Abstract
Animals live in close association with microorganisms, mostly prokaryotes, living in or on them as commensals, mutualists or parasites, and profoundly affecting host fitness. Most animal–microbe studies focus on microbial community structure; for this project, allometry (scaling of animal attributes with animal size) was applied to animal–microbe relationships across a range of species spanning 12 orders of magnitude in animal mass, from nematodes to whales. Microbial abundances per individual animal were gleaned from published literature and also microscopically counted in three species. Abundance of prokaryotes/individual versus animal mass scales as a nearly linear power function (exponent = 1.07, R2 = 0.94). Combining this power function with allometry of animal abundance indicates that macrofauna have an outsized share of animal-associated microorganisms. The total number of animal-associated prokaryotes in Earth's land animals was calculated to be 1.3–1.4 × 1025 cells and the total of marine animal-associated microbes was calculated to be 8.6–9.0 × 1024 cells. Animal-associated microbes thus total 2.1–2.3 × 1025 of the approximately 1030 prokaryotes on the Earth. Microbes associated with humans comprise 3.3–3.5% of Earth's animal-associated microbes, and domestic animals harbour 14–20% of all animal-associated microbes, adding a new dimension to the scale of human impact on the biosphere. This novel allometric power function may reflect underlying mechanisms involving the transfer of energy and materials between microorganisms and their animal hosts. Microbial diversity indices of animal gut communities and gut microbial species richness for 60 mammals did not indicate significant scaling relationships with animal body mass; however, further research in this area is warranted.
Keywords: allometry, animals, microbiota, prokaryotes, symbiosis
1. Introduction
Virtually all animals associate with smaller organisms, primarily prokaryotes (bacteria and archaea) that digest complex organic substrates, fix CO2, fix N2, stimulate ontogeny, affect behaviour, compete against pathogens, synthesize growth factors or serve as prey [1–3]. Molecular analyses of microbial metagenomes have revealed diverse health-related interactions between humans and their microbiota [4]. The most common approaches for studying animal-associated microbiota, e.g. small-subunit rRNA gene-based methods and metagenomic analyses, reveal percentages of individual species; quantification of total microbial abundance, e.g. by direct microscopic counting or qPCR, is performed less frequently. Allometry is the study of how various attributes, e.g. metabolic rate, organ size, abundance, etc. scale with body size, usually expressed as body mass. This approach is usually used to compare a broad range of animal species and in many cases provides clues to underlying physical/chemical mechanisms that control this scaling [5–7]. The allometric approach has not previously been applied to animal–microbe interactions except for the more narrow case of animal parasites [8–10].
This study was, to our knowledge, undertaken as the first ever allometric study of the relationship between animal body mass (wet weight) and abundance of microbes per individual animal across a wide range of vertebrate and invertebrate species and of the animal-associated microbial abundance in the biosphere estimated from this relationship. Whitman et al. [11] conducted a planet-wide census of prokaryotes in various habitats, including the guts of humans, domestic animals and termites. Their total for all Earth prokaryotes was 4.2–6.4 × 1030 cells, which was revised by Kallmeyer et al. [12] to 9.2–31.7 × 1029 cells. Clearly, microbes living within animal habitats form only a small fraction of the Earth's total inventory of prokaryotes, yet they are vitally important to animal well-being and by extension to the functioning of the biosphere. It was hypothesized here that microbial abundance scales with animal body mass across a wide range of vertebrate and invertebrate species. The existence of a consistent allometric relationship between microbes and animals could have implications for the energetic dependence of animals on microbes and for the total abundance of animal-associated microbes. It was further hypothesized that the diversity of animal-associated microbes also scales with body mass, with larger animals harbouring a greater diversity. This hypothesis was based on well-established species–area relationships, in which species diversity within a habitat generally increases monotonically as the size of the habitat increases [13]. One might also postulate such a relationship based on the finding that herbivores generally have a greater diversity of gut microbes than carnivores and omnivores [14], and the most massive animals are herbivores [14].
