Abstract
Macroautophagy (hereafter autophagy) is the process by which cytosolic material destined for degradation is enclosed inside a double-membrane cisterna known as the autophagosome and processed for secretion and/or recycling. This process requires a large collection of proteins that converge on certain sites of the ER membrane to generate the autophagosome membrane. Recently, it was shown that actin accumulates around autophagosome precursors and could play a role in this process, but the mechanism and role of actin polymerization in autophagy were unknown. Here, we discuss our recent finding that the nucleation-promoting factor (NPF) WHAMM recruits and activates the Arp2/3 complex for actin assembly at sites of autophagosome formation on the ER. Using high-resolution, live-cell imaging, we showed that WHAMM forms dynamic puncta on the ER that comigrate with several autophagy markers, and propels the spiral movement of these puncta by an Arp2/3 complex-dependent actin comet tail mechanism. In starved cells, WHAMM accumulates at the interface between neighboring autophagosomes, whose number and size increases with WHAMM expression. Conversely, knocking down WHAMM, inhibiting the Arp2/3 complex or interfering with actin polymerization reduces the size and number of autophagosomes. These findings establish a link between Arp2/3 complex-mediated actin assembly and autophagy.
Keywords: actin cytoskeleton, Arp2/3 complex, macroautophagy, nucleation promoting factor
The 7-subunit Arp2/3 complex (ARPC1, ARPC2, ARPC3, ARPC4, ARPC5, ARP2, ARP3) nucleates actin filament polymerization and branching in countless cellular processes, including cell motility, endocytosis, and cytokinesis. The omnipresence of actin- and Arp2/3 complex-driven movement extends to several pathogens, which have evolved mechanisms to hijack this system for infection and/or to propel their movement within and between cells. In isolation, however, the Arp2/3 complex is inactive. This creates an opportunity for the complex to be exquisitely regulated in time and space by a group of proteins known as nucleation-promoting factors, which activate the complex for different activities and at different subcellular locations (Fig. 1A). These proteins share little in common, other than their C-terminal Proline-rich, WH2 domain, Central, Acidic (PWCA) region, which mediates interactions with PFN/profilin-actin, actin, and the Arp2/3 complex, respectively. The PWCA region is in itself sufficient to activate the complex in vitro. The other domains of NPFs are responsible for their regulation and localization, as these proteins tend to be found at specific subcellular locations (Fig. 1A). Thus, 2 NPFs, WASL/N-WASP and the WASF/WAVE complex, recruit the Arp2/3 complex to the leading edge of the cell, where they promote the formation of dendritic actin networks in lamellipodia necessary for cell motility and the invagination of endosomes. Another NPF, the WASH complex, links the Arp2/3 complex to endosomal sorting, transport and maturation. Finally, 2 NPFs, WHAMM and JMY, are related in sequence but have been implicated in different processes. JMY is found in the nucleus, where it regulates transcription, and in the ER and the trans-Golgi network, where it drives vesicle trafficking. WHAMM was initially localized to the ER-Golgi intermediate compartment and the cis-Golgi apparatus and was implicated in ER-to-Golgi transport.
Figure 1.
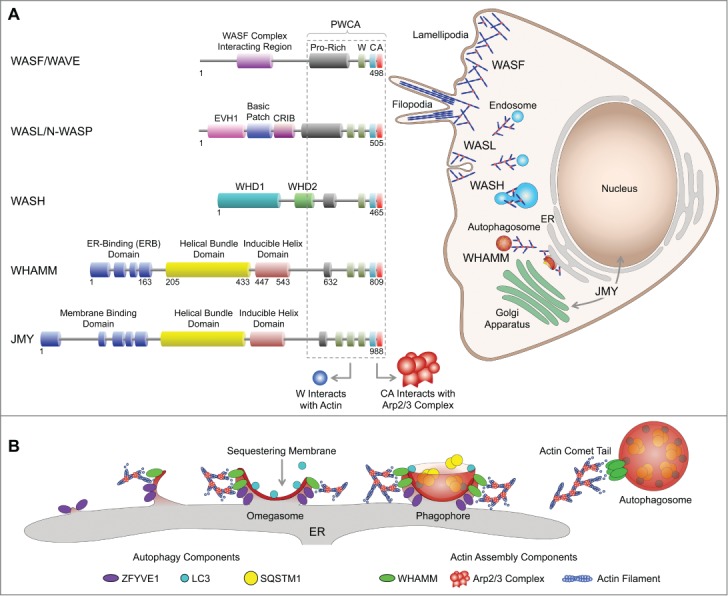
WHAMM links actin assembly via the Arp2/3 complex to autophagy. (A) A family of NPFs controls the activity of the Arp2/3 complex in time and space. NPFs share a C-terminal PWCA region necessary for Arp2/3 complex activation, whereas their variable N-terminal domains are implicated in regulation and localization. The WASF/WAVE complex and WASL/N-WASP are involved in lamellipodia formation and endocytosis. The WASH complex is involved in endosomal trafficking. JMY regulates transcription in the nucleus and vesicle trafficking between the ER and trans-Golgi network. WHAMM was initially implicated in ER to cis-Golgi transport. We recently found that WHAMM also participates in autophagosome biogenesis. (B) Autophagosome formation is initiated at specific ER loci that give rise to ZFYVE1-coated omegasomes. LC3 is lipidated and anchored to the developing sequestering membrane, and recruits SQSTM1 that mediates the binding of ubiquitinated proteins destined for degradation. The phagophore closes off, detaches from the ER membrane, and moves through an actin comet tail mechanism, mediated by WHAMM and the Arp2/3 complex.
While the cellular roles of NPFs are diverse, they typically link the Arp2/3 complex and actin polymerization to membrane remodeling events. For example, during clathrin-mediated endocytosis, the plasma membrane first invaginates, then expands to form the endocytic cup, and finally closes off to form the endosome, which can in some cases be transported by an actin comet tail mechanism. WASL/N-WASP and the Arp2/3 complex have been implicated in each of these steps. Autophagosome biogenesis follows a pathway that is in some ways reminiscent of endocytosis, including the nucleation, expansion and closure of the sequestering membrane, its detachment from the ER membrane, and the transport and fusion of autophagosomes with lysosomes for degradation (Fig. 1B). Adding to this parallel, it was recently shown that actin colocalizes with early autophagosome markers, and inhibition of actin assembly downregulates starvation-mediated autophagy. However, the mechanism for actin assembly during autophagosome formation and its role in this process remained mysterious.
Our findings indicate that WHAMM plays a role in autophagosome biogenesis that is somewhat analogous to that of WASL/N-WASP in endocytosis. Live-cell imaging in ARPE-19 and HeLa cells shows that WHAMM forms dynamic puncta on the surface of the ER membrane. These puncta most commonly display spiral motions, and appear to pull on ER tubules. Occasionally, these tubules are coated along their length by WHAMM molecules. Interestingly, the WHAMM-coated tubules do not appear to colocalize with general ER markers such as SEC61B/Sec61β and CANX/calnexin, suggesting that they represent a unique loci within the ER membrane, probably characterized by a distinct lipid composition. The WHAMM puncta colocalize and comigrate with autophagy markers, including ZFYVE1/DFCP1, LC3, and SQSTM1/p62. However, WHAMM does not colocalize with the early marker ATG14 or the lysosomal marker LAMP1, suggesting that WHAMM is not present during the very early stages of autophagosome biogenesis nor at the late stage when autophagosomes merge with lysosomes. The motility of WHAMM puncta depends on actin filament assembly and the Arp2/3 complex. Thus, inhibiting actin polymerization or Arp2/3 complex activation halts the motility of the WHAMM puncta. Moreover, knocking down WHAMM significantly reduces the size and number of autophagosomes. Taken together, these findings suggest that WHAMM recruits the Arp2/3 complex to sites of autophagosome formation on the ER, harnessing the forces of actin polymerization to form the sequestering membrane, and to detach and move the autophagosomes (Fig. 1B).
Consistent with the general idea that the N-terminal region of NPFs controls their localization, and thus that of the Arp2/3 complex, we found that the region comprising the first ˜170 residues of WHAMM is absolutely required for ER localization. Interestingly, this so-called ER-binding (ERB) domain is also the most variable region between the 2 related NPFs WHAMM and JMY. Yet, perhaps surprisingly, we found that JMY puncta also colocalize with LC3, suggesting that these 2 NPFs could have similar and/or complementary roles in autophagy.
An important observation of this study is the presence of actin comet tails that invariably follow WHAMM puncta when these are associated with autophagy markers. This behavior was observed in both ARPE-19 and HeLa cells cotransfected with plasmids encoding WHAMM, LifeAct, and either LC3 or ZFYVE1. Actin comet tails are also observed in association with LC3- and ZFYVE1-coated vesicles in the absence of exogenous WHAMM expression, whereas knocking down WHAMM almost entirely eliminates the formation of comet tails in association with autophagy markers. Together, these results suggest that an actin comet tail motility mechanism is a fundamental component of the autophagosome maturation pathway.
In summary, we have demonstrated a new role for actin, the Apr2/3 complex, and its activator WHAMM in autophagosome biogenesis during starvation-induced autophagy.
Funding
This work was supported by the National Institutes of Health grants P01 GM087253 and R01 MH087950. D.J.K. was additionally supported by American Cancer Society grant PF-13-033-01-DMC.


