Abstract
Purpose.
To explore different molecular factors impairing the activities of superoxide dismutase (SOD) isoforms in senile cataractous lenses.
Methods.
Enzyme activity of SOD isoforms, levels of their corresponding cofactors copper (Cu), manganese (Mn), zinc (Zn), and expression of mRNA transcripts and proteins were determined in the lenses of human subjects with and without cataract. DNA from lens epithelium (LE) and peripheral blood was isolated. Polymerase chain reaction–single strand conformation polymorphism (PCR-SSCP) followed by sequencing was carried out to screen somatic mutations. The impact of intronic insertion/deletion (INDEL) variations on the splicing process and on the resultant transcript was evaluated. Genotyping of IVS4+42delG polymorphism of SOD1 gene was done by PCR–restriction fragment length polymorphism (RFLP).
Results.
A significant decrease in Cu/Zn- and Mn-SOD activity (P < 0.001) and in Cu/Zn-SOD transcript (P < 0.001) and its protein (P < 0.05) were found in cataractous lenses. No significant change in the level of copper (P = 0.36) and an increase in the level of manganese (P = 0.01) and zinc (P = 0.02) were observed in cataractous lenses. A significant positive correlation between the level of Cu/Zn-SOD activity and the levels of Cu (P = 0.003) and Zn (P = 0.005) was found in the cataractous lenses. DNA sequencing revealed three intronic INDEL variations in exon4 of SOD1 gene. Splice-junction analysis showed the potential of IVS4+42delG in creating a new cryptic acceptor site. If it is involved in alternate splicing, it could result in generation of SOD1 mRNA transcripts lacking exon4 region. Transcript analysis revealed the presence of complete SOD1 mRNA transcripts. Genotyping revealed the presence of IVS4+42delG polymorphism in all subjects.
Conclusions.
The decrease in the activity of SOD1 isoform in cataractous lenses was associated with the decreased level of mRNA transcripts and their protein expression and was not associated with either modulation in the level of enzyme cofactors or with INDEL variations.
Keywords: superoxide dismutase, cofactors, somatic mutation, splice-transcript variant, senile cataract
Superoxide dismutase is an important defense enzyme against oxidative stress. Impaired enzyme activity during cataractogenesis was not owing to modulation in enzyme cofactors and genetic polymorphisms.
Introduction
Despite having a powerful antioxidant system, the aging human lens often falls prey to oxidative stress (OS) owing to constant exposure to factors producing reactive oxygen species (ROS).1 Superoxide dismutase (SOD) is most important in this milieu as it provides the first line of defense against deleterious ROS. Three isoforms of SOD, cytosolic copper/zinc–dependent Cu/Zn-SOD (or SOD1), mitochondrial manganese-dependent Mn-SOD (or SOD2), and an extracellular (EC) Cu/Zn-dependent EC-SOD (or SOD3), have been identified. All SOD isoforms catalyze the conversion of highly reactive, more dangerous superoxide anion (O2.-) into less reactive hydrogen peroxide (H2O2) and molecular oxygen. Hydrogen peroxide is then converted into water by either catalase (CAT) or glutathione peroxidase (GPX).2 However, failure in any of the conversion processes or imbalance between the production of ROS and the antioxidant enzymes results in tissue damage, which leads to OS-induced cataract.3 Cataract is defined as a gradual, painless loss of lens transparency that impedes the passage of light to the retina and impairs vision. It is one of the leading causes of blindness in elderly individuals over the age of 50 years. It is estimated that there are 16 million cases of cataract worldwide, with approximately 50% of the cases originating from Africa and Asia.4 Surgical intervention is the only available remedy at present.
Activity levels of antioxidant enzymes SOD, CAT, and GPX are impaired in human senile cataractous lenses.5–11 A few studies have suggested that impaired SOD isoforms activity in cataractous lenses could be associated with physiological and molecular factors such as abnormal levels of cofactors,12 diabetes-induced glycation,13,14 and genetic polymorphisms of SOD genes.15 Other molecular factors such as production of aberrant SOD transcripts as a result of alternate splicing,16–18 mutations in coding and noncoding regions,19,20 and epigenetic control through promoter hypermethylation21 have also been reported in other diseases. However, the precise molecular mechanisms and/or factors impairing SOD isoforms activity in senile cataractous lenses have not been well established to date.