2. Material and methods
(a). Animal masses and microbial counts from published literature
Most of the data used for calculating the allometric relationship between animal mass and microbial abundance were drawn from published sources (see the electronic supplementary material). For most animal species, this study focused on microbes inhabiting the gut and especially the most active fermentative organs. These organs contain approximately 107–1012 microbes per millilitre or gram of digesta in many species of insects [15]; fishes [16], birds [11,16] and mammals [11,16], and thus comprise the bulk of microbial symbionts in most animals. In some invertebrates, the bulk of the microbes are associated with other organs or tissues, e.g. the spongocoel in sponges, the trophosome in the deep sea hydrothermal vent worm Riftia pachyptila, and the epidermal and endodermal epithelium in Hydra vulgaris; microbial counts from these sites were used for this study. The microbial counts for humans, domestic animals, chickens, ducks and termites were obtained from Whitman et al. [11], but required division of the total number of microbes associated with the entire population of a species by the total abundance of that species. Microbial counts for some animals were derived from the published volumes of the gut organs and direct microscopic counts of microbes per gram or millilitre of gut contents; this approach was used for oxen [16,17], Balaenoptera acutorostrata (minke whales) [16,18,19], horses [16,17], Xestospongia muta (giant sponges) [20], dogs [16,21], guinea pigs [22], R. pachyptila (hydrothermal vent tube worm) [23], hamsters [16,22,24], Apostichopus japonicas (sea cucumbers) [25–27], mice [16,22], Hirudo medicinalis (medicinal leeches) [28], Lumbricus rubellus (earthworms) [29,30] and Euphausia superba (Antarctic krill) [31]. Microbial abundances for several insect species were published as microscopic counts per individual animal [15], and these were used directly. Fluorescent in situ hybridization was used to count bacteria associated with Hydra vulgaris (S. Fraune 2014, personal communication). Heterotrophic plate count data were used for the nematode Caenorhabditis elegans [32]; this may have underestimated the abundance, but the nematodes were raised on bacteria cultivated in the laboratory, and so the majority of these bacteria were probably cultivable.
(b). Direct microscopic counts of microbes
Live brown planaria (Dugesia tigrina) and vinegar eels (Turbatrix aceti) were obtained from Carolina Biological Supply Company. Planaria and vinegar eels were fixed in Ringers solution (6.5 g NaCl, 0.42 g KCl, 0.25 g CaCl2 and 0.2 g NaHCO3 l−1 H2O) containing 3.7% formaldehyde. Fixed animals were homogenized using a ground glass tissue homogenizer. The resulting slurry was then filtered through a 0.45 µm pore-size polycarbonate filter. Filters were then stained with 0.3% acridine orange. The filters were dried at room temperature and placed onto a microscope slide. A drop of immersion oil and a coverslip were placed onto the filter. Microbial cells were counted using epifluorescence microscopy. Live zebrafish (Danio rerio) were purchased from Walmart in Socorro, NM, and euthanized by placing them into an ice-water bath. The zebrafish were dissected and the gastrointestinal tracts were removed and weighed. The gastrointestinal tracts were homogenized and diluted 1 : 100 in Ringers solution, and the associated microbes were quantified by acridine-orange counting as described for the planaria and vinegar eels.
(c). Total animal biomass and associated microbes
Published estimates of total land and animal biomasses were combined with the allometric relationship of this study to calculate total abundances of animal-associated microbes on land, in the oceans, in humans and in domestic animals (table 1). Whittaker & Likens [33] published an estimate of the total biomass of land animals, exclusive of humans and domestic animals, in gram dry weight; dry weights were converted to wet weights assuming 60% water content. Whitman et al.'s [11] estimate for total human-associated microbes was updated to a current population of 7.1 billion people. Two different sources were used for domestic animals. Barnosky et al. (fig. 5 in [34]) calculated a modern wet weight of domestic animals (land calculation 1); Whitman et al.'s [11] estimate for microbes associated with domestic animals was also used (land calculation 2). Two different published values for marine animal biomass were used: Whittaker & Likens's [33] estimate in gram dry weight, converted to wet weight using 60% water (marine calculation 1), and Jennings et al.'s estimate in gram wet weight [35] (marine calculation 2). A low estimate of the total animal-associated microbes on the Earth was calculated by adding the lowest of the land and ocean estimates; a high estimate was calculated as the sum of the high land and marine estimates. These high and low estimates were then compared to Kallmeyer et al.'s estimate for total prokaryotes on the Earth [12]; the totals of human-associated and domestic animal-associated microbes were also used to estimate the percentages of Earth's animal-associated microbes that they comprise.
Table 1.
Calculation of the Earth's total animal biomass and abundance of animal-associated microbes. (Sources of published data are shown in brackets.)
| category | animal biomass (gram dry weight) | animal biomass (gram wet weight) | total no. animal-associated microbes |
|---|---|---|---|
| land animals | |||
| calculation 1 | |||
| all except humans and domestic animals | 1.01 × 1015 [33] | 2.53 × 1015 | 8.69 × 1024 |
| humans | 7.49 × 1023 [11] | ||
| domestic animals | 9.50 × 1014 [11] | 3.27 × 1024 | |
| total | 3.48 × 1015 [34] | 1.27 × 1025 | |
| calculation 2 | |||
| all except humans and domestic animals | 1.01 × 1015 [33] | 2.53 × 1015 | 8.69 × 1024 |
| humans | 7.49 × 1023 [11] | ||
| domestic animals | 4.26 × 1024 [11] | ||
| total | 1.37 × 1025 | ||
| marine animals | |||
| calculation 1 | |||
| total | 9.97 × 1015 [33] | 2.49 × 1015 | 8.57 × 1024 |
| calculation 2 | |||
| total | 2.62 × 1015 [35] | 9.01 × 1024 | |
| total land and marine | |||
| low estimate | 2.13 × 1025 | ||
| high estimate | 2.27 × 1025 | ||
(d). Diversity indices
Diversity indices for animal-associated microbial communities were calculated based on metagenomic data from MG-RAST [36]. Publically available metagenomes were sorted by biome and then one example for each species was selected from the microbial metagenomes of animal-associated habitats and human-associated habitats. The selected metagenomes represented the microbial communities in the portion of the gastrointestinal tract having the largest number of microbes, e.g. the mouse caecum, the human large intestine, etc. For projects with multiple treatments involving healthy and diseased animals, unusual diets, etc., a metagenome was selected that represented healthy animals. MG-RAST standardly calculates alpha diversity, which has units of number of species and is defined as the antilog of the Shannon–Wiener index. Shannon–Wiener, evenness, Simpson and Chao diversity indices [37] were calculated from the MG-RAST genus-level identifications, similarly to the diversity calculation approach used by the Human Genome Project Consortium [4]. Animal-associated microbial diversity data for 60 species of mammals in the form of species richness (total operational taxonomic units, OTUs taken from Ley et al.'s [14] electronic supplementary material) were also analysed in relation to animal mass.
