 bp for which data were available for all samples. The bi-allelic genotypes for each individual were used to identify single-nucleotide polymorphisms (SNPs) and export them with Adegenet for R [38]. An SNP-based species tree of marine iguana populations using the multispecies coalescent method [39] was inferred using the Snapp algorithm in Beast v. 2.0, grouping samples according to dataset E.
(c). Analysis of morphological characters
Measurements and scale counts were taken from a total of 143 microsatellite-genotyped marine iguanas from San Cristóbal to simply demonstrate morphological differences between diverged lineages. With the exception of body size, none of these morphological characters are currently known to be directly affected by environmental conditions. The following were measured in the field: snout–vent length (SVL) and total length to the nearest 10 mm; width, length and height of the head (HW, HL and HH), and length of fourth toe (TOEL) to the nearest 0.1 mm; and weight to the nearest 0.01 kg. Scale counts were performed on digital photos taken in the field. For a complete list of counts, see electronic supplementary material; values are reported here for infralabials (INFL), series of scales below INFL (infra-infralabials, INFINF) and number of dorsal crest spines (DORSC1) in anteriormost part of dorsal crest, and in addition we counted lamellae under third and fourth toe (LAM3T and LAM4T), INFL, supralabials and supra-supralabials. Multivariate analyses of variance and Mann–Whitney U-tests were performed in Statistica v. 7.1 (StatSoft Inc.).
3. Results
In a phylogeny based on four single-copy protein-coding nuclear genes from 78 squamate species covering all iguanine genera [23], Amblyrhynchus and Conolophus formed a clade diverging from Ctenosaura around 8.25 Ma (95% credibility intervals, 5.85–11.06 Ma), and diverging from each other at 4.52 (CI 2.76–6.67) Ma (figure 1a).
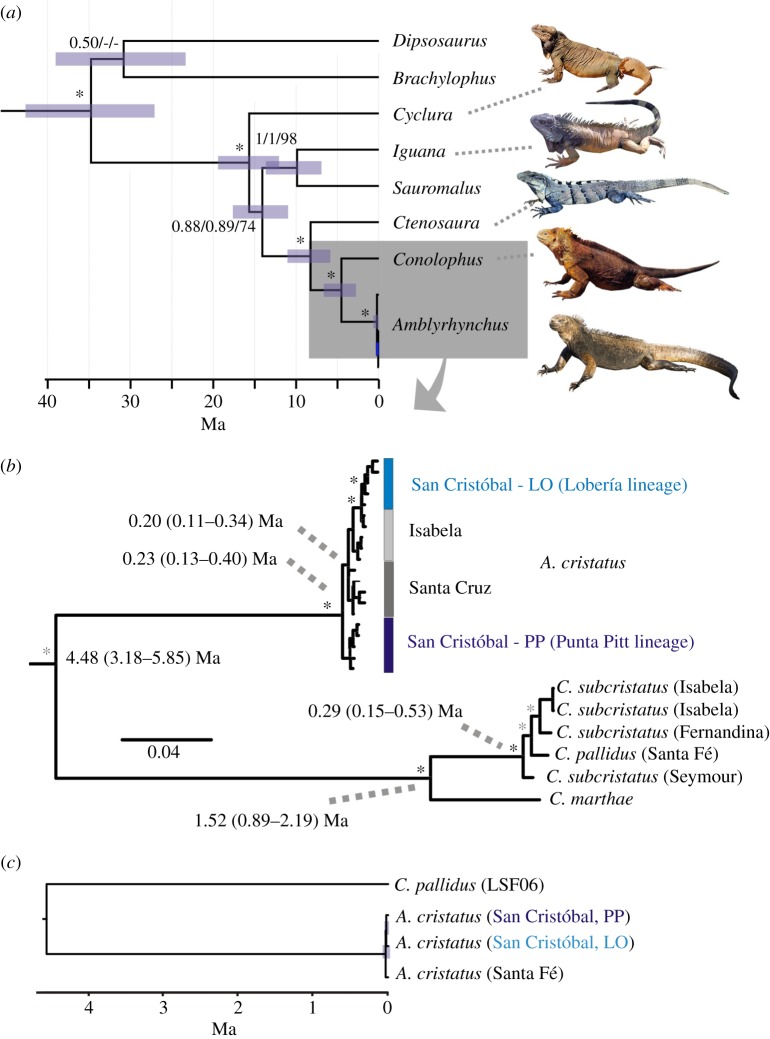
Figure 1.
