Abstract
In addition to its established role in inflammation, the stress-activated p38 MAP kinase pathway plays major roles in the regulation of cell cycle, senescence, and autophagy. Robust studies could establish mechanistic links between MAPK11-MAPK14/p38 signaling and macroautophagy converging at ATG9-trafficking and BECN1 phosphorylation. However, several reports seem to monitor MAPK11-MAPK14/p38-dependence of autophagy exclusively by the use of the SB203580/SB202190 class of MAPK14/MAPK11/p38α/β inhibitors. In this “Letter to the editor” we present data to support our claim that these inhibitors interfere with autophagic flux in a MAPK11-MAPK14/p38-independent manner and hence should no longer be used as pharmacological tools in the analysis of MAPK11-MAPK14/p38-dependence of autophagy. We propose a general guideline from Autophagy with regard to this issue to avoid such misinterpretations in the future.
Keywords: autophagy, LC3, MAPK11-MAPK14/p38, SB202190, SB203580, SQSTM1, vacuoles
Dear Editor,
An article by Zhong et al.1 analyzed the role played by stress-induced MAPK11-MAPK14/p38 signaling in the expression of autophagy-related (ATG) genes and concludes that MAPK11-MAPK14/p38 isoforms α, β, γ, and δ (MAPK14/11/12/13) are involved in the transcription of ATG genes in response to a novel anticancer copper complex. We have serious concerns regarding the title and conclusions of this publication, which should be discussed to preserve the high standards of Autophagy.
Our major point concerns the analysis of the role of the MAPK11-MAPK14/p38 pathway in the regulation of autophagy by the pyridinyl imidazole class MAPK14/p38α-MAPK11/p38β-inhibitor SB203580. Several technically robust publications in the past decade have conclusively established a context-dependent role for the stress-activated MAPK11-MAPK14/p38 pathway in the regulation of MTOR signaling and autophagy.2-4 Furthermore, a connection between the MAPK14/p38α-MAPK11/p38β-activated protein kinase MAPKAPK2/MK2 and autophagy was established recently via demonstrating phosphorylation of BECN1/Beclin-1 at serine 90, using a dominant-negative mutant of MAPK14/p38α instead of MAPK11-MAPK14/p38 inhibitors.5 However, we are deeply concerned about the use of a class of pyridinyl imidazole inhibitors, such as SB203580 and SB202190, in monitoring the role of MAPK14/p38α-MAPK11/p38β signaling in autophagy, because we had previously reported that these compounds alter autophagic flux and pro-autophagic gene expression in a cell type-specific, MAPK14/p38α-MAPK11/p38β-independent manner.6 In the figure panels (Fig. 1A-H), we provide additional data to support our claims that:
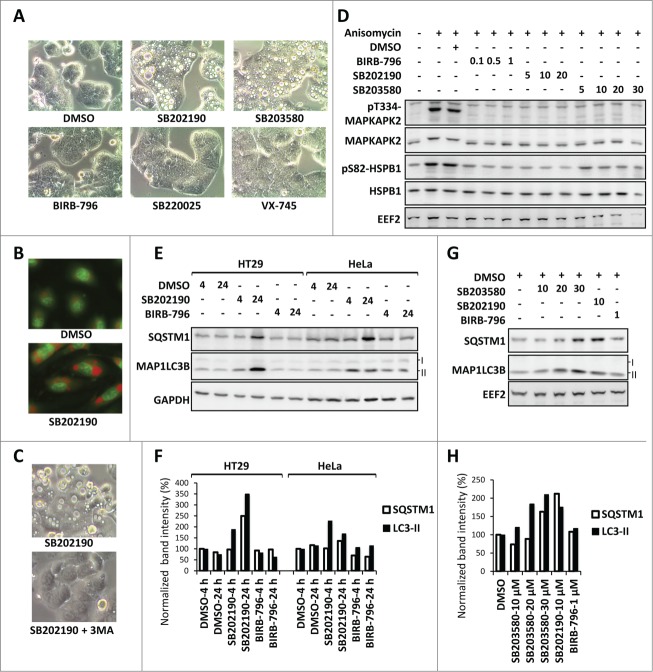
Figure 1.
