Abstract
Lipid droplets (LDs) are the conserved organelles for the deposit of neutral lipids, and function as reservoirs of membrane and energy sources. To date, functional links between autophagy and LD dynamics have not been fully elucidated. Here, we report that a vacuolar putative lipase, Atg15, required for degradation of autophagic bodies, is crucial for the maintenance of LD amount in the yeast Saccharomyces cerevisiae in the stationary phase. Mutant analyses revealed that the putative lipase motif and vacuolar localization of Atg15 are important for the maintenance of LD amount. Loss of autophagosome formation by simultaneous deletion of core ATG genes cancelled the reduction in the LD amount in ATG15-deleted cells, indicating that degradation of autophagic bodies accounts for the functional involvement of Atg15 in LD dynamics. The reduced level of LDs in the mutant strain was dependent on Tgl3 and Tgl4, major lipases for lipolysis in S. cerevisiae. An altered phosphorylation status of Tgl3, higher accumulation of Tgl4, and closer associations of Tgl3 and Tgl4 with LDs were detected in the ATG15-deleted cells. Furthermore, increased levels of downstream metabolites of lipolysis in the mutant strain strongly suggested enhanced lipolytic activity caused by loss of ATG15. Our data provide evidence for a novel link between autophagic flux and LD dynamics integrated with Atg15 activity.
Keywords: Atg15, autophagy, lipid droplet, lipolysis, Tgl3, Tgl4, triacylglycerol
Abbreviations
- DAG
diacylglycerol
- ER
endoplasmic reticulum
- FFA
free fatty acid
- GFP
green fluorescent protein
- LDs
lipid droplets
- mCherry
monomeric cherry fluorescent protein
- PL
phospholipid
- PNPLA2/ATGL
patatin-like phospholipase domain containing 2
- SE
sterol ester
- TAG
triacylglycerol
- WT
wild type.
Introduction
The lipid droplet (LD) functions as a main organelle for storage of neutral lipids such as triacylglycerol (TAG) and sterol ester (SE). These neutral lipids are packed into the core of LD, and the surface of LD is surrounded by a phospholipid monolayer.1 Although LD had been simply considered as an energy-storage organelle, recent studies, most of which addressed yeast LD dynamics, revealed other functions of LD such as those that supply membrane sources and in maintaining lipid homeostasis.2-4 Intriguingly, this organelle was found connected with other organelles, such as the endoplasmic reticulum (ER), mitochondria, peroxisomes, vacuoles/lysosomes, and also with autophagosomes, which implies functional associations of LDs with lipid metabolisms in these organelles.5-7 It is thought that LDs are generated from the ER, where several enzymes with acyltransferase activity (Dga1, Lro1, Are1 and Are2) synthesize neutral lipids, leading to the budding of a nascent LD.8 In fact, LDs are not formed in an are1Δ are2Δ dga1Δ lro1Δ strain.9,10
Degradation of LDs is mainly accomplished by lipases localized on LDs, through a process known as lipolysis. In mammalian cells, 2 types of enzymes function in lipolysis, one termed LIPE/HSL (lipase, hormone-sensitive) controlled by a signaling pathway through cyclic AMP, and PNPLA2/ATGL (patatin-like phospholipase domain containing 2).11 There are no homologs of LIPE in yeast, where most of the triacylglycerol is degraded by 2 lipases, Tgl3, Tgl4, which are localized on LDs in S. cerevisiae.12,13 Tgl4 is the functional ortholog of PNPLA2 in yeast, harboring a characteristic patatin domain. Lipolytic activity by Tgl4 plays an important role in supplying membrane sources when quiescent (G0) cells are transferred to fresh medium.14 Both Tgl3 and Tgl4 possess enzymatic activities in addition to those as triacylglycerol lipases; Tgl3 has acyltransferase activity15 and Tgl4 exhibits a lipase activity toward SE as well as acyltransferase activity.16
Recently, a degradation pathway of LD by autophagy (lipophagy) was discovered.17 The relationship between autophagy and LD in mammalian cells was originally reported in a study of hepatocytes,18 and subsequent findings based on electron microscopy confirmed that LDs are surrounded by autophagosomes in mice.19 In yeast cells, a similar phenomenon (incorporation of LDs into the vacuole) was already reported in 1979.20 Recently, it was reported that LDs are directly incorporated into the vacuole through a microautophagic pathway, when cells are grown on oleate and then transferred to a nitrogen starvation medium, or when the cells are grown in synthetic complete (SC) medium to stationary phase.21,22 This process, termed microlipophagy, was reported to require many ATG genes that are needed for macroautophagy. However, to date, little is known about the functional relationship between LD dynamics and autophagy except for the existence of lipophagy. In this study we carried out a comprehensive analysis of LDs in atg mutants, and found that Atg15 is specifically required for the maintenance of LD amount in post-diauxic to stationary phases.
In yeast, vesicles transported into the vacuole through macroautophagy (macroautophagic bodies) are lysed by Atg15, and then the released contents including proteins are degraded mainly by intravacuolar proteases.23 Atg15 is also essential for the degradation of the cytoplasm to vacuole targeting (Cvt) bodies and multivesicular body vesicles in S. cerevisiae.24-26 In the denoted yeast microlipophagy, Atg15 is required for the vacuolar degradation of the neutral lipids in LDs.21 In this study, we revealed that Atg15 involvement in the maintenance of LD amount was dependent on the protein's putative lipase motif, and was canceled in multiple atg mutants defective in macroautophagy, suggesting that the enzymatic activity of Atg15 toward macroautophagic bodies was critical for the maintenance of LD amount. We further discovered that the Atg15 contribution to LD amount was dependent on the lipolytic enzymes Tgl3 and Tgl4, whose in vivo activities were strongly suggested to be elevated by loss of Atg15. Altogether, this study provides a novel insight to autophagic flux and LD dynamics.