Thus, the present study was designed to explore the molecular factors impairing SOD isoform activity in senile cataracts. The level of SOD isoforms activity and their cofactors such as copper, zinc, and manganese, and their mRNA transcripts and proteins were assessed. Screening of mutations and/or single nucleotide polymorphisms in the coding and noncoding regions of SOD1 was carried out. Further, an attempt was also made to find the association of IVS4+42delG polymorphism of SOD1 with senile cataracts.
Methods
Subjects and Samples
The present study adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Ethical Committee. All patients of age ≥ 50 years were clinically examined and diagnosed by the chief ophthalmologist (ARV). Using slit-lamp observation, the type of cataract was categorized by the zone of opacification and the grade was categorized by the degree and color of opalescence using the lens opacification classification system (LOCS) III.22 Patients with primary cataract were included. Written informed consent was obtained from all the participants.
Anterior lens epithelium (LE) and peripheral cortical fibers along with the lens nucleus (LN) were obtained from the cataract patients recruited for extracapsular cataract extraction surgery at Raghudeep Eye Clinic, Ahmedabad (n = 134: nuclear [47], cortical [46], and posterior subcapsular [41]; mean age, 67.1 ± 11.8 years). Human donor eyes were obtained from C.S. Samaria Red Cross Eye Bank, Ahmedabad. From these donor eyes, whole lenses were extracted within 6 to 8 hours of death and examined for the presence of opacity. Clear lenses thus obtained (n = 37; mean age, 61.4 ± 10.1 years) were included in the study. Lens epithelium and LN were separated from the clear lenses under an operating microscope (Carl Zeiss Microscopy GmbH, Göttingen, Germany). Peripheral venous blood (2–5 mL) was collected from the cataract (n = 100; mean age, 65.4 ± 9.2 years) and noncataract control (n = 100; mean age, 66.4 ± 8.7 years) subjects.
Determination of SOD Activity
Each of the LE and LN samples was homogenized separately in a homogenization buffer (100 mM sodium pyrophosphate buffer, pH 7.4). The enzyme source was obtained by centrifugation at 10,000g for 15 minutes at 4°C. The total protein level was determined using the Micro BCA protein estimation kit (Pierce Biotechnology, Rockford, IL). The levels of total Cu/Zn-SOD and Mn-SOD activities were determined using SOD Activity Assay Kit (BioVision Inc., Milpitas, CA) according to the manufacturer's instruction, with slight modifications. The activity of Mn-dependent SOD was discriminated from Cu/Zn-dependent SOD by incubating the enzyme source with 5 mM sodium cyanide because Cu/Zn-dependent SODs are sensitive to cyanide.11 SOD activity was expressed as a unit activity per milligram of protein.
Estimation of Cofactors
Lens epithelium and LN of the same lens samples were pooled separately in acid-washed amber glass vials and dried under reduced pressure at 37°C in a hot air oven for the first 1 hour, and then dried overnight at 60°C (for approximately 16 hours). The dry weight of the samples was obtained, and digested in a 5-mL mix of nitric acid and perchloric acid (3:1) in a closed chamber overnight or until the solid particle was no longer visible. The digested contents were preheated at 100°C in a microwave oven for the first hour and then at 200°C until the contents became colorless and the acid content was reduced to 2 to 3 mL. The digested contents were cooled, mixed with the required volume of distilled water, filtered through a Whatman No. 42 filter paper (GE Healthcare Life Sciences, Piscataway, NJ), and made up to 50 mL with HPLC-grade distilled water. The quantities of Cu, Zn, and Mn were estimated using Spectra AA220 Zeeman flame atomic absorption spectrophotometer (Varian Australia Pty Ltd., Mulgrave, Victoria) and expressed as micrograms per gram of dry tissue weight.