Temporal framework of iguana evolution on the Galápagos Islands. (a) Partial timetree based on four nuclear genes (3000 bp) time-calibrated using multiple time constraints applied to a total dataset of 78 squamates (full tree in electronic supplementary material, figure S1). Numbers at nodes indicate support from partitioned BI analyses (posterior probability values; PP) and maximum-parsimony (MP) bootstrapping (bootstrapping values; BS); asterisks indicate maximum support. Bars are 95% credibility intervals of time estimates. (b) Maximum credibility tree from a partitioned BI analysis of 5557 bp of mtDNA. Black asterisks indicate concordant maximum support from a partitioned BI analysis, timetree analysis and MP bootstrap analysis. Small grey asterisks indicate high support (PP > 0.94; BS > 70%) from at least two of these analyses. Time estimates and 95% credibility intervals from a timetree analysis are given at selected nodes. (c) Timetree based on a complete matrix of 1 793 845 nucDNA sequences obtained by RADSeq of three Galápagos marine iguanas (selected to represent the deepest splits within the species) and one land iguana (Conolophus pallidus), showing the extremely shallow divergences within Amblyrhynchus.
Further phylogenetic analysis of 5557 bp of mtDNA corroborated reciprocal monophyly of Conolophus and Amblyrhynchus, but revealed differences in their diversification patterns. In land iguanas, speciation resulted in the evolutionary branching of Conolophus marthae around 1.52 (CI 0.89–2.19) Ma, followed by further diversification around 0.29 (CI 0.15–0.53) Ma. By contrast, marine iguanas form a single long branch leading to a very recent and phylogenetically unresolved divergence of major lineages around 0.23 (CI 0.13–0.40) Ma (figure 1b). Genome-wide analysis of 1800 kb of nucDNA derived from RADSeq suggests that this diversification is even younger, occurring at 0.03 (CI 0.02–0.06) Ma (figure 1c).
Analysis of 12 nuclear microsatellite loci revealed at least 10 major genetically distinct population clusters of Amblyrhynchus across the archipelago, and typically identified one cluster per island (figure 2a). One striking exception was San Cristóbal, where PP and LO clusters represented genetically distinct lineages. Phylogenetic relationships between major genetic clusters, reconstructed from nucDNA-derived SNPs, provided strong support for a northern clade including populations from Pinta, Marchena and Genovesa, as well as a San Cristóbal clade containing the PP and LO populations (figure 2b; electronic supplementary material, figure S6).
Figure 2.
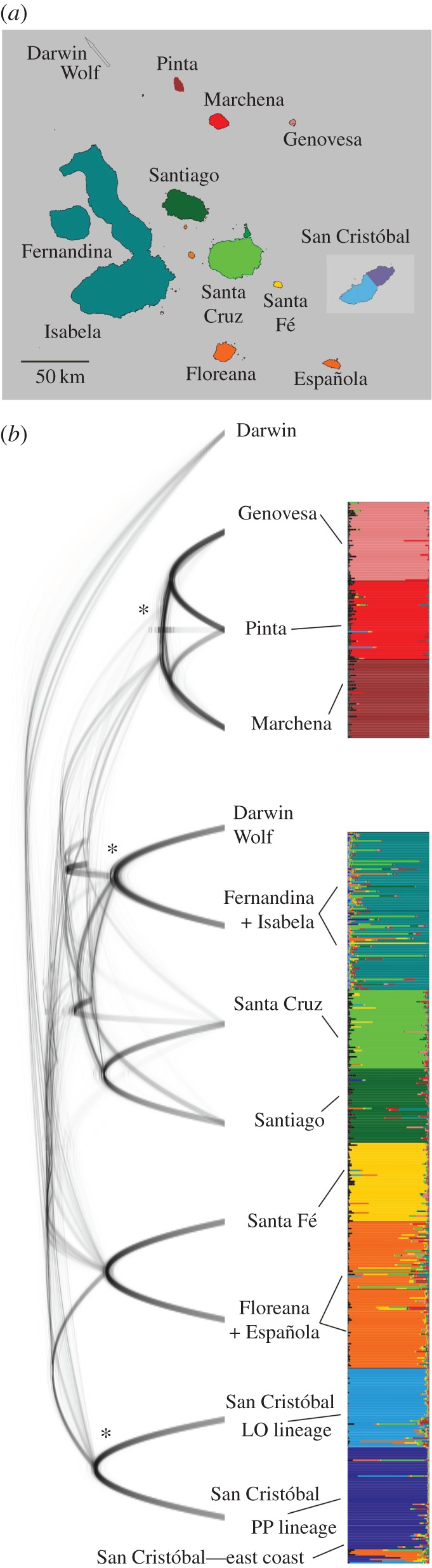
Marine iguana population clusters and phylogenetic relationships. (a) Map of the Galápagos archipelago with major islands colour-coded according to their marine iguana population cluster assignment inferred from structure analysis of 614 individuals genotyped for 12 microsatellite loci (vertical panel in (b)). (b) Species tree cloudogram based on an analysis of 6257 RADSeq-derived SNPs in 33 marine iguanas from across the archipelago, including both San Cristóbal lineages. The graph shows the posterior distribution of consensus trees. Asterisks mark nodes with posterior probability = 1.0 (all other nodes less than 0.9). Specimens were grouped according to population assignment based on structure analysis.