Results
Deletion of ATG15 causes a decrease in LD in the stationary phase
To study the effects of autophagy on the dynamics of LD, we carried out the quantification of TAG levels in the set of core atg mutants (atg1Δ through atg18Δ),27 grown in SC medium for 3 d. We found that the TAG amounts (standardized with the optical density [OD600] of the culture) varied depending on the mutant strains. The TAG level was notably higher in atg2Δ, vps30Δ/atg6Δ, and atg14Δ cells (Fig. 1 A). This result seems to correlate with several lines of data in previous studies: One report analyzing the functions of a mammalian Atg2 homolog showed that this protein localizes on LDs and reduces LD amount,28 and the other study reported that knockdown of the mammalian Atg14 homolog, a component of the class III phosphatidylinositol 3-kinase complex together with BECN1 (Vps30/Atg6 homolog), elevated TAG levels in liver and serum.29
Figure 1.
For figure legend, see page 1250.
Interestingly, unlike other atg mutants, the TAG amount in atg15Δ cells was significantly lower than that in its parental wild-type cells (Fig. 1A). We also conducted fluorescence microscopy of LDs by staining the organelle with BODIPY 493/503. The number of LDs in the atg15Δ strain was comparable to that in the wild-type strain when the cells were grown in SC medium for 12 h to a mid-log growth phase (Fig. 1B–D). In contrast, the number of LDs was found to be markedly smaller at the post-diauxic phase in the atg15Δ cells relative to that in the wild-type cells (Fig. 1B–D). This drop in the number of LDs in the atg15Δ cells was evident throughout the culture period thereafter (Fig. 1C and D). In accordance with the microscopy analysis, the amounts of TAG and SE, the main components of LDs, in the atg15Δ cells were about half those in the wild-type cells when they were cultured in SC medium for 48 h (Fig. 1E and F). We confirmed the induction of autophagy under this culture period by the detection of GFP-Atg8 fluorescence in the vacuole (fluorescence microscopy), as well as the detection of a cleaved band from GFP-Atg8 (immunoblot analysis), in the cells cultured in SC medium for 48 h (Fig. S1A and B). In contrast, fluorescence microscopy of the vacuolar membrane (stained with FM 4–64 dye) and LD (stained with BODIPY 493/503) visualized few LDs inside the vacuole of the wild-type and atg15Δ cells 48 h after the start of the culture (Fig. S1C). These results demonstrate that Atg15 functions in the maintenance of LD amount in cells in post-diauxic to stationary phases, when autophagic activities, but not the morphological signs of stationary-phase lipophagy,22 are detected.
Vacuolar localization and the putative lipase motif of Atg15 are important for the maintenance of LD amount
Fluorescence microscopy observation of cells with GFP-tagged Atg15 showed the diffusion of GFP fluorescence in the vacuole, confirming that Atg15 was accumulated in this compartment (Fig. 2A). A previous work showed that Atg15 harbors a GXSXG putative lipase motif which is required for the degradation of autophagic bodies.25 Additionally, another study reported that when the endogenous Atg15 was replaced by an Atg15 derivative with an HDEL motif (an ER retention/retrieval signal in yeast) at its C terminus, the maturation of precursor Ape1, the main cargo of Cvt vesicles, was diminished.24 To address whether the putative lipase activity and vacuolar localization of Atg15 are required for its functional involvement in LD dynamics, we expressed in atg15Δ cells an Atg15 variant mutated within the putative lipase motif, Atg15S332A, or another Atg15 variant fused to the HDEL sequence (Atg15-HDEL). Both the number of LD and TAG level were markedly reduced in the mutants expressing Atg15S332A than in the corresponding wild-type cells when they were cultured in SC medium for 48 h (Fig. 2B and C). We confirmed ER localization of Atg15-GFP-HDEL expressed with its native promoter in atg15Δ cells under the same culture conditions (data not shown). The LD number and TAG level were also found decreased in the mutant cells with Atg15-HDEL cultured in SC medium for 48 h (Fig. 2B and C). Thus, these results indicate that the putative lipase activity of Atg15 in the vacuole is important for the LD maintenance in stationary phase.
Figure 2.

Enzymatic activity and vacuolar localization of Atg15 are required for the maintenance of LD dynamics. (A) ATG15-deleted cells co-expressing GFP-tagged Atg15 and Vph1 tagged with mCherry (a vacuole marker) under their own promoter regulations were grown in SC medium for 48 h. The cells were harvested and subjected to fluorescence microscopy. The merged image consists of a green image representing the GFP signal and a red image for the mCherry signal. The corresponding lightfield (DIC) image is also shown. Scale bar: 5 µm. (B) Visualization of LDs in atg15 mutants by fluorescence microscopy. LDs were stained with BODIPY 493/503 after the cells were grown in SC medium for 48 h. Vector means the backbone plasmid for the expression of Atg15 variants (pRS316). The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (C) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culture in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100% and other values were normalized. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells shown in (B) were counted and presented with SD values. At least 100 cells were used for the calculation. Statistical significance of the differences relative to the values of the atg15Δ strain harboring the vector was shown with asterisks (P< 0.01) and dagger symbols (P < 0.05).
Absence of macroautophagy suppresses the deficiency of LD in atg15Δ cells
Atg15 activity in the vacuole disintegrates multiple types of vesicles including those generated by the multivesicular body pathway and autophagic bodies. In order to investigate whether the involvement of Atg15 in LD dynamics indeed requires autophagic flux, we generated atg1Δ atg15Δ cells (Fig. 3A). The number of LDs and TAG level in atg1Δ atg15Δ cells after culturing the cells in SC medium for 48 h was found to be comparable to those in wild-type cells (Fig. 3B and C), indicating a cancellation of the phenotype of the ATG15-deleted strain by the simultaneous deletion of the ATG1 gene. This suggested that an Atg1-dependent activity is a prerequisite for the Atg15 involvement in LD dynamics. Similar results were obtained for other atg mutants defective in macroautophagy, as well as for a vac8Δ mutant that exhibits a significant drop in macroautophagic activity (Fig. 3D).30 In contrast, in deletion mutants of ATG11, ATG19, ATG21, or ATG32, the genes for selective autophagic pathways,31-35 the number of LD was decreased by additional disruption of ATG15 (Fig. 3D). These results indicated that the function of Atg15 in LD dynamics is dependent on macroautophagic flux to the vacuole, but not on selective autophagy.