Quantitative Real-Time PCR Analysis
Lens epithelium and LN of the same lenses were pooled and washed with sterile PBS, pH 7.4. The lenses were homogenized in 2 mL TriZol reagent (Invitrogen Inc., Carlsbad, CA) and total RNA was extracted. cDNA was synthesized using First Strand cDNA Synthesis Kit (Invitrogen Inc.) according to the manufacturer's instructions in a 20-μL reaction volume containing 1.0 μg total RNA). SOD gene isoforms were amplified in a 20-μL reaction volume containing 1X of SYBR green master mix (Roche Diagnostics GmbH, Mannheim, Germany), 100 ng cDNA, and 10 pmol each of forward and reverse primers (Supplementary Table S1) using standard operating program, which consists of preincubation at 95°C for 10 minutes, amplification at 95°C for 10 seconds, annealing at 60°C for 10 seconds, and extension at 72°C for 10 seconds in a LightCycler 480II real-time PCR system (Roche Diagnostics GmbH). Level of β actin (ACTB) gene expression was kept as a control to normalize the expression status. The difference in the level of relative expression between the clear and the cataract samples was calculated based on the 2−ΔΔCp function using the Relative Expression Software Tool (REST) 2009.23
Western Blot Analysis
Lens epithelium and LN of the same lenses were pooled and lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (10 mmol/L tris-HCl, 150 mmol/L NaCl, 1.0% NP40, 0.25% deoxycholate, 0.1 mmol/L phenylmethanesulfonyl fluoride (PMSF), 0.1 mmol/L iodoacetamide, 10 μg/mL leupeptin, and 10 μg/mL aprotinin; pH 7.4) at 4°C for 30 minutes. The supernatant was obtained by centrifugation at 12,000g for 10 minutes. Approximately 50 μg total protein was separated by 12% SDS-PAGE, and subsequently blotted onto a nitrocellulose (NC) membrane (100 V for 1 hour). The membrane was blocked in an incubation buffer (10 mmol/L tris-HCl, 50 mmol/L NaCl, 0.05% Tween 20, pH 7.5, containing 5% BSA) at room temperature (RT) for 1 hour followed by incubation with anti-SOD polyclonal antibodies (rabbit anti-SOD1; GenScript, Piscataway, NJ; and rabbit anti-SOD2 and anti-SOD3; Abcam Inc., Cambridge, MA) overnight at 4°C. Nitrocellulose membrane was washed thrice, incubated with goat anti-rabbit IgG-horse-radish peroxidase conjugate for 2 hours at RT, and then visualized using diaminobenzidine (DAB)/H2O2 detection method. The band intensity of SOD1 protein levels was normalized using β-actin protein levels. The relative band intensity between SOD1 and β-actin was calculated using ImageJ (provided in the public domain by http://rsbweb.nih.gov/ij/) program.
Screening of Mutations and Single Nucleotide Polymorphisms (SNP)
DNA was extracted from LE and peripheral blood using the NucleoSpin Blood Genomic DNA Extraction Kit (Macherey-Nagel GmbH & Co., Düren, Germany) according to the manufacturer's instruction. Primers for all exons of SOD1 gene were designed19 (Supplementary Table S2) to amplify 5′ and 3′ splice sites along with coding sequences. Polymerase chain reaction was performed using 1X SappireAmp Fast PCR Master Mix (TaKaRa Bio Inc., Shiga, Japan) in a 50-μL reaction mix consisting of 100 ng genomic DNA and 50 pmol each of forward and reverse primers. The thermal reaction comprised 1 cycle of initial denaturation at 94°C/1 min, and 40 cycles of second denaturation at 98°C/5 s; annealing at 55°C to 58°C for 5 seconds; extension at 72°C/10 s, and a final extension at 72°C/3 min. All amplicons were resolved using 2% agarose gel and visualized by UV-transilluminator upon ethidium bromide staining. Single strand conformation polymorphism (SSCP) analysis24 using 10% polyacrylamide gel followed by silver staining25 was done for all amplicons. The gels were photographed, and the bands that showed a mobility shift were sequenced (First BASE Laboratories, Selangor Darul Ehsan, Malaysia) using the BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), in an Applied Biosystems Prism 3130xl Genetic Analyzer. Sequence homology and/or nucleotide polymorphisms in the coding and noncoding regions were assessed using the Basic Alignment Sequence Tool (BLAST) (provided in the public domain by www.ncbi.nlm.nih.gov/blast) program.