3. Results
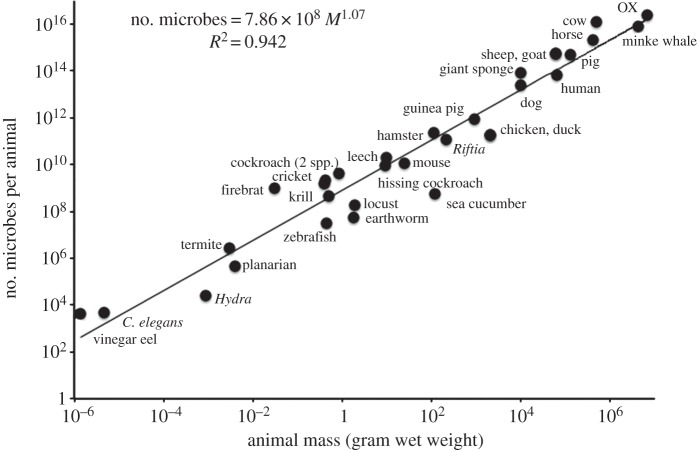
Plotting data for the abundances of microbial cells per individual animal versus masses of those animal species (wet weight) (figure 1) yielded a power function:
| 3.1 |
where M = body mass (wet weight) in grams. The coefficient of determination (R2) = 0.94 for this power function. This novel allometric relationship extends downward in body mass and complexity to include even nematodes, for example, approximately 1000 cell C. elegans, itself nearly microscopic. Plotting the data linearly yields
| 3.2 |
with R2 = 0.80. Each gram of an animal thus averages approximately 3.4 × 109 of associated prokaryotes. Converting microbial abundance to microbial biomass using 1 × 10−12 g wet weight prokaryotic cell−1, animals are typically approximately 0.34% prokaryotes by weight.
Figure 1.
Microbial counts of microorganisms per individual animal versus individual animal body mass (M, wet weight in gram), log–log plot.
Using this allometric relationship between animal masses and abundances of associated microbes, the abundances of animal-associated microbes were estimated. Beginning with land animals, the size–density relationship [38],
| 3.3 |
was combined with equation (3.1) to produce the relationship between animal mass and abundance of microbes on land:
| 3.4 |
Thus, although there are fewer large animals than small ones, the positive exponent of this power function means macrofauna have far more of the ecosystems' animal-associated microbes. Total biomass of land animals on the Earth has been estimated as 3.9 × 1015 g wet weight [11,33,34]; associated microbes should total 2.1–2.3 × 1025 cells (table 1). Animal biomass in the oceans scales with individual body mass [35] as
| 3.5 |
The product of equations (3.1) and (3.4) produces the relationship between the total number of animal-associated microbial cells in the oceans and individual animal body mass:
| 3.6 |
The scaling factor is twice that for land animals, meaning that marine macrofauna account for an even greater proportion of marine animal-associated microbes than terrestrial macrofauna do among land animals. Estimates of total marine animal biomass, 2.5 × 1015 g [33] and 2.6 × 1015 g [35], yield an associated microbial abundance of 8.6–9.0 × 1024 cells (table 1). The estimate for all of Earth's animal-associated microbes (land plus marine) is 1.9–2.3 × 1025 cells, a trifling proportion (0.00067–0.0025%) of the 9.2–31.7 × 1029 total prokaryotes on the Earth [12]. The low percentage is unsurprising, given the animals' roles as consumers and the inefficiency of energy and material transport between trophic levels.