Figure 3.
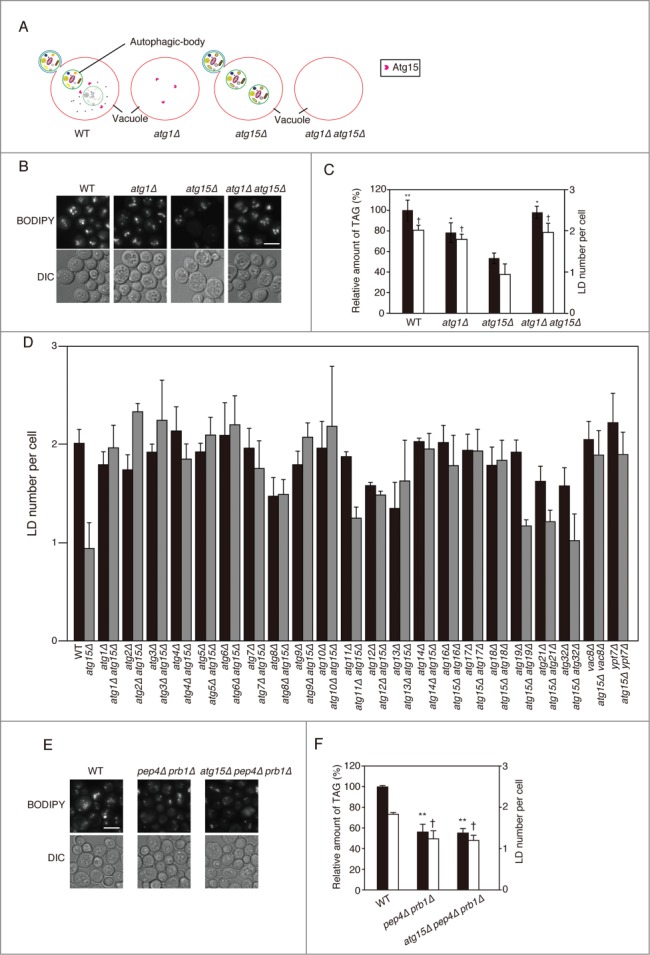
Effects of simultaneous disruptions of autophagy-related genes on LD abundance in the atg15Δ strain. (A) Schematic drawing of autophagosomes, autophagic bodies, and Atg15 in the denoted strains. (B) Fluorescence microscopy of LDs stained with BODIPY493/503 in the designated strains after culturing in SC medium for 48 h. The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (C) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culture in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100%. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells shown in (B) were presented with SD values. At least 100 cells were used for the calculation. The asterisk and dagger symbols indicate P-values < 0.05, and the double asterisk indicates a P-value < 0.005, in comparison with the values of the atg15Δ strain. (D) Quantifications of LD numbers in various deletion mutants of autophagy-related genes. The numbers of LDs in the denoted strains cultured in SC medium for 48 h were counted after the staining of LDs with BODIPY493/503 and fluorescence microscopy. The error bars show SD values. At least 100 cells were used for the calculation. (E) Fluorescence microscopy of LDs in the denoted mutant strains stained with BODIPY 493/503 after culturing in SC medium for 48 h. The corresponding lightfield (DIC) images are also shown. Scale bar: 5 µm. (F) (Left columns) The relative amounts of TAG in the denoted strains after 48 h culturing in SC medium were determined and shown so that the TAG level in the wild-type strain was presented as 100% and the other values were normalized. The error bars indicate SD values. (Right columns) The numbers of LDs in the cells were counted and presented with SD values. At least 100 cells were used for the calculation. Statistical significance of the differences was shown with the asterisk and dagger symbols indicating P-values < 0.05, and the double asterisk indicating a P-value < 0.01.
Next, we investigated whether accumulation of macroautophagic bodies caused by another approach led to the same phenotype as that seen in the atg15Δ cells. For this purpose we generated a pep4Δ prb1Δ strain that is devoid of the main proteases in the vacuole and accumulates macroautophagic bodies.23 After culture in SC medium for 48 h, this mutant strain exhibited a significant reduction in the amount of TAG and the number of LD in comparison with the wild-type strain, similar to atg15Δ cells (Fig. 3E and F). Further deletion of the ATG15 gene in the pep4Δ prb1Δ strain had no affect on the TAG level and the number of LD (Fig. 3E and F), strongly suggesting that the proteases and Atg15 act in the same process, that is, degradation of macroautophagic bodies.
In order to examine whether the LD amount was affected by blockade of the autophagy flux at another step, namely the fusion of autophagosomes with the vacuolar membrane, we generated ypt7Δ and atg15Δ ypt7Δ strains lacking a small GTPase protein that is required for the fusion step.36 We found that 1) the number of LDs in ypt7Δ cells was slightly increased similar to that in the wild-type cells, and unlike the decreased number of LDs seen in the atg15Δ strain, and 2) further deletion of the ATG15 gene in the ypt7Δ cells did not cause a remarkable decrease in the number of LDs (Fig. 3D). From these data we concluded that the accumulation of autophagic bodies in the vacuole, not that of autophagosomes in the cytoplasm, caused the decrease in the amount of LDs. And the latter result was consistent with our finding that the autophagic flux to the vacuole is a prerequisite of Atg15 involvement in LD dynamics.