Splice-Junction and Alternate-Splice Transcript Analysis
Splice-junction analysis was done using algorithms, SCAN and RI of SEQUENCE WALKER26,27 program to determine the potential effect of insertion/deletion (INDEL) variations on native donor and acceptor sites. Further, the information contribution at each newly formed cryptic acceptor and donor site was determined. Reverse Transcription PCR was performed using three specific primer pairs (Supplementary Table S3)28 to check the presence of alternate splice-transcripts. Amplicons were resolved using 2% agarose gel.
Genotyping by PCR-Restriction Fragment Length Polymorphism (RFLP)
The potential loss or gain of restriction sites owing to INDEL variations on exon4 was determined using the restriction mapper online tool (provided in the public domain by www.restrictionmapper.org). DNA was isolated from LE of cataract and cadaveric donor eyes and from peripheral blood of cataract and control subjects. Exon4 was amplified, digested with 0.1 to 1 U of TatI enzyme at 65°C overnight. Digested products were resolved using 4% agarose gel.
Statistical Analysis
Statistical analyses were performed with SPSS. Differences between the means of the two variables were evaluated by the Student's t-test and the Mann-Whitney U test. The Pearson correlation analysis was done to check the influence of modulated levels of cofactors on corresponding enzymatic activity in the cataractous lenses. A P value of <0.05 was considered statistically significant.
Results
SOD Isoforms Activity
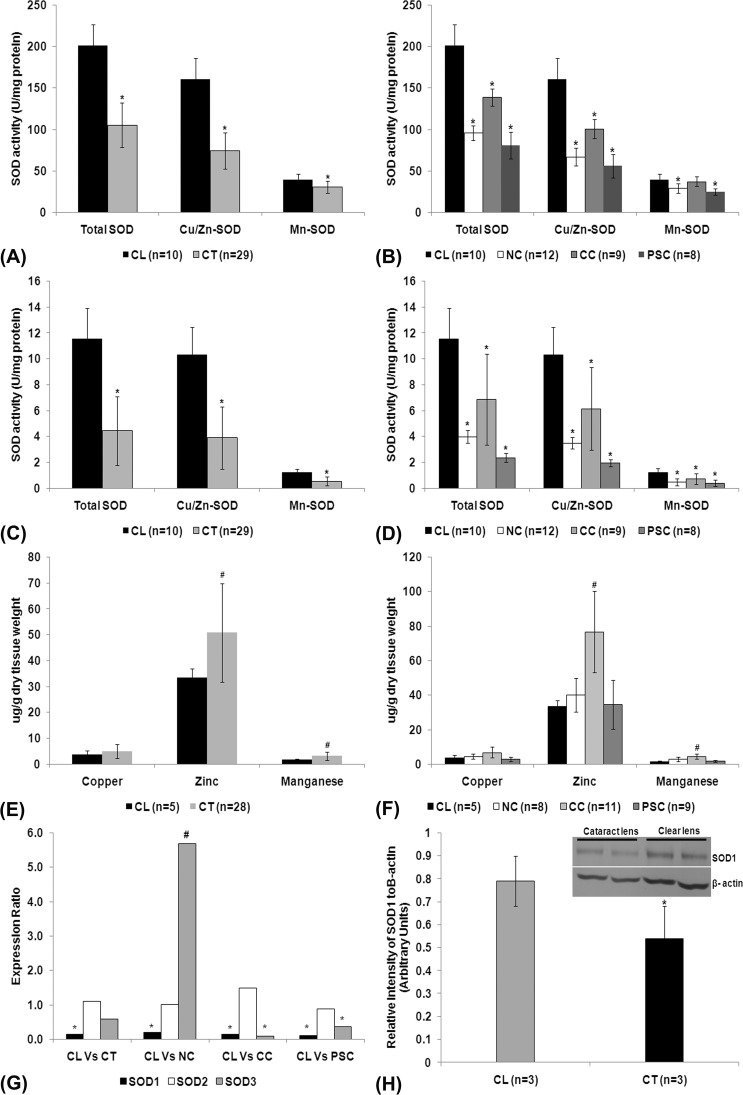
The LE showed a higher level of SOD activity, almost 15- to 20-fold, as compared with the LN of both clear (n = 10) and cataractous lenses (n = 29). However, when compared with clear lenses, cataract lenses showed a decreased level of total Cu/Zn- and Mn-SOD (P < 0.05; Figs. 1A, 1C, Supplementary Table S4) in both LE and LN. There was a significant decrease in the level of total Cu/Zn- and Mn-SOD activity in LE and LN of nuclear, cortical, and posterior subcapsular cataracts (P < 0.05; Figs. 1B, 1D) except Mn-SOD in LE of cortical cataract (P = 0.45). Further, the level of total and Cu/Zn-SOD activity in LE and LN of cataractous lenses was approximately 50% less than in the clear lenses. The level of Mn-SOD activity was negligible when compared with total activity in both clear and cataract lenses.
Figure 1.
SOD activity in lens epithelium (A), lens nucleus (C), and lens epithelium and lens nucleus of different types of cataract (B, D). Values are expressed as mean ± SD; * indicates significant decrease in the level of activity (P < 0.05). Estimation of cofactors in lenses (E) and in different types of cataracts (F); values were expressed as mean ± SD; # indicates a significant increase in the level of cofactors (P < 0.05). (G) Real-time quantitative PCR analysis of SOD isoform transcripts in clear (n = 12) and cataractous (n = 54) lenses and also in different types of cataracts. Values were expressed as means expression ratio. * indicates significant downregulation (P < 0.05) and # indicates significant upregulation (P < 0.05). (H) Western blot (inlet) and densitometric analysis of SOD1 protein levels in lenses. Relative band intensity analysis revealed a significant decrease in the level of SOD1 protein (*P < 0.05) in the cataractous lenses. CL, clear lens; CT, cataract lens; NC, nuclear cataract; CC, cortical cataract; PSC, posterior subcapsular cataract; Vs, versus.
Cofactors
There was an increase in the level of manganese and zinc (P < 0.05) and no change in the level of copper (P = 0.36) in cataractous lenses (Fig. 1E, Supplementary Table S5). Further, the increase in Zn and Mn was solely contributed by cortical cataract (P < 0.05; Fig. 1F). Pearson correlation analysis showed a positive correlation between the level of Cu/Zn-SOD activity and the levels of Cu (P = 0.003) and Zn (P = 0.005) and no correlation between the levels of Mn-SOD activity and Mn (P > 0.05) in the cataractous lenses.
Transcript Expression and Protein Level
The level of SOD1 transcript expression (P < 0.001) was significantly decreased in cataractous lenses. However, the levels of SOD2 (P = 0.73) and SOD3 (P = 0.33) expression were not significantly altered (Fig. 1G, Supplementary Table S6). Downregulation of SOD1 and no change in SOD2 was observed in nuclear, cortical, and posterior subcapsular cataract (PSC) cataracts (P < 0.05); however, an upregulation of SOD3 was observed in nuclear cataract (P < 0.05). Further, Western blot analysis revealed a significant decrease (P < 0.05) in the level of SOD1 protein in cataractous lenses (Fig. 1H). However, immunoreactivity against specific antibodies for SOD2 and SOD3 proteins in clear and cataractous lenses could not be detected even after developing the blot with Enhanced Chemi-Luminescence Detection System (Thermo Fisher Scientific Inc., Rockford, IL).
SNPs and Mutation Screening