MAPK11-MAPK14/p38-independent effects of SB202190/SB203580 in autophagy. (A) SB202190 and SB203580 (10 µM each) but not the other more specific MAPK11-MAPK14/p38 inhibitors (SB220025, 10 µM; BIRB-796, 1 µM; or VX-745, 10 µM) induce large vacuoles in HT29 cells (24 h treated). (B) The SB202190-induced vacuoles are acidic compartments as shown by strong acridine orange staining in primary HUVECs. (C) Autophagy inhibitor 3-MA suppresses SB-induced vacuolation in HT29 cells. (D) The efficacy of BIRB-796, SB202190, and SB203580 to inhibit MAPK14/p38α-MAPK11/p38β signaling in HeLa cells was compared by monitoring their effect on stress-induced phosphorylation of the direct MAPK14/p38α-MAPK11/p38β substrate MAPKAPK2 at Thr334 (T334) and of the downstream target HSPB1/HSP27 at Ser82 (S82). The membrane was reprobed with MAPKAPK2, HSPB1 and EEF2 (eukaryotic translation elongation factor 2) antibodies as loading controls. Cells were treated with the indicated concentrations of inhibitors (µM) prior to 30 min anisomycin (10 µg/ml) stimulation. (E and F) The off-target effect of SB202190 in autophagy is independent of cell-type specific vacuolation. In both, vacuole-positive HT29 and vacuole-negative HeLa cells (see Table 1), long-term SB202190 treatment (10 µM for 4 or 24 h) leads to the accumulation of autophagy substrates SQSTM1 and lipid conjugated MAP1LC3B (LC3-II) (E). Quantified band intensities for LC3B-II and SQSTM1 normalized to that of the loading control (GAPDH) are shown (F). (G and H) Dose-dependent (10-30 µM) effect of SB203580 on autophagy in HeLa cells demonstrated by monitoring the levels of SQSTM1 and MAP1LC3B (LC3-II) at 24 h treatment (G). Quantified band intensities for lipid conjugated MAP1LC3B (LC3-II) and SQSTM1 normalized to the loading control (EEF2) are shown (H).
1. SB202190 and SB203580, but not the structurally nonrelated and more potent MAPK11-MAPK14/p38 inhibitor BIRB-796,7 induce vacuoles (Fig. 1A) characterized as acidic compartments (Fig. 1B) in HT29 cells in a 3-methyladenine (3MA)-sensitive manner (Fig. 1C) indicating a compound-specific, MAPK11-MAPK14/p38-independent autophagic response.6,8,9
2. SB202190 does induce vacuole formation in about 70% of the cell lines analyzed when used at very low concentrations (Table 1), but induces accumulation of the autophagy substrate SQSTM1/p62 and lipid-conjugated MAP1LC3B (LC3-II) also in cells, which display no vacuole formation, in a compound-specific, MAPK11-MAPK14/p38-independent manner (Fig. 1E and F). As expected from the structural similarity, SB203580 gave results very similar to SB202190 albeit with less potency (Fig. 1G and H). In contrast, BIRB-796 did not affect the levels of autophagy substrates (Fig. 1E–H), although it effectively blocked MAPK14/p38α-MAPK11/p38β signaling as monitored by stress-induced downstream phosphorylation events (Fig. 1D) already at low concentrations.
Table 1.
Cell-type specificity of SB202190-induced vacuole formation.
| No | Cell line | Species | Cell type | Vacuoles |
|---|---|---|---|---|
| 1 | AGS | Human | gastric adenocarcinoma | + |
| 2 | A549 | Human | lung carcinoma | + |
| 3 | BHK21 | Hamster | adult kidney fibroblast | + |
| 4 | C2C12 | Mouse | myoblast | − |
| 5 | Caco-2 Bbe | Human | colorectal adenocarcinoma | + |
| 6 | HCT 116 | Human | colorectal adenocarcinoma | + |
| 7 | HEK293T | Human | embryonic kidney | − |
| 8 | HeLa | Human | cervical adenocarcinoma | − |
| 9 | hMSC | Human | primary mesenchymal stem cells | + |
| 10 | HT29 | Human | colorectal adenocarcinoma | + |
| 11 | HUVEC | Human | primary endothelial cells | + |
| 12 | IEC6 | Rat | small intestinal epithelium | + |
| 13 | L929 | Mouse | fibrosarcoma | + |
| 14 | MCF-10A | Human | mammary epithelial | + |
| 15 | MEF-T | Mouse | embryonic fibroblast | + |
| 16 | NIH 3T3 | Mouse | embryonic fibroblast | − |
| 17 | NMuMG | Mouse | mammary epithelial | + |
| 18 | RAW 264.7 | Mouse | monocytic | + |
| 19 | RGM1 | Rat | gastric epithelium | + |
| 20 | Sh-SY5Y | Human | neuroblastoma | − |
| 21 | SW480 | Human | colorectal adenocarcinoma | + |
| 22 | WM1617 | Human | melanoma | − |
| 23 | WM793 | Human | melanoma | − |
The table depicts the cell-type specificity of SB202190-induced autophagy-dependent vacuole formation. Cells were treated with 5 µM SB202190 for 12 h. Vacuoles were clearly visible in most of the cell lines after approximately 2 h of SB202190 treatment.