Spatially fine-scale genetic analysis of 454 individuals from 17 sites on San Cristóbal, using 18 microsatellite loci, combined with mtDNA D-loop haplotype assignment, suggested generalized genetic differentiation between PP and LO lineages—a congruence which usually characterizes reproductively isolated species (figure 3). Island-wide, 20 individuals were identified as either migrants from other islands or hybrids (figure 3; electronic supplementary material, table S3). Genetic assignment identified migrants as from either Santa Cruz or Española islands. On the east coast, an entire colony of iguanas with genetic signatures of Española island was identified (site SRECA in figure 3; electronic supplementary material, figure S2). Individuals of the LO and PP lineages hybridized more frequently with these animals or with migrants from Santa Cruz (in total eight occurrences) than with each other, as evidenced by just two PP/LO hybrids in almost 500 analysed animals.
Demographic modelling, based on 18 microsatellite loci, suggested that both PP and LO have experienced dramatic reductions in their effective population sizes approximately 1800–3000 years ago (electronic supplementary material, tables S4 and S6, and figure S3), largely concordant with the most recent lava formations on this island (figure 3c).
Despite their young evolutionary age, we found morphological differences between PP and LO (details in electronic supplementary material, tables S9–S18 and figure S7). PP specimens were on average smaller (SVL in adult males and females: Mann–Whitney U-test; n = 67, p = 0.0006; and n = 24, p = 0.015), had more INFL, INFINFL and spines in the first portion of the dorsal crest (n = 122, p = 0.002; n = 112, p = 0.0002; n = 95, p = 0.0017) as well as relatively longer heads (residuals of head length: n = 81, p = 0.0053) when compared with those of the LO cluster.
4. Discussion
(a). Evolutionary age of Galápagos iguanas coincides with the subaerial age of present islands
Our phylogeny of protein-coding nuclear genes (figure 1) confirms a sister group relationship between Galápagos iguanas and the Central American Ctenosaura [41]. Amblyrhynchus and Conolophus diverged around 4.5 Ma (figure 1b), whereas previous estimates based solely on mtDNA [22] suggested a much older divergence of around 10 Ma on the now-sunken islands of the archipelago. Our results reconcile Galápagos iguana divergence with the geological age of the oldest extant islands (Española and San Cristóbal; figure 1b) and are consistent with similar estimates for other Galápagos fauna [12], including giant tortoises (3–4 Ma [42]), lava lizards (2.8 Ma [43]) and Darwin's finches (2.0–2.3 Ma [44]); though, conversely, the radiation of leaf-toed geckos may have occurred far earlier (13.2 Ma [45]).