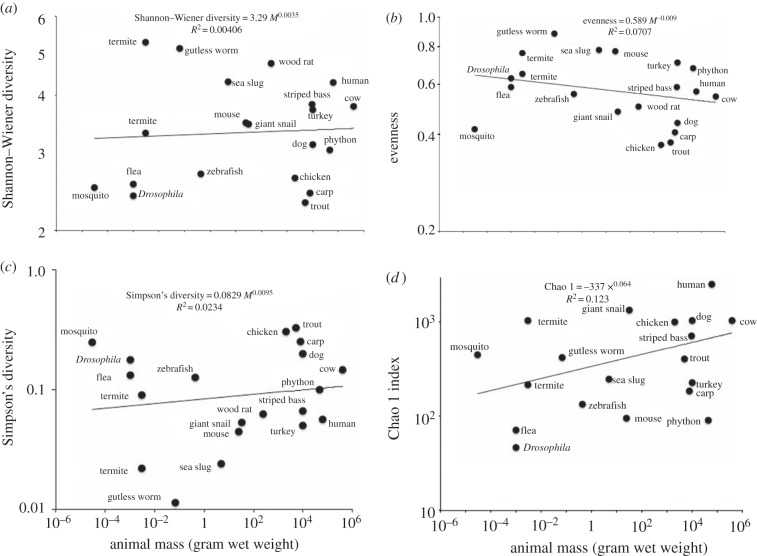
The microbial diversity indices calculated from MG-RAST data (see the electronic supplementary material) did not show significant scaling relationships with animal mass (figure 2). The diversity indices were mostly correlated to each other and none was correlated with animal body mass (table 2). Some diversity indices (alpha, evenness and Chao indices) showed correlations with the total number of base pairs of sequence in MG-RAST; the Shannon–Wiener and Simpson indices were not correlated with the sizes of the metagenomic surveys. Animal-associated species richness for 60 mammal species, reported as total OTUs by Ley et al. [14], plotted against animal mass also failed to show a consistent pattern (figure 3).
Figure 2.
Diversity indices versus individual animal body mass (M, wet weight in grams), log–log plots. (a) Shannon–Wiener index; (b) evenness; (c) Simpson's index and (d) Chao 1 index.
Table 2.
Spearman's rank correlation coefficient (upper right) and Pearson product-moment correlations (lower left) of animal wet weight, total number of base pairs (after quality control) and various diversity indices for microbial communities in animal-associated habitats, calculated from MG-RAST metagenomic data.
| wet weight | base pairsa | alpha diversity indexb | Shannon–Wiener index | evenness | Simpson index | Chao 1 index | |
|---|---|---|---|---|---|---|---|
| wet weight | 0.308 (0.187) |
0.299 (0.201) | 0.165 (0.487) | −0.222 (0.347) |
0.100 (0.675) |
0.330 (0.155) |
|
| base pairs | −0.0164 | 0.481 (0.0317) |
0.174 (0.462) | −0.607 (0.0045) |
0.161 (0.498) |
0.871 (<0.00001) |
|
| alpha diversity | 0.0001 (>0.999) |
0.599 (0.00527) |
0.869 (<0.00001) |
0.232 (0.324) |
−0.615 (0.0039) |
0.785 (0.00004) |
|
| Shannon–Wiener | 0.102 (0.668) |
0.394 (0.0859) |
0.868 (<0.00001) |
0.576 (0.00078) |
−0.836 (<0.00001) |
0.543 (0.0134) |
|
| evenness | −0.0661 (0.782) |
−0.175 (0.460) |
0.307 (0.189) |
0.624 (0.00328) |
−0.826 (<0.00001) |
−0.329 (0.156) |
|
| Simpson | 0.0305 (0.898) |
−0.208 (0.378) |
−0.522 (0.0184) |
0.781 (0.00005) |
−0.816 (0.00327) |
−0.164 (0.490) |
|
| Chao 1 | 0.0023 (0.992) |
0.920 (<0.0001) |
0.626 (0.00315) |
0.413 (0.0703) | −0.193 (0.415) |
−0.174 (0.463) |
aMG-RAST post-quality-control total base pair count.
bAlpha diversity as calculated by MG-RAST is the antilog of the Shannon–Wiener index. It has units of number of species.
Figure 3.
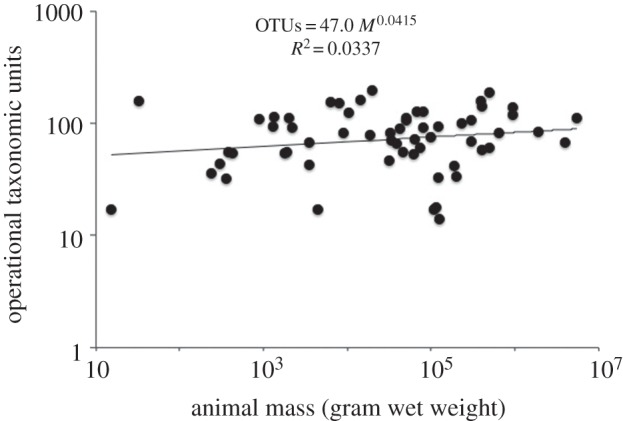
Total animal-associated microbial operational units (OTUs) as reported by Ley et al. [14], electronic supplementary material, for 60 species of mammals versus animal mass in gram wet weight, log–log plot.