Because of the MAPK11-MAPK14/p38-independent interference with autophagy, the SB-compounds should no longer be used as pharmacological tools in the analysis of MAPK11-MAPK14/p38-dependence of autophagy.
Another concern regards the findings and title of the paper, the latter of which explicitly states that MAPK11/12/13/14 are involved in the transcriptional response induced by the copper complex. These conclusions are exclusively based on the use of the inhibitor SB203580, which targets only MAPK14/p38α and MAPK11/p38β.10 Hence, the title statement about MAPK13/p38δ and MAPK12/p38γ is not justified by the data presented and should be corrected.
Materials and methods
SB203580 (Calbiochem, 559389), SB220025 (Sigma, S9070), BIRB-796 (Axon Medchem, 1358), VX-745 (Philip Cohen, University of Dundee) and SB202190 (Axon Medchem, 1364) stocks were prepared in DMSO (Carl Roth, 4720.4). Primary antibodies used were: MAP1LC3B (Cell Signaling Technology, 3868), pT334-MAPKAPK2 (Cell Signaling Technology, 3007), MAPKAPK2 (Cell Signaling Technology, 3042), pS82-HSPB1 (Cell Signaling Technology, 2401), SQSTM1 (BD Biosciences, 610833), HSPB1 (Santa Cruz Biotechnology, sc-1048) and GAPDH (Chemicon international, MAB374). Secondary antibodies used were goat anti-mouse IgG-HRP (Santa Cruz Biotechnology, sc-2005), donkey anti-goat IgG-HRP (Santa Cruz Biotechnology, sc-2033) and goat anti-rabbit IgG (Dianova, 111-035-003). Acridine orange (Sigma, 158550), anisomycin (Sigma, A9789) and 3-methyladenine (Calbiochem, 189490) were purchased as indicated. Cell culture, microscopy and immunoblotting procedures were followed as described previously.6
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dr. Philip Cohen (University of Dundee) for the gift of VX-745 and discussion of the results.
Funding
This work was supported by Deutsche Forschungsgemeinschaft.
References
- 1.Zhong W, Zhu H, Sheng F, Tian Y, Zhou J, Chen Y, Li S, Lin J. Activation of the MAPK11/12/13/14 (p38 MAPK) pathway regulates the transcription of autophagy genes in response to oxidative stress induced by a novel copper complex in HeLa cells. Autophagy 2014; 10:1285-300; PMID:24905917; http://dx.doi.org/ 10.4161/auto.28789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38[alpha] MAPK through mAtg9 and p38IP. EMBO J 2010; 29:27-40; PMID:19893488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moruno-Manchon JF, Perez-Jimenez E, Knecht E. Glucose induces autophagy under starvation conditions by a p38 MAPK-dependent pathway. Biochem J 2013; 449:497-506; PMID:23116132; http://dx.doi.org/ 10.1042/BJ20121122 [DOI] [PubMed] [Google Scholar]
- 4.Cully M, Genevet A, Warne P, Treins C, Liu T, Bastien J, Baum B, Tapon N, Leevers SJ, Downward J. A role for p38 stress-activated protein kinase in regulation of cell growth via TORC1. Mol Cell Biol 2010; 30:481-95; PMID:19917724; http://dx.doi.org/ 10.1128/MCB.00688-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, Sinha S, Gaestel M, Levine B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. Elife 2015; 4; PMID:25693418; http://dx.doi.org/ 10.7554/eLife.05289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon MB, Kotlyarov A, Gaestel M. SB202190-induced cell type-specific vacuole formation and defective autophagy do not depend on p38 MAP kinase inhibition. PLoS One 2011; 6:e23054; PMID:21853067; http://dx.doi.org/ 10.1371/journal.pone.0023054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J 2007; 408:297-315; PMID:17850214; http://dx.doi.org/ 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, Ingravallo G, Modica S, Lo Sasso G, Moschetta A, et al.. A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ 2007; 14:693-702; PMID:17159917; http://dx.doi.org/ 10.1038/sj.cdd.4402076 [DOI] [PubMed] [Google Scholar]
- 9.Chiacchiera F, Matrone A, Ferrari E, Ingravallo G, Lo Sasso G, Murzilli S, Petruzzelli M, Salvatore L, Moschetta A, Simone C. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ 2009; 16:1203-14; PMID:19343039;http://dx.doi.org/ 10.1038/cdd.2009.36 [DOI] [PubMed] [Google Scholar]
- 10.Eyers PA, Craxton M, Morrice N, Cohen P, Goedert M. Conversion of SB 203580-insensitive MAP kinase family members to drug-sensitive forms by a single amino-acid substitution. Chem Biol 1998; 5:321-8; PMID:9653550; http://dx.doi.org/ 10.1016/S1074-5521(98)90170-3 [DOI] [PubMed] [Google Scholar]