Marine iguanas existed as a monospecific lineage for several million years, only diverging as recently as the Late Pleistocene (less than or equal to 0.23 Ma according to mtDNA) or even later (0.03 Ma based on nucDNA SNP loci). Possible male-biased dispersal [21] and the earlier coalescence time of mtDNA [46] might account for these between-marker differences. Regardless of the discrepancy, divergences within Amblyrhynchus are remarkably recent, especially compared with Conolophus. This could imply that marine iguanas experienced a massive archipelago-wide decline in the Pleistocene. Despite records of catastrophic crashes of Amblyrhynchus populations through El Niño events [47], the effects vary greatly between islands [48], making an archipelago-wide extinction rather unlikely. Therefore, an alternative scenario, whereby regular dispersal facilitates gene flow between islands, is a more probable explanation for this weak phylogeographic structure; a similar situation is found for another semi-aquatic Galápagos organism, the Galápagos sea lion [49].
(b). Incipient speciation of marine iguanas on San Cristóbal
The sharp genetic differentiation observed between the PP and LO lineages on San Cristóbal is in stark contrast to the overall pattern of a single genetic cluster per island (figure 2a). From a mitochondrial perspective, PP and LO do not form a monophyletic group (figure 1b). Conversely, phylogenetic analyses based on genome-wide SNP loci unambiguously reconstruct them as sister lineages (figure 2b; electronic supplementary material, figure S6), indicating that a considerable portion of their current genomic differentiation must have occurred on San Cristóbal. This mismatch could be due to the mitochondrial genome reflecting a complex history of introgressive gene flow in the species' past [20,21].
Typically, speciation within the geographical boundaries of one island occurs when gene flow is prevented by a large island size, low vagility of a species, or both [50]. The area effect is paramount, and therefore complete within-island speciation is exceedingly rare on small islands [1]. In adaptive island radiations of Anolis, for example, which comprise small- to medium-sized lizards, no in situ diversification was detected in Caribbean islands smaller than 3000 km2 [51]. In birds, most phylogenetic studies of species occurring on single islands rejected sister-species relationships, thereby ruling out within-island speciation [52]. Only small-sized or less mobile organisms, such as palms, provide unambiguous examples of sympatric speciation on small volcanic islands [6]. Given that marine iguanas are large and vagile reptiles, the observed incipient speciation event, occurring in less than 30 000 years on a small island, is unexpected. This process was probably facilitated by an interaction of geological and environmental factors which separated populations spatially, and, in parallel, reduced their sizes. The current geographical distribution of PP and LO (figure 3c) and their recent phylogenetic divergence match remarkably well with the distribution of contemporary volcanism on the island. Lava flows recurrently occurred in central San Cristóbal during the last 0.1 Myr [40], and the severe bottlenecks evident in both lineages around approximately 1800–3000 years ago coincide with the most recent lava formation (figure 3c).
Accordingly, speciation might have initially followed a micro-parapatric pattern, where repeated volcanic events caused geographical disruption of marine iguana habitat and isolated colonies on the northeastern and southwestern extremes of the island. Yet these events cannot fully account for their significant morphological differentiation and current lack of genetic admixture. Local adaptation and/or prezygotic mating barriers may have also contributed to the consolidation of the divergence process. Difference in body size, as observed between PP and LO, could be a sign of differential habitat adaptation. In marine iguanas, body size is under strong natural selection, and largely depends on the occurrence and abundance of preferred algae [53]. Migrants and hybrids found in sampling sites within the PP lineage (electronic supplementary material, table S3) are larger than pure PP animals, suggesting that smaller body size in PP is to some extent genetically determined.