4. Discussion
Abundances of animal-associated microbes showed a consistent allometric scaling relationship with animal mass, thereby supporting the first hypothesis. The scaling of microbial abundance is consistent with previous scaling of animal gut size. Total gut volume has been estimated to scale with animal body mass with exponents of 1.0 to 1.08, depending on the animals selected [5,39–41]. However, counts of microbes per unit volume or mass of gut contents vary over several orders of magnitude, and the proportion of the gut devoted to intensive microbial activities also varies extensively [16,17]. None of the previous considerations of animal gut allometry has considered microbial abundance and none has extended to the smallest multicellular animals.
The allometric relationships and global estimates of biological abundance reported here are necessarily based on limited datasets: in this case, the number of animal species whose microbes have been quantified. Insects might be over-represented, but they comprise a substantial proportion of animal species [42] and even of terrestrial animal biomass when one considers ants [43,44]. Humans and their domestic animals are commonly studied and so are well represented here, but this is also justified by their large populations and abundant microbiota [11]. The Earth's total number of animal species, approximately 8.7 million [42], is approximately 4 × 105-fold higher than those included here, leaving abundant room for further development of animal–microbe allometry. This paucity of data points to the need for quantification of microbes associated with a greater number of animal species to complement the extensive studies of microbial diversity. Development and analyses of larger datasets may reveal subtle differences among animals with different lifestyles, e.g. among herbivores, carnivores and omnivores; among ruminants, foregut fermenters, hindgut fermenters; between animals with dominantly heterotrophic microbes and those with autotrophs; or between vertebrate and invertebrate animals. Quantification of microbes within various taxa, for example, bacteria and archaea, across many animal species may reveal novel scaling properties, too.
Humans and their domestic animals, being highly successful macrofauna, may harbour an inordinately large proportion of the Earth's animal-associated microbial cells. Humans (7.1 × 109 individuals, 7.5 × 1023 microbes, updated from Whitman et al. [11], table 1) account for 3.3–3.5% of all animal-associated microbes. Estimates of microbes associated with domestic animals range from 3.3 × 1024 to 4.3 × 1024 cells (table 1), thus representing 14–20% of all animal-associated microbes. Productivity of gut microbes in mammals is prodigious (approx. 4 × 1027 cells year−1 for humans and domestic animals) [11], and they have high rates of mutation [11], and thus high potential for metabolic innovation and, in high densities, a high potential for horizontal gene transfer. These estimates further underscore human impacts on the biosphere. The phenomenal success of humans [34,45] has favoured many thousands of associated microbial species. The scales of human manipulation of those microbes, for example, by dosing humans and domestic animals with antibiotics, may have an even greater impact than previously considered.
Production of methane among non-ruminant mammals ranging in size from guinea pigs to elephants was previously found to scale with body size nearly linearly (exponent = 0.97) [41]. This scaling is consistent with the scaling of both gut volume and microbial abundance. Considering the relationships in equations (3.4) and (3.6), the scaling of methane production with animal mass also confirms the importance of macrofauna in the production of methane, a potent greenhouse gas. Macrofauna, especially domestic animals, generate an extravagant share of animal-generated methane.
Some successful animal groups other than humans and domestic animals also have significant gut microbiota. The world's fish biomass (9.0 × 1014 g [35] to 2.0 × 1015 g [46]) should have a total fish-associated microbial abundance of 3.1–6.9 × 1024 cells. Antarctic krill (E. superba) may be one of the most abundant metazoan species on the Earth, with an estimated biomass of 3.79 × 1014 g [47] and a bacterial load of approximately 5 × 108 microbes g−1 [31], so its global share of animal-associated microbes is approximately 1.9 × 1023 cells.
Microbial abundance per individual animal appears to scale as the approximate 1-power of body mass rather than as one of the common ‘quarter-power’ functions (±0.75, ±0.25), which are thought to be dictated by physical/chemical phenomena, e.g. the fractal geometry of resource distribution systems (blood vessels, bronchi) [6,7]. Metabolic rate, quantified in units of power, e.g. watts, scales with body mass as M0.75 [5–7,48]. Dividing the metabolic rate function by the microbial abundance function gives an exponent of −0.32 for the metabolic rate or power expended by the animal per microbe:
| 4.1 |
The surface area of the gut scales with body mass as M0.75 [49] and gut volume scales as M1.0 to 1.08 [5], so the surface area-to-volume ratio of the gut scales as M−0.25 to –0.33:
| 4.2 |
It is suggestive that the scaling of animal metabolic rate per microbe (exponent = −0.32) is very similar to the scaling of the gut surface area-to-volume ratio (exponent = −0.25 to −0.33). The animal metabolic rate per microbe declines as body mass increases, perhaps in part due to declining ability to transfer energy and materials through the interface between the microbial compartment and the rest of the animal. This finding does not exclude other explanations for the scaling of metabolic rate, e.g. fractal theory [6,7]. Animals have developed much more elaborate morphological adaptations for distributing materials, including relatively insoluble O2, throughout animal tissues than for transferring metabolites through the animal–microbe interface. The gut surface areas are actually greater than calculated owing to microvilli and other convolutions; however, microvilli are relatively invariant in size and density over a wide range of animal sizes [39] and the absorption by microvilli is concentrated at the distal ends [49]. These convolutions in the gut surface area may thus raise the intercept for the scaling of gut area with body mass, but have little effect on the slope. Mixing of the gut contents, for example, by muscle action on the rumen, can speed rates of diffusion, but this only partially alleviates the problem of transferring metabolites to the animal. Probably, multiple interacting factors govern the nearly linear relationship of animal mass to gut volume and to microbial abundance.