The decrease of LDs in atg15Δ cells is dependent on Tgl3 and Tgl4
For elucidating the molecular mechanism underlying Atg15 function in the maintenance of LD dynamics, we hypothesized 2 possibilities responsible for the reduction of LD: 1) the synthesis of LDs is reduced in atg15Δ cells, or 2) the lipolytic activity is augmented in atg15Δ cells. To investigate the biosynthetic activity of LDs, we focused on 2 major acyltransferases, Dga1 and Lro1, acting in the final step of TAG synthesis.37 We generated Dga1-3 × HA and Lro1-3 × HA to compare the amounts of enzymes in cells in the steady state (48 h after the onset of the culture), but found no detectable differences between the mutant and wild-type cells (Fig. S2A). We also generated strains expressing Dga1-mCherry and Lro1-mCherry. Dga1 moves from the ER to LDs to synthesize TAG from DAG in the early stationary phase.38 In both the wild-type and atg15Δ cells, the Dga1-mCherry signal exhibited ring-shaped patterns characteristic of its ER localization, and it was not colocalized with the signal from BODIPY 493/503 representing LD localization, when the cells were cultured in SC medium for 48 h (Fig. S2B). In a previous study Lro1 was found localized adjacent to LDs at the diauxic phase.39 In contrast, fluorescence from Lro1-mCherry in the 48 h-cultured cells was not detected juxtaposed to LDs represented by the BODIPY 493/503 signal (Fig. S2B). Based on these observations, the biosynthesis process of LDs was shown to be inactivated both in the wild-type and atg15Δ cells under the conditions used for our analysis.
To investigate whether loss of Atg15 caused a reduction in LDs through lipolysis, we generated several mutants lacking the lipolysis enzymes. Fluorescence microscopy of LDs in these mutant strains indicated that while the numbers of the LDs in tgl3Δ and/or tgl4Δ mutants were comparable to that in the wild-type strain, the sizes of the LDs were augmented in the mutant strains. Notably, fluorescence microscopy with BODIPY 493/503-stained cells showed that the numbers of LDs in atg15Δ tgl3Δ, atg15Δ tgl4Δ and atg15Δ tgl3Δ tgl4Δ were similar to that in the tgl3Δ, tgl4Δ, and tgl3Δ tgl4Δ strains, respectively (Fig. 4A and B). The determination of TAG levels also showed that deletion of TGL3 and/or TGL4 cancelled the diminishment of LD abundance caused by disruption of ATG15 (Fig. 4B). Thus, Tgl3 and Tgl4 were found responsible for the degradation of LDs in atg15Δ cells.
Figure 4.
For figure legend, see page 1254.
Next we compared the expression levels of Tgl3 and Tgl4 between the wild-type and atg15Δ cells. Immunoblot analysis of Tgl3-3 × HA after normal SDS-PAGE showed no differences in the protein amount between wild-type and atg15Δ cells at any growth phases (Fig. 4C). However, when the cell lysates were applied to Phos-tag SDS-PAGE for better separating the phosphorylated and nonphosphorylated populations of Tgl3-3 × HA, we detected only the phosphorylated form (possessing less mobility during the electrophoresis) in the wild-type, and both phosphorylated and nonphosphorylated forms of the protein in the atg15Δ samples derived from 18-h and 24-h cultured cells (Fig. 4D). Since these time periods corresponded to those when a more remarkable decrease in the LD amount was induced in atg15Δ cells (Fig. 1D), this result implied that loss of ATG15 affected the phosphorylation status of Tgl3, leading to further activation of the enzyme.
While the protein abundance of Tgl4-3 × HA was found gradually decreased during the culture period in both the wild-type and atg15Δ cells, the decline of the Tgl4-3 × HA amount was delayed in the atg15Δ cells (Fig. 4C). Thus, the enhanced degradation of LDs in atg15Δ cells correlates with a higher amount of Tgl4 in the mutant than in the wild-type cells, as well as with a different phosphorylation status of Tgl3.
In order to address the localizations of the lipolytic enzymes, we carried out fluorescence microscopy of Tgl3 and Tgl4 tagged with mCherry after 48 h culture in SC medium (Fig. 4E). The signals from Tgl3-mCherry and Tgl4-mCherry were found as grain-like patterns in both wild-type and atg15Δ cells, but the colocalization frequencies with LDs (stained with BODIPY 493/503) were significantly higher in the atg15Δ cells than in the wild-type cells (Fig. 4E). This result showed consistency with the decrease in LD amounts in the atg15Δ cells.
Enhancement of lipolysis affects lipid metabolism in atg15Δ cells
In order to verify the upregulation of lipolytic activity in atg15Δ cells, we next carried out quantitative comparisons of metabolites related to mobilization of TAG, between the wild-type and atg15Δ cells. As shown in Figure 5A, atg15Δ cells cultured in SC medium for 48 h contained higher levels of diacylglycerol (DAG) than wild-type cells under the same culture conditions. The amount of free fatty acids (FFAs) was also higher in atg15Δ cells than in the wild-type cells (Fig. 5B).
Figure 5.

Increased levels of DAG, FFAs, and phospholipids in atg15Δ cells. (A) Quantification of DAG amount in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (B) Quantification of FFA amounts in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (C) Quantification of phospholipid amounts in wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The double asterisk indicates that the difference is statistically significant (P-value <0.005). (D) Quantification of phospholipid amounts in microsome fractions of wild-type and atg15Δ cells grown in SC medium for 48 h. The values from 3 independent experiments were plotted as mean ± SD. The asterisk indicates that the difference is statistically significant (P-value <0.05). (E) Wild-type and atg15Δ cells expressing Opi1 tagged with GFP under its own promoter regulation were cultured in SC medium for 48 h. Cells were harvested and subjected to fluorescence microscopy. The GFP images along with the corresponding lightfield (DIC) images are shown. The arrows indicate Opi1-GFP signals exhibiting an ER-localization pattern. Scale bar: 5 µm. (F) Cell viabilities of the denoted strains cultured in SC medium for the indicated time periods were calculated by fluorescence microscopy of dead cells stained with phloxine B. The error bars represent SD values.
Next, we speculated that the increase in the products of lipolysis (DAG and FFAs) could lead to enhanced production of phospholipids in the ER. To address this question, we quantified phospholipids in wild-type and atg15Δ cells. We found that the cellular phospholipid amount was higher in atg15Δ cells than that in wild-type cells (Fig. 5C). Since we could not exclude the possibility that the higher level of phospholipid amount in atg15Δ cells was caused by accumulation of autophagic bodies inside the vacuole, we also analyzed the phospholipid amount in the microsomal (ER) fraction prepared by subcellular fractionation. The phospholipid amount in the fraction was also higher in atg15Δ cells than in wild-type cells (Fig. 5D). Additionally, we observed the localization of Opi1, a negative regulator of enzymes for phospholipid synthesis.40 This protein is localized on the ER when phosphatidic acid is abundantly present in the organelle. Opi1-GFP was observed on ER membrane only in the atg15Δ cells, 48 h after culturing in SC medium (Fig. 5E). These results indicated that the mobilization of neutral lipids and the synthesis of phospholipids are activated in atg15Δ cells.