(c). Within-island speciation in parallel with between-island hybridization
Populations of PP and LO are geographically proximate (approx. 12 km coastline between LO-SRCB and PP-SRBS; figure 3b), and PP populations show no significant isolation by distance over distances of more than 30 km (electronic supplementary material, figure S4), indicating that they could potentially migrate into the nearest LO populations. It is surprising that of the 474 individuals sampled around the islands' coastline, not one full migrant individual of either lineage was detected in the range of the other (figure 3a). Furthermore, given reports of occasional hybridization occurring even between marine and land iguanas [54], the discovery of only two unambiguous PP/LO hybrids (electronic supplementary material, table S3) is also remarkable. By contrast, between-island hybridization, evidenced by eight occurrences, was more common; a notable result, as only 10 migrants from neighbouring islands (Santa Cruz and Española) were found on San Cristóbal, making opportunities for this type of hybridization rare. Therefore, it seems that two distinct evolutionary processes are acting in parallel on San Cristóbal. Incipient within-island speciation is evident, but at the same time, introgressive hybridization with individuals from other islands prevents the completion of this process on an archipelago-wide scale. We hypothesize that these contrary processes have influenced the evolutionary history of marine iguanas on the Galápagos archipelago.
Although introgressive hybridization is now increasingly viewed as a driving force in speciation [9,10], the overall pattern observed in marine iguanas resembles more the process of despeciation, described for Darwin's finches [14], where one species is genetically absorbed into another via hybridization [15], or lineage fusion, as seen in Galápagos giant tortoises from Volcano Wolf on Isabela [17]. By contrast, the phylogeny of Galápagos land iguanas reflects ancient and fully completed speciation, at least in the case of C. marthae [55], which diverged around 1.52 Ma (figure 1b). Furthermore, Darwin's finches diversified into 14 species and subspecies within 1.6 Myr [14]. This is in clear contrast to the lack of phylogenetic bifurcation of the marine iguana branch for almost 4.5 Myr. Such a pattern would commonly be interpreted as an absence of speciation processes, an assumption contradicted in this case by the strong differentiation of island populations and the case of incipient speciation on San Cristóbal. Thus, although A. cristatus appears as a single phylogenetic species, incipient speciation events, made visible here via the contemporary snapshot provided by highly variable markers, may well have also occurred in the evolutionary past of this species.
Marine iguana populations regularly experience strong selective pressure during climatic El Niño oscillations [47,48], which vary in strength between locations and disproportionately remove larger individuals; such selection may partly explain the morphological variation among island populations, especially in terms of body size [53]. Nevertheless, marine iguanas are highly successful and occur archipelago-wide, whereas land iguanas currently occur on only four major islands [55]. The geography of the Galápagos archipelago is particularly conducive to the emergence of novel local adaptation in geographical isolation, and in a mobile species like the marine iguana, new genetic variants are rapidly assimilated into a common gene pool via introgressive hybridization. The establishment of such a gene pool, incorporating events of local adaptation and speciation, might be an important mechanism underlying the evolutionary success of marine iguanas. Insight from these processes might enhance our understanding of how a species can persist despite frequent and severe climatic oscillations, such as El Niño events, which can induce severe population crashes.
Supplementary Material
Acknowledgements
We are grateful to the Galápagos National Park authority for research permission; to S. Rea for help with paperwork; to S. Pasachnik for providing samples; to L. Unsworth, Maryuri Yépez, L. Cardas, M. D. Astudillo, L. Cruz, D. Toninho and T. Reinhardt for field assistance; to G. Jimenez for granting access to preserved specimens; to D. J. Geist for discussion; to J. C. Marshall for comments on the manuscript; to C.-K. Baillie, E. Hippauf, G. Keunecke, M. Kondermann, S. Künzel and S. Weißelberg for help with laboratory work; and to S. Herzog for morphometric measurements. This publication is contribution number 2115 of the Charles Darwin Foundation for the Galápagos Islands.
Ethics
Sampling of marine iguanas was done with permissions of the Galápagos National Park (permit numbers PC-26-12, PC-23-13 and PC-22-14).
Data accessibility
All sequences generated for this study have been deposited in GenBank under accession numbers KR350691-KR351205. Full alignments, SNP and microsatellite genotypes have been deposited in the Dryad data repository (http://dx.doi.org/10.5061/dryad.pp6bm). Further data, extended methods, results and discussion supporting this manuscript have been submitted as part of the electronic supplementary material.
Authors' contributions
S.S., A.M., F.T. and M.V. designed the study. A.M., C.G. and G.Q. collected field data. G.G. and A.C. contributed data. A.M., A.R., P.O. and M.V. analysed data. S.S., M.V. and A.M. wrote the manuscript.