The apparent lack of pattern to the relationship between animal-associated microbial species diversity and animal mass is surprising, considering the well-known relationship between habitat size and species diversity [13]. However, a much larger dataset is needed before a firm conclusion can be made about the existence or absence of a relationship between microbial diversity and animal mass. Ideally, the microbial diversity data should be generated by comparable methods. In the case of this study, although the MG-RAST data were analysed using the same algorithms, the original data were generated using a variety of sequencing instruments and the depth of coverage varied extensively. As shown in this study, it is especially important that the sequencing coverage be sufficient and comparable. Nearly all of Ley et al.'s species diversity data [14] were generated by the same method as part of a single study; however, the number of replicate animals was small and variable and the clone library approach is limited compared with current high-throughput sequencing methods. The microbiota of increasing numbers of animal species are being extensively characterized using modern sequencing approaches, so further analyses of the scaling of diversity should be forthcoming.
Implications of animal mass-microbial abundance scaling appear wide-reaching, especially for macrofauna and their abundant microbes. At the other end of the mass spectrum, the fact that even minuscule metazoans follow this allometric relationship suggests an early evolutionary dependence of animals on microorganisms. If this allometric relationship withstands the test of time and additional data, then one presumably could even extrapolate upward in size and backward in time to the dinosaurs and other extinct animals. One can also consider the ongoing sixth mass extinction [50,51]. The loss of a large number of animal species is accompanied by the loss of the microbial species that are uniquely adapted to those animals. The findings of this study provide a new measure of the interdependence of animals and microbes.
5. Conclusion
The number of microbial cells associated with a single animal scales with body mass as a power function with an exponent of 1.07. This relationship applies over 12 orders of magnitude, from minute invertebrates to extremely large mammals. Based on this relationship, the total number of animal-associated microbes on the Earth is estimated to be 2.1–2.3 × 1025 cells. Despite the lower overall abundances of large animals compared with invertebrates and other small animals, the macrofauna harbour an outsized proportion of Earth's animal-associated microbes. The microbes of humans and their domestic animals comprise 3.3–3.5 and 14–20% of all animal-associated microbes on the Earth, respectively. The very large numbers of microbes in humans and domestic animals are especially concerning when one considers the widespread use of antibiotics and the high rates of production, mutation and horizontal gene transfer of the gut microbiota. The apparent lack of a scaling relationship between microbial species diversity and animal mass requires further testing.
Supplementary Material
Acknowledgements
We thank James Brown, Mitchell Sogin, Kevin Kirk, Jamie Voyles and two anonymous reviewers for helpful comments on the manuscript.
Ethics
Euthanization protocols were in accordance with an approved New Mexico Tech IACUC (Institutional Animal Care and Use Committee) protocol (no. 2014–2).
Data accessibility
Data used for calculating allometric relationships are presented in the electronic supplementary material.
Authors' contributions
T.K. conceived of the study, designed the study, performed all literature searching including acquisition of published microbial count data, performed all calculations and interpretations, and wrote the manuscript. K.S. suggested using MG-RAST for diversity data and performed all of the new direct microscopic counting of microbes. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by a Focus grant from the New Mexico IDeA Networks of Biomedical Research Excellence.