Finally, we investigated whether the changes in lipid metabolism by loss of Atg15 affected the physiological state of the cells, by comparing cell viabilities of the strains lacking ATG1, ATG15 and/or TGL3. While the cell viabilities of all the tested strains were comparable up to 5 d after the start of the culture in SC medium, the atg15Δ strain exhibited a more remarkable loss of viability than the wild type or even atg1Δ cells, after a prolonged culture for 7 to 9 d (Fig. 5F). This result was consistent with a previous study that investigated the cell viability of the atg15Δ strain under a nitrogen-starved condition.26 Of note, the loss of viability found in the atg15Δ strain was partially suppressed by further deletion of the TGL3 gene (atg15Δ tgl3Δ; Fig. 5F), suggesting that the enhanced lipolysis in the atg15Δ strain contributed to the loss of viability of this mutant strain to some extent.
Discussion
Atg15 was previously identified as the sole putative vacuolar lipase for degradation of autophagic bodies in S. cerevisiae.24,25 To our knowledge, the only functional association of Atg15 with LD metabolism unveiled in S. cerevisiae was the enzyme's participation in the final step of lipophagy.21 This previous study had shown higher accumulation of TAG and lowered lipase activities in the vacuoles of atg15Δ cells.21 In the present work, we revealed that the number of LDs and the amount of neutral lipids were remarkably lower in the atg15Δ strain, in comparison to the wild type and the other tested atg mutant strains cultured to stationary phase (Fig. 1). Results from these 2 studies are not contradictory to each other, since impairment of lipase activities in the vacuole of the mutant may provide a signal resulting in enhancement of lipolytic activity outside the organelle, leading to reduction in the LD amount as a whole. It is also notable that under our culture conditions, we were not able to detect the intravacuole population of LDs reported in a previous report on lipophagy in the stationary phase,22 in the wild-type nor atg15Δ cells (Fig. S1C). Since we utilized a different medium from that used in the previous study and observed the LD dynamics at earlier time points (up to 3 d) instead of the observation up to 7 d after the start of the culture in the previous study, the phenotype of the atg15Δ strain found in this study was thought to be independent from the outcome of lipophagy in the stationary phase.
Our findings uncovered the putative lipase's important role in the degradation of macroautophagic bodies in the stationary phase, as the phenotype of the atg15Δ cells was suppressed by the simultaneous deletion of ATG genes required for macroautophagy (Fig. 3). The failure to degrade macroautophagic bodies is thought to cause pleiotropic effects on the cell status. The most plausible consequence from this failure is deficiencies of compounds for the biosynthesis of cellular components, due to lack of recycling flow from the vacuole. Another possibility is that the remnant autophagic bodies inside the vacuoles of the mutant cells alter the intravacuolar environments, rendering the cells exposed to more stresses. Indeed, we detected a higher ROS accumulation in atg15Δ cells than in the wild-type cells grown to stationary phase (data not shown). These outcomes may cause signaling events to enhance lipolytic activities as discussed below.
Atg15 function in LD dynamics was investigated from the viewpoints of biosynthesis and degradation of the organelle. We could not detect any active populations of the TAG-synthesising enzymes (Dga1 and Lro1) under our culture conditions, or any differences in protein abundance and localization of these enzymes between the wild-type and atg15Δ cells (Fig. S2). In contrast, deletion of genes for lipolysis, TGL3 and/or TGL4, suppressed the reduction in LD and TAG amount in atg15Δ cells (Fig. 4A and B). Although the abundance of Tgl3 was found constant during the culturing to stationary phase and no difference in the amount of the enzyme was detected between the wild-type and atg15Δ cells (Fig. 4C), the phosphorylation status and localization of the protein were affected by loss of Atg15: in the atg15Δ strain, a nonphosphorylated population of the enzyme was detected, but not in the wild-type cells, and Tgl3-mCherry was found more closely associated with LDs in atg15Δ cells (Fig. 4D and E). A pioneering study on Tgl4 identified phosphorylation events leading to activation of the enzyme.14 A similar regulation may also exist for Tgl3 in the cells cultured to stationary phase, and the detailed mechanism should be addressed in the future.
In contrast to the constant expression of Tgl3, the intracellular amount of Tgl4 declined after the diauxic shift (Fig. 4C), and the kinetics of Tgl4 diminishment were found to be slower in atg15Δ cells than in the wild-type cells (Fig. 4C). In a mammalian experimental system, the amount of PNPLA2 (the ortholog of Tgl4) is regulated by degradation via the ubiquitin-proteasome pathway.41 Thus, it is possible that yeast Tgl4 is also degraded by the ubiquitin-proteasome pathway, whose activity is downregulated in atg15Δ cells after the diauxic phase.
A previous study revealed that mobilization of TAG by lipolysis supported efficient biosynthesis of phospholipids in the ER.42 Indeed, our data indicated a higher accumulation of phospholipids in the ER of atg15Δ cells, concomitant with the enhancement of mobilization of TAG in the mutant (Fig. 5A–D). Opi1, a negative regulator of many phospholipid synthesis enzymes that acts by sensing the abundance of phosphatidic acid at the ER,40 was localized on ER membranes in atg15Δ cells, but not in the wild-type cells (Fig. 5E), validating the accumulation of the phospholipid in atg15Δ cells. While the enhanced accumulation of phospholipids was deemed to be a consequence from the upregulation of lipolysis, we could not exclude the possibility that the other properties of Tgl3 and Tgl4, namely the acyltransferase activities of these enzymes,15,16 contributed to the elevated levels of phospholipids in atg15Δ cells. Such alternations in lipid metabolism by loss of Atg15 may have significant physiological impacts, since atg15Δ cells lose their viability during prolonged culture in SC medium more quickly than the wild-type, or even than atg1Δ cells, which was partially reversed by the loss of Tgl3 (Fig. 5F).