Competing interests
All authors hereby declare that they have no competing interests.
Funding
This work was supported by grants from the Swiss Friends of the Galápagos, the Galápagos Conservation Trust and the National Geographic Society (grant no. GEFNE99-13).
References
- 1.Losos JB, Ricklefs RE. 2009. Adaptation and diversification on islands. Nature 457, 830–836. ( 10.1038/nature07893) [DOI] [PubMed] [Google Scholar]
- 2.Kambysellis M, Craddock E. 1997. Ecological and reproductive shifts in the diversification of the endemic Hawaiian Drosophila. In Molecular evolution and adaptive radiation (eds Givnish TJ, Sytsma KJ.), pp. 475–509. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 3.Gillespie R. 2004. Community assembly through adaptive radiation in Hawaiian spiders. Science 303, 356–359. ( 10.1126/science.1091875) [DOI] [PubMed] [Google Scholar]
- 4.Losos JB. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Oakland, CA: University of California Press. [Google Scholar]
- 5.Mayr E. 1963. Animal species and evolution. London, UK: Oxford University Press. [Google Scholar]
- 6.Savolainen V, et al. 2006. Sympatric speciation in palms on an oceanic island. Nature 441, 210–213. ( 10.1038/nature04566) [DOI] [PubMed] [Google Scholar]
- 7.Ryan PG, Bloomer P, Moloney CL, Grant TJ, Delport W. 2007. Ecological speciation in South Atlantic island finches. Science 315, 1420–1423. ( 10.1126/science.1138829) [DOI] [PubMed] [Google Scholar]
- 8.Friesen V, Smith A, Gomez-Diaz E, Bolton M, Furness R, González-Solis J, Monteiro L. 2007. Sympatric speciation by allochrony in a seabird. Proc. Natl Acad. Sci. USA 104, 18 589–18 594. ( 10.1073/pnas.0700446104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwenk K, Brede N, Streit B. 2008. Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Phil. Trans. R. Soc. B 363, 2805–2811. ( 10.1098/rstb.2008.0055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbott R, et al. 2013. Hybridization and speciation . J. Evol. Biol. 26, 229–246. ( 10.1111/j.1420-9101.2012.02599.x) [DOI] [PubMed] [Google Scholar]
- 11.Geist DJ, Snell H, Snell H, Goddard C, Kurz MD. 2014. A paleogeographic model of the Galápagos Islands and biogeographical and evolutionary implications. In The Galápagos: a natural laboratory for the earth sciences (eds Karen SH, Mittelstaedt E, d'Ozouville N, Graham DW.), pp. 145–166. New York, NY: John Wiley and Sons. [Google Scholar]
- 12.Parent CE, Caccone A, Petren K. 2008. Colonization and diversification of Galápagos terrestrial fauna: a phylogenetic and biogeographical synthesis. Phil. Trans. R. Soc. B 363, 3347–3361. ( 10.1098/rstb.2008.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant PR, Grant BR. 2008. How and why species multiply: the radiation of Darwin’s finches. Princeton, NJ: Princeton University Press. [Google Scholar]
- 14.Grant PR, Grant BR. 2014. 40 years of evolution: Darwin’s finches on Daphne Major Island. Princeton, NJ: Princeton University Press. [Google Scholar]
- 15.Grant PR, Grant BR, Markert JA, Keller LF, Petren K. 2004. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution 58, 1588–1599. ( 10.1111/j.0014-3820.2004.tb01738.x) [DOI] [PubMed] [Google Scholar]
- 16.Kleindorfer S, O'Connor JA, Dudaniec RY, Myers SA, Robertson J, Sulloway FJ. 2014. Species collapse via hybridization in Darwin's tree finches. Am. Nat. 183 325–341. ( 10.1086/674899) [DOI] [PubMed] [Google Scholar]
- 17.Garrick RC, et al. 2014. Lineage fusion in Galápagos giant tortoises. Mol. Ecol. 23, 5276–5290. ( 10.1111/mec.12919) [DOI] [PubMed] [Google Scholar]
- 18.Boersma PD. 1983. An ecological study of the Galapagos marine iguana. In Patterns of evolution in Galápagos organisms (eds Bowman RI, Berson M, Leviton AE.), pp. 157–176. San Francisco, CA: AAAS, Pacific Division. [Google Scholar]