References
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human microbe interactions. Nature 449, 811–818. ( 10.1038/nature06245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl Acad. Sci. USA 110, 3229–3236. ( 10.1073/pnas.1218525110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fraune S, Bosch TCG. 2010. Why bacteria matter in animal development and evolution. Bioessays 32, 571–580. ( 10.1002/bies.200900192) [DOI] [PubMed] [Google Scholar]
- 4.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. ( 10.1002/bies.200900192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peters RH. 1983. The ecological implications of body size. Oxford, UK: Cambridge University Press. [Google Scholar]
- 6.West GB, Brown JH. 2005. The origin of allometric scaling laws in biology from genomes to ecosystems: toward a quantitative unifying theory of biological structure and organization. J. Exp. Biol. 208, 1575–1592. ( 10.1242/jeb.01589) [DOI] [PubMed] [Google Scholar]
- 7.West GB, Brown JH, Engquist BJ. 1997. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126. ( 10.1126/science.276.5309.122) [DOI] [PubMed] [Google Scholar]
- 8.Arneberg P, Skorping A, Grenfell B, Read AF. 1998. Host densities as determinants of abundance in parasite communities. Proc. R. Soc. Lond. B 265, 1283–1289. ( 10.1098/rspb.1998.0431) [DOI] [Google Scholar]
- 9.Hechinger RF, Lafferty KD, Dobson AP, Brown JH, Kuris AM. 2011. A common scaling rule for abundance, energetics, and production of parasitic and free-living species. Science 333, 445–448. ( 10.1126/science.1204337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagrue C, Poulina R, Cohen JE. 2015. Parasitism alters three power laws of scaling in a metazoan community: Taylor's law, density-mass allometry, and variance-mass allometry. Proc. Natl Acad. Sci. USA 112, 1791–1796. ( 10.1073/pnas.1422475112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes, the unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583. ( 10.1073/pnas.95.12.6578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. 2012. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl Acad. Sci. USA 109, 16 213–16 216. ( 10.1073/pnas.1203849109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenzweig ML. 1995. Species diversity in space and time. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 14.Ley RE, et al. 2008. Evolution of mammals and their gut microbes. Science 320, 1647–1651. ( 10.1126/science.1155725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazemier AE, Hackstein JHP, den Camp O, Rosenberg J, van der Drift C. 1997. Bacteria in the intestinal tract of different species of arthropods. Microb. Ecol. 33, 189–197. ( 10.1007/s002489900021) [DOI] [PubMed] [Google Scholar]
- 16.Stevens CE, Hume ID. 1998. Contributions of microbes in vertebrate gastrointestinal tracts to production and conservation of nutrients. Physiol. Rev. 79, 393–427. [DOI] [PubMed] [Google Scholar]
- 17.Stevens CE. 1977. Comparative physiology of the digestive system. In Duke‘s physiology of domestic animals (ed. Swenson MJ.), pp. 216–232, 9th edn Ithaca, NY: Cornell Univ. Press. [Google Scholar]
- 18.Olsen MA, Aagnes TH, Mathiesen SD. 1994. Digestion of herring by indigenous bacteria in the minke whale stomach. Appl. Environ. Microbiol. 60, 4445–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen MA, Blix AS, Utsi THA, Sørmo W, Mathiesen SD. 2000. Chitinolytic bacteria in the minke whale forestomach. Can. J. Microbiol. 46, 85–94. ( 10.1139/cjm-46-1-85) [DOI] [PubMed] [Google Scholar]
- 20.Hentschel U, Usher KM, Taylor MW. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55, 167–177. ( 10.1111/j.1574-6941.2005.00046.x) [DOI] [PubMed] [Google Scholar]
- 21.Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. 2010. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microb. Ecol. 71, 304–312. ( 10.1111/j.1574-6941.2009.00812.x) [DOI] [PubMed] [Google Scholar]
- 22.Gibbons RJ, Kapsimalis B. 1967. Estimates of the overall rate of growth of the intestinal microflora of hamsters, guinea pigs, and mice. J. Bacteriol. 93, 510–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavanagh CM, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. 1981. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila . Science 213, 340–342. ( 10.1126/science.213.4505.340) [DOI] [PubMed] [Google Scholar]
- 24.Sonoyama K, Fujiwara R, Tekemura N, Ogasawara T, Watanabe J, Ito H, Morita T. 2009. Response of the gut microbiota to fasting and hibernation in Syrian hampsters. Appl. Environ. Microbiol. 75, 6451–6456. ( 10.1128/AEM.00692-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, Yang HS, Xu Q, Wang FY, Liu GB, German DP. 2008. Phenotypic plasticity of gut structure and function during periods of inactivity in Apostichopus japonicus. Comp. Biochem. Physiol. A 150, 255–262. ( 10.1016/j.cbpb.2008.03.011) [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Yang H, Xu Q, Wang F, Liu G. 2009. Effect of temperature on digestive enzyme activity and gut mass in sea cucumber Apostichopus japonicas (Selenka), with special reference to aestivation. Chin. J. Oceanol. Limnol. 27, 714–722. ( 10.1007/s00343-009-9202-3) [DOI] [Google Scholar]