Although the association of Atg15 in lipophagy was recently analyzed,21 few studies have addressed the relation between general autophagy and lipolysis. Altogether, our data presented in this study suggested that degradation of autophagic bodies in the vacuole by Atg15 suppresses lipolysis after the diauxic shift. These findings provide novel insight into the regulatory network of lipid metabolism and autophagy.
Materials and Methods
Yeast strains, plasmids, and culture conditions
Saccharomyces cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used as the parental wild-type strain. Cells were grown in SC medium containing 0.67% yeast nitrogen base without amino acids (BD, 291940), 2% glucose, and 0.192% (w/v) yeast synthetic drop-out medium supplements without tryptophan (Sigma-Aldrich, Y1876-20G). All the strains and plasmids used in this study are listed in Table S1 and Table S2. The construction of plasmids for expressing Atg15, Atg15S332A or Atg15-HDEL in yeast cells was done as follows: The genomic sequence covering the 5′ promoter, open reading frame, and 3′-untranslated regions of ATG15 was amplified by PCR using primers as follows, forward; 5′-GCGTTCACAGTTCTCATTTACACTGTAC-3′, reverse; 5′–GCATCCATTAGTTGATCTAGACCAGAAG-3′, then digested with XbaI and XhoI, and ligated into XbaI/XhoI-digested pRS316, yielding the pMB100 plasmid. Plasmids for expressing Atg15S332A or Atg15-HDEL were generated by the inverse PCR method using pMB100 as a template, which yielded pMB101 (encoding Atg15S332A) and pMB102 (encoding Atg15-HDEL). The plasmid for expression of Atg15-GFP was generated by the gap-repair cloning method:43 a GFP-coding DNA fragment was amplified by PCR and cotransformed into yeast cells with a gene fragment that was amplified with pMB100 as a template. The resultant plasmid encoding Atg15-GFP (pMB103) was retrieved form the yeast cells. The yeast gene-deletion strains and the strains expressing the epitope-tagged proteins other than Atg15-GFP were obtained from EUROSCARF or generated by a PCR-based transformation method.44 The construction of the double gene-deletion strain was performed as described previously.45 Successful gene disruptions were verified by PCR.
The culturing of the cells in SC medium was done at 28°C with a starting OD600 = 0.1.
Lipid extraction and lipid analyses
Two OD600 units of the cells grown in SC medium were harvested by centrifugation, washed once with distilled water, resuspended in 280 µL of 50 mM Tris-HCl buffer (pH 7.0), and subjected to glass-bead disruption. Extraction of lipids from the cell lysate was performed by the Bligh and Dyer method with some modifications.46 Briefly, to the cell lysate obtained after the glass-bead disruption, 1 ml of chloroform/methanol (1:2), 350 µl of chloroform, and 200 µl of distilled water were successively added and mixed. The samples were centrifuged at 1,000 × g for 10 min and the organic phase was retrieved and dried. For separations of neutral lipids, the dried samples were dissolved in chloroform/methanol (2:1) and applied to thin layer chromatography. TAG and SE were separated using silica gel 60 plates (Merck Millipore, 1.05721.0001) and solvent systems of light petroleum:diethyl ether:acetic acid (30:70:2) followed by light petroleum:diethyl ether (49:1).47 For separations of DAG and FFA, a mixture of chloroform:acetone:acetic acid (90:8:1) was utilized as the solvent system. The separated lipids were visualized by immersing the plates in 10% CuSO4 (w/v) solution containing 10% H3PO4 (v/v) and by heating them to 120°C for 30 min. The quantifications of the lipid amounts were done by densitometry using ImageJ software (NIH).48
Quantifications of total phospholipids were conducted according to the method established by Broekhuyse.49 Briefly, total cell lipid extracts were applied to 2-dimensional thin layer chromatography using chloroform:methanol:25% ammonia (65:35:5) as the solvent system for the first dimension, and chloroform:acetone:methanol:acetic acid:water (50:20:10:10:5) for the second dimension. The spots for 5 phospholipids (phosphatidic acid, phosphatidylcholine, phosphatidylethanolamine, phosphatidylinositol, and phosphatidylserine) visualized by staining with iodine vapor were scraped off from the plate, and subjected to determinations of phosphorus.
Fluorescence microscopy
Cells were stained with BODIPY 493/503 (Invitrogen, D-3922) in the dark for 10 min at room temperature, washed twice with phosphate-buffered saline (137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, and 1.8 mM KH2PO4, pH adjusted to 7.0 with HCl), and immediately subjected to microscopy. Fluorescence microscopy was performed using an IX81 inverted microscope (Olympus) equipped with Uplan Apo 100 × 1.35 oil iris objective lens. A XF104-3 filter set (Omega Optics) was used for acquiring signals from BODIPY 493/503 and GFP, and mCherry was visualized with a 546/512-nm bandpass excitation filter, a 560-nm dichromatic mirror and a 575/640-nm bandpass emission filter (Omega Optics). Images were processed by Adobe Photoshop software.
For determinations of LD numbers, we manually counted the grain-shaped signals from BODIPY 493/503 staining; when we could discern several grain-shaped structures in one chunk of the signal, we counted the chunk as several LDs.