- 19.Trillmich KG, Trillmich F. 1986. Foraging strategies of the marine iguana, Amblyrhynchus cristatus. Behav. Ecol. Sociobiol. 18, 259–266. ( 10.1007/BF00300002) [DOI] [Google Scholar]
- 20.Steinfartz S, et al. 2009. Progressive colonization and restricted gene flow shape island-dependent population structure in Galápagos marine iguanas (Amblyrhynchus cristatus). BMC Evol. Biol. 9, 297 ( 10.1186/1471-2148-9-297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rassmann K, Tautz D, Trillmich F, Gliddon C. 1997. The microevolution of the Galápagos marine iguana Amblyrhynchus cristatus assessed by nuclear and mitochondrial genetic analyses. Mol. Ecol. 6, 437–452. ( 10.1046/j.1365-294X.1997.00209.x) [DOI] [Google Scholar]
- 22.Rassmann K. 1997. Evolutionary age of the Galápagos iguanas predates the age of the present Galápagos Islands. Mol. Phylogenet. Evol. 7, 158–172. ( 10.1006/mpev.1996.0386) [DOI] [PubMed] [Google Scholar]
- 23.Townsend TM, Mulcahy DG, Noonan BP, Sites JW, Kuczynski CA, Wiens JJ, Reeder TW. 2011. Phylogeny of iguanian lizards inferred from 29 nuclear loci, and a comparison of concatenated and species-tree approaches for an ancient, rapid radiation. Mol. Phylogenet. Evol. 61, 363–380. ( 10.1016/j.ympev.2011.07.008) [DOI] [PubMed] [Google Scholar]
- 24.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. 2010. The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186, 983–995. ( 10.1534/genetics.110.118661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. ( 10.1111/j.1471-8286.2007.01758.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLeod A, Koch V, Garcia-Parra C, Trillmich F, Steinfartz S. 2012. New highly polymorphic microsatellite loci for the Galápagos marine iguana, Amblyrhynchus cristatus. Amphib. Reptil. 33, 533–536. ( 10.1163/15685381-00002854) [DOI] [Google Scholar]
- 29.Baudouin L, Lebrun P. 2001. An operational Bayesian approach for the identification of sexually reproduced cross-fertilized populations using molecular markers. Acta Hortic. 546, 81–94. [Google Scholar]
- 30.Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A. 2004. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539. ( 10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- 31.Piry S, Luikart G, Cornuet JM. 1999. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 90, 502–503. ( 10.1093/jhered/90.4.502) [DOI] [Google Scholar]
- 32.Beaumont MA. 1999. Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciofi C, Beaumontf MA, Swingland IR, Bruford MW. 1999. Genetic divergence and units for conservation in the Komodo dragon Varanus komodoensis. Proc. R. Soc. Lond. B 266, 2269–2274. ( 10.1098/rspb.1999.0918) [DOI] [Google Scholar]
- 34.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton DA. 2014. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 30, 1844–1849. ( 10.1093/bioinformatics/btu121) [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jombart T, Ahmed I. 2011. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. ( 10.1093/bioinformatics/btr521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. 2012. Inferring species trees directly from biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 29, 1917–1932. ( 10.1093/molbev/mss086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geist DJ, McBirney AR, Duncan RA. 1986. Geology and petrogenesis of lavas from San Cristobal Island, Galapagos archipelago. Geol. Soc. Am. Bull. 97, 555–566. ( 10.1130/0016-7606) [DOI] [Google Scholar]