- 27.Enomoto M, Nakagawa S, Sawabe T. 2012. Microbial communities associated with Holothurians: presence of unique bacteria in the coelomic fluid. Microb. Environ. 27, 300–305. ( 10.1264/jsme2.ME12020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikuchi Y, Bomar L, Graf J. 2009. Stratified bacterial community in the bladder of the medicinal leech, Hirudo verbena. Environ. Microbiol. 11, 2758–2770. ( 10.1111/j.1462-2920.2009.02004.x) [DOI] [PubMed] [Google Scholar]
- 29.Daniel O, Anderson JM. 1992. Microbial biomass and activity in contrasting soil materials after passage through the gut of the earthworm Lumbricus rubellus Hoffmeister. Soil. Biol. Biochem. 24, 465–470. ( 10.1016/0038-0717(92)90209-G) [DOI] [Google Scholar]
- 30.Kristufek V, Ravasz K, Pizl V. 1992. Changes in densities of bacteria and microfungi during gut transit in Lumbricus rubellus and Aporrectodea caliginosa (Oligochaeta: Lumbricidae). Soil. Biol. Biochem. 24, 1499–1500. ( 10.1016/0038-0717(92)90139-O) [DOI] [Google Scholar]
- 31.Donachie SP, Zdanowski MK. 1998. Potential digestive function of bacteria in krill Euphausia superba stomach. Aquat. Microb. Ecol. 14, 129–136. ( 10.3354/ame014129) [DOI] [Google Scholar]
- 32.Portal-Celhay C, Bradlkey ER, Blaser MJ. 2013. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12, 49 ( 10.1186/1471-2180-12-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whittaker RH, Likens GE. 1975. The biosphere and man. In Ecological studies vol. 14: primary productivity of the biosphere (eds Lieth H, Whittaker RH.), pp. 305–328. Berlin, Germany: Springer. [Google Scholar]
- 34.Barnosky AD. 2008. Megafauna biomass tradeoff as a driver of quaternary and future extinctions. Proc. Natl Acad. Sci. USA 105, 11 543–11 548. ( 10.1073/pnas.0801918105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jennings S, Melin F, Blanchard JL, Forster RM, Dulvy NK, Wilson RW. 2008. Global-scale predictions of community and ecosystem properties from simple ecological theory. Proc. R. Soc. B 275, 1375–1383. ( 10.1098/rspb.2008.0192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer F, et al. 2008. The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 9, 386 ( 10.1186/1471-2105-9-386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill TCJ, Walsh KA, Harris JA, Moffett BF. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43, 1–11. ( 10.1016/S0168-6496(02)00449-X) [DOI] [PubMed] [Google Scholar]
- 38.Damuth J. 1981. Population density and body size in mammals. Nature 290, 699–700. ( 10.1038/290699a0) [DOI] [Google Scholar]
- 39.Franz R, Hummel J, Kienzle E, Kolle P, Gunga HC, Class M. 2009. Allometry of visceral organs in living amniotes and its implications for sauropod dinosaurs. Proc. R. Soc. B 276, 1731–1776. ( 10.1098/rspb.2008.1735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franz R, Hummel J, Muller DWH, Bauert M, Hatt JM, Clauss M. 2011. Herbivorous reptiles and body mass: effects on food intake, digesta retention, digestibility and gut capacity, and a comparison with mammals. Comp. Biochem. Physiol. A 158, 94–101. ( 10.1016/j.cbpa.2010.09.007) [DOI] [PubMed] [Google Scholar]
- 41.Franz R, Soliva CR, Kreuzer M, Hummel J, Clauss M. 2011. Methane output of rabbits (Oryctolagus cuniculus) and guinea pigs (Cavia porcellus) fed a hay-only diet: implications for the scaling of methane production with body mass in non-ruminant mammalian herbivores. Comp. Biochem. Physiol. A 158, 177–181. ( 10.1016/j.cbpa.2010.10.019) [DOI] [PubMed] [Google Scholar]
- 42.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. 2011. How many species are there on Earth and in the ocean? PLoS Biol. 9, e1001127 ( 10.1371/journal.pbio.1001127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poulsen M, Sapountzis P. 2012. Behind every great ant, there is a great gut. Mol. Ecol. 21, 2054–2057. ( 10.1111/j.1365-294X.2012.05510.x) [DOI] [PubMed] [Google Scholar]
- 44.Zeintz E, Feldhaar H, Stoll S, Gross R. 2005. Insights into the microbial world associated with ants. Arch. Microbiol. 184, 199–206. ( 10.1007/s00203-005-0041-0) [DOI] [PubMed] [Google Scholar]
- 45.Vitousek PM, Mooney HA. 1997. Human domination of Earth's ecosystems. Science 277, 494–499. ( 10.1126/science.277.5325.494) [DOI] [Google Scholar]
- 46.Wilson RW, et al. 2009. Contribution of fish to the marine inorganic carbon cycle. Science 323, 359–362. ( 10.1126/science.1157972) [DOI] [PubMed] [Google Scholar]
- 47.Atkinson A, Siegel V, Pakhomov EA, Jessopp MJ, Loeb V. 2009. A re-appraisal of the total biomass and annual production of Antarctic krill. Deep-Sea Res. I 56, 727–740. ( 10.1016/j.dsr.2008.12.007) [DOI] [Google Scholar]
- 48.Kleiber M. 1932. Body size and metabolism. Hilgardia 16, 315–353. ( 10.3733/hilg.v06n11p315) [DOI] [Google Scholar]
- 49.Clauss M, Hummel J. 2005. The digestive performance of mammalian herbivores: why big may not be that much better. Mammal. Rev. 35, 174–187. ( 10.1111/j.1365-2907.2005.00062.x) [DOI] [Google Scholar]
- 50.Wake DB, Vredenburg VT. 2008. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl Acad. Sci. USA 105, 11 466–11 473. ( 10.1073/pnas.0801921105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnosky AD, et al. 2011. Has the Earth's sixth mass extinction already arrived? Nature 471, 51–57. ( 10.1038/nature09678) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for calculating allometric relationships are presented in the electronic supplementary material.