Immunoblot analyses
Total cell lysates for the immunoblot analyses were prepared as follows: Briefly, the cells were centrifuged, washed with distilled water, and resuspended in 0.2 M NaOH solution containing 100 mM dithiothreitol (DTT) for 10 min on ice. The samples were mixed with 1/9 volume of trichloroacetic acid (100% w/v), and after an additional incubation for 15 min on ice, they were subjected to centrifugation at 13,000 × g for 5 min at 4°C. The pellet fractions were resuspended in acetone and subjected to centrifugation at 13,000 × g for 5 min at 4°C. The resultant pellet fractions were dried and resuspended in 1 × SDS sample buffer (0.1 M Tris-HCl, pH 7.5, 2% SDS [Wako, 191-07145], 10% glycerol, 20 mM DTT, trace amount of bromophenol blue), incubated for 5 min at 60°C, and then boiled for 3 min. After centrifugation at 10,000 × g for 1 min at room temperature, the supernatant fractions were resolved by SDS-PAGE. To detect phosphorylation-dependent mobility shifts of Tgl3-3 × HA, the whole cell extracts were loaded onto 5.5% SDS-PAGE gels containing 20 µM Phos-tag acrylamide (Wako, 304-93521) and 100 µM MnCl2. The proteins separated by SDS-PAGE were transferred to Immobilon P polyvinylidene difluoride membrane (Merck Millipore, IPVH00010) using a Trans-Blot SD semi-dry transfer cell (Bio-Rad, 170-3940JA), and detected with the indicated antibodies: anti-HA antibody (F-7) (Santa Crutz Biotechnology, sc-7392); anti-Act1 antibody (Abcam, ab8224); anti-GFP monoclonal antiserum (Clontech, 632381); anti–mouse IgG conjugated with HRP (MBL, PM009-7).
Chemiluminescence detection was done with the Western Lightning kit (Perkin-Elmer Life Science, NEL105001EA) and the signals were analyzed with Light-Capture II (ATTO, 2104451).
Cell fractionation
For the preparation of spheroplasts, cells were harvested by centrifugation at 4,000 × g for 5 min and washed twice with sterilized water. The cells (0.5 g wet weight/ml) were resuspended in buffer A (Tris-HCl, pH 9.4, 10 mM DTT) and incubated at 30°C for 20 min. Afterwards cells were washed once with 1.2 M sorbitol (Wako, 198-03755) and suspended in buffer B (1.2 M sorbitol, 20 mM phosphate potassium buffer, pH 7.4). Zymolyase-100T (Nacalai Tesque, 07665-55) was added at a concentration of 2 mg/g wet weight of the cells. The suspension was incubated at 30°C with gentle shaking for 60 min. Spheroplasts were collected by centrifugation for 5 min at 1,500 × g, and washed twice with 1.2 M sorbitol.
For microsome isolation, the spheroplasts were resuspended in buffer C (5 mM MES-KOH, pH 6.0, 0.6 M sorbitol, 1 mM KCl, 0.5 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail [Roche, 11873580001]) and chilled on ice. The spheroplasts were homogenized with a Dounce homogenizer by applying 20 strokes using a tight fitting pistil. After centrifugation at 3,000 × g for 5 min at 4°C to remove cell debris and nuclei, the supernatant fractions were collected, and homogenized again, and subjected to ultracentrifugation at 27,000 × g for 15 min at 4°C in a SW41Ti rotor (Beckman). The floating layers were transferred to a new tube, and were subjected to ultracentrifugation at 40,000 × g for 30 min at 4°C in the SW41Ti rotor. The pellet fractions consisting of highly enriched microsomes were collected, resuspended in 100 mM phosphate-potassium buffer (pH 7.9) and reserved at −80°C until use.
Assessment of cell viability
The cell viabilities were assayed by staining the cultured cells with phloxine B (Nacalai Tesque, 27514-92) and fluorescence microscopy, as described previously.50
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Grant-in-Aid for Scientific Research on Priority Areas 26111511 (to YS) from the Ministry of Education, Culture, Sports, Science and Technology, Japan and by Grant-in-Aid for Young Scientists (B) 26850064 (to MO) from the Japan Society for the Promotion of Science.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem 2002; 277:44507-12; PMID:12221100; http://dx.doi.org/ 10.1074/jbc.M207712200 [DOI] [PubMed] [Google Scholar]
- 2.Gaspar ML, Hofbauer HF, Kohlwein SD, Henry SA. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J Biol Chem 2011; 286:1696-708; PMID:20972264; http://dx.doi.org/ 10.1074/jbc.M110.172296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horvath SE, Wagner A, Steyrer E, Daum G. Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 2011; 1811:1030-7; PMID:21875690; http://dx.doi.org/ 10.1016/j.bbalip.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanghellini J, Natter K, Jungreuthmayer C, Thalhammer A, Kurat CF, Gogg-Fassolter G, Kohlwein SD, von Grünberg H-H. Quantitative modeling of triacylglycerol homeostasis in yeast–metabolic requirement for lipolysis to promote membrane lipid synthesis and cellular growth. FEBS J 2008; 275:5552-63; PMID:18959743; http://dx.doi.org/ 10.1111/j.1742-4658.2008.06681.x [DOI] [PubMed] [Google Scholar]
- 5.Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RGW, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol 2006; 173:719-31; PMID:16735577; http://dx.doi.org/ 10.1083/jcb.200511125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman JM. The Gregarious Lipid Droplet. J Biol Chem 2008; 283:28005-9; PMID:18611863; http://dx.doi.org/ 10.1074/jbc.R800042200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfisterer SG, Bakula D, Cezanne A, Robenek H, Proikas-Cezanne T. WIPI beta-propellers at the crossroads of autophagosome and lipid droplet dynamics. Biochem Soc Trans 2014; 42:1414-7; PMID:25233424; http://dx.doi.org/ 10.1042/BST20140152 [DOI] [PubMed] [Google Scholar]
- 8.Kassan A, Herms A, Fernández-Vidal A, Bosch M, Schieber NL, Reddy BJN, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, et al.. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol 2013; 203:985-1001; PMID:24368806; http://dx.doi.org/ 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem 2002; 277:6478-82; PMID:11741946; http://dx.doi.org/ 10.1074/jbc.M109109200 [DOI] [PubMed] [Google Scholar]
- 10.Walther TC, Farese RV. The life of lipid droplets. Biochim Biophys Acta 2009; 1791:459-66; PMID:19041421; http://dx.doi.org/ 10.1016/j.bbalip.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 2009; 48:275-97; PMID:19464318; http://dx.doi.org/ 10.1016/j.plipres.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Athenstaedt K, Daum G. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J Biol Chem 2003; 278:23317-23; PMID:12682047; http://dx.doi.org/ 10.1074/jbc.M302577200 [DOI] [PubMed] [Google Scholar]
- 13.Athenstaedt K, Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J Biol Chem 2005; 280:37301-9; PMID:16135509; http://dx.doi.org/ 10.1074/jbc.M507261200 [DOI] [PubMed] [Google Scholar]
- 14.Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell 2009; 33:53-63; PMID:19150427; http://dx.doi.org/ 10.1016/j.molcel.2008.12.019 [DOI] [PubMed] [Google Scholar]
- 15.Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell 2010; 21:501-10; PMID:20016004; http://dx.doi.org/ 10.1091/mbc.E09-09-0775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajakumari S, Daum G. Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J Biol Chem 2010; 285:15769-76; PMID:20332534; http://dx.doi.org/ 10.1074/jbc.M109.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H, Czaja MJ. Regulation of lipid droplets by autophagy. Trends Endocrinol Metab 2011; 22:234-40; PMID:21419642; http://dx.doi.org/ 10.1016/j.tem.2011.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; PMID:19339967; http://dx.doi.org/ 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest 2009; 119:3329-39; PMID:19855132; http://dx.doi.org/ 10.1172/JCI35541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moeller CH, Thomson WW. Uptake of lipid bodies by the yeast vacuole involving areas of the tonoplast depleted of intramembranous particles. J Ultrastruct Res 1979; 68:38-45; PMID:379363; http://dx.doi.org/ 10.1016/S0022-5320(79)90140-0 [DOI] [PubMed] [Google Scholar]
- 21.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2014; 25:290-301; PMID:24258026; http://dx.doi.org/ 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C-W, Miao Y-H, Chang Y-S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 2014; 206:357-66; PMID:25070953; http://dx.doi.org/ 10.1083/jcb.201404115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992; 119:301-11; PMID:1400575; http://dx.doi.org/ 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epple UD, Eskelinen E-L, Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J Biol Chem 2003; 278:7810-21; PMID:12499386; http://dx.doi.org/ 10.1074/jbc.M209309200 [DOI] [PubMed] [Google Scholar]
- 25.Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. Aut5/Cvt17p, a Putative Lipase Essential for Disintegration of Autophagic Bodies inside the Vacuole. J Bacteriol 2001; 183:5942-55; PMID:11566994; http://dx.doi.org/ 10.1128/JB.183.20.5942-5955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem 2001; 276:2083-7; PMID:11085977; http://dx.doi.org/ 10.1074/jbc.C000739200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 28.Velikkakath AKG, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 2012; 23:896-909; PMID:22219374; http://dx.doi.org/ 10.1091/mbc.E11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem 2012; 287:39107-14; PMID:22992773; http://dx.doi.org/ 10.1074/jbc.M112.412569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott SV, Nice DC 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem 2000; 275:25840-9; PMID:10837477; http://dx.doi.org/ 10.1074/jbc.M002813200 [DOI] [PubMed] [Google Scholar]
- 31.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 2009; 17:98-109; PMID:19619495; http://dx.doi.org/ 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol 2001; 153:381-96; PMID:11309418; http://dx.doi.org/ 10.1083/jcb.153.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 2009; 17:87-97; PMID:19619494; http://dx.doi.org/ 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 34.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell 2001; 7:1131-41; PMID:11430817; http://dx.doi.org/ 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell 2004; 15:3553-66; PMID:15155809; http://dx.doi.org/ 10.1091/mbc.E04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435-46; PMID:10525546; http://dx.doi.org/ 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorger D, Daum G. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J Bacteriol 2002; 184:519-24; PMID:11751830; http://dx.doi.org/ 10.1128/JB.184.2.519-524.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, Rapoport TA. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep 2014; 6:44-55; PMID:24373967; http://dx.doi.org/ 10.1016/j.celrep.2013.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C-W, Lee S-C. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J Cell Sci 2012; 125:2930-9; PMID:22454508; http://dx.doi.org/ 10.1242/jcs.100230 [DOI] [PubMed] [Google Scholar]
- 40.Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem 2007; 282:37293-7; PMID:17981800; http://dx.doi.org/ 10.1074/jbc.R700038200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai Z, Qi W, Li C, Lu J, Mao Y, Yao Y, Li L, Zhang T, Hong H, Li S, et al.. Dual regulation of adipose triglyceride lipase by pigment epithelium-derived factor: a novel mechanistic insight into progressive obesity. Mol Cell Endocrinol 2013; 377:123-34; PMID:23850519; http://dx.doi.org/ 10.1016/j.mce.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 42.Rajakumari S, Rajasekharan R, Daum G. Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 2010; 1801:1314-22; PMID:20727985; http://dx.doi.org/ 10.1016/j.bbalip.2010.08.004 [DOI] [PubMed] [Google Scholar]
- 43.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res 1997; 25:451-2; PMID:9016579; http://dx.doi.org/ 10.1093/nar/25.2.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/ 10.1002/(SICI)1097-0061(199807)14:10%3c953::AID-YEA293%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 45.Chinen T, Ota Y, Nagumo Y, Masumoto H, Usui T. Construction of multidrug-sensitive yeast with high sporulation efficiency. Biosci Biotechnol Biochem 2011; 75:1588-93; PMID:21821930; http://dx.doi.org/ 10.1271/bbb.110311 [DOI] [PubMed] [Google Scholar]
- 46.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959; 37:911-7; PMID:13671378; http://dx.doi.org/ 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 47.Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast 1994; 10:1421-8; PMID:7871881; http://dx.doi.org/ 10.1002/yea.320101105 [DOI] [PubMed] [Google Scholar]
- 48.Girish V, Vijayalakshmi A. Affordable image analysis using NIH Image/ImageJ. Indian J Cancer 2004; 47. [PubMed] [Google Scholar]
- 49.Broekhuyse RM. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim Biophys Acta 1968; 152:307-15; PMID:4296335; http://dx.doi.org/ 10.1016/0005-2760(68)90038-6 [DOI] [PubMed] [Google Scholar]
- 50.Noda T. Viability assays to monitor yeast autophagy. Methods in enzymology 2008; 451:27-32; PMID:19185710 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