- 41.Townsend TM, Leavitt DH, Reeder TW. 2011. Intercontinental dispersal by a microendemic burrowing reptile (Dibamidae). Proc. R. Soc. B 282, 2568–2574. ( 10.1098/rspb.2010.2598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulakakis N, Russello M, Geist D, Caccone A. 2012. Unravelling the peculiarities of island life: vicariance, dispersal and the diversification of the extinct and extant giant Galápagos tortoises. Mol. Ecol. 21, 160–173. ( 10.1111/j.1365-294X.2011.05370.x) [DOI] [PubMed] [Google Scholar]
- 43.Benavides E, Baum R, Snell HM, Snell HL, Sites JW., Jr 2009. Island biogeography of Galapagos lava lizards (Tropiduridae: Microlophus): species diversity and colonization of the archipelago. Evolution 63, 1606–1626. ( 10.1111/j.1558-5646.2009.00617.x) [DOI] [PubMed] [Google Scholar]
- 44.Sato A, Tichy H, O'hUigin C, Grant PR, Grant BR, Klein J. 2001. On the origin of Darwin's finches. Mol. Biol. Evol. 18, 299–311. ( 10.1093/oxfordjournals.molbev.a003806) [DOI] [PubMed] [Google Scholar]
- 45.Torres-Carvajal O, Barnes CW, Pozo-Andrade MJ, Tapia W, Nicholls G. 2014. Older than the islands: origin and diversification of Galápagos leaf-toed geckos (Phyllodactylidae: Phyllodactylus) by multiple colonizations. J. Biogeogr. 41, 1883–1894. ( 10.1111/jbi.12375) [DOI] [Google Scholar]
- 46.Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744. ( 10.1046/j.1365-294X.2003.02063.x) [DOI] [PubMed] [Google Scholar]
- 47.Laurie WA. 1990. Effects of the 1982–83 El Niño-Southern oscillation event on marine iguana (Amblyrhynchus cristatus Bell, 1825) populations on Galapagos. Elsev. Oceanogr. Ser. 52, 361–380. ( 10.1016/S0422-9894(08)70041-2) [DOI] [Google Scholar]
- 48.Steinfartz S, Glaberman S, Lanterbecq D, Marquez C, Rassmann K, Caccone A. 2007. Genetic impact of a severe El Nino event on Galápagos marine iguanas (Amblyrhynchus cristatus). PLoS ONE 2, e1285 ( 10.1371/journal.pone.0001285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf JB, Harrod C, Brunner S, Salazar S, Trillmich F, Tautz D. 2008. Tracing early stages of species differentiation: ecological, morphological and genetic divergence of Galápagos sea lion populations. BMC Evol. Biol. 8, 150 ( 10.1186/1471-2148-8-150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kisel Y, Barraclough TG. 2010. Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 175, 316–334. ( 10.1086/650369) [DOI] [PubMed] [Google Scholar]
- 51.Losos JB, Schluter D. 2000. Analysis of an evolutionary species–area relationship. Nature 408, 847–850. ( 10.1038/35048558) [DOI] [PubMed] [Google Scholar]
- 52.Coyne JA, Price TD. 2000. Little evidence for sympatric speciation in island birds. Evolution 54, 2166–2171. ( 10.1111/j.0014-3820.2000.tb01260.x) [DOI] [PubMed] [Google Scholar]
- 53.Wikelski M. 2005. Evolution of body size in Galapagos marine iguanas. Proc. R. Soc. B 272, 1985–1993. ( 10.1098/rspb.2005.3205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rassmann K, Trillmich F, Tautz D. 1997. Hybridization between the Galápagos land and marine iguana (Conolophus subcristatus and Amblyrhynchus cristatus) on Plaza Sur. J. Zool. 242, 729–739. ( 10.1111/j.1469-7998.1997.tb05822.x) [DOI] [Google Scholar]
- 55.Gentile G, Fabiani A, Marquez C, Snell HL, Snell HM, Tapia W, Sbordoni V. 2009. An overlooked pink species of land iguana in the Galápagos. Proc. Natl Acad. Sci. USA 106, 507–511. ( 10.1073/pnas.0806339106) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated for this study have been deposited in GenBank under accession numbers KR350691-KR351205. Full alignments, SNP and microsatellite genotypes have been deposited in the Dryad data repository (http://dx.doi.org/10.5061/dryad.pp6bm). Further data, extended methods, results and discussion supporting this manuscript have been submitted as part of the electronic supplementary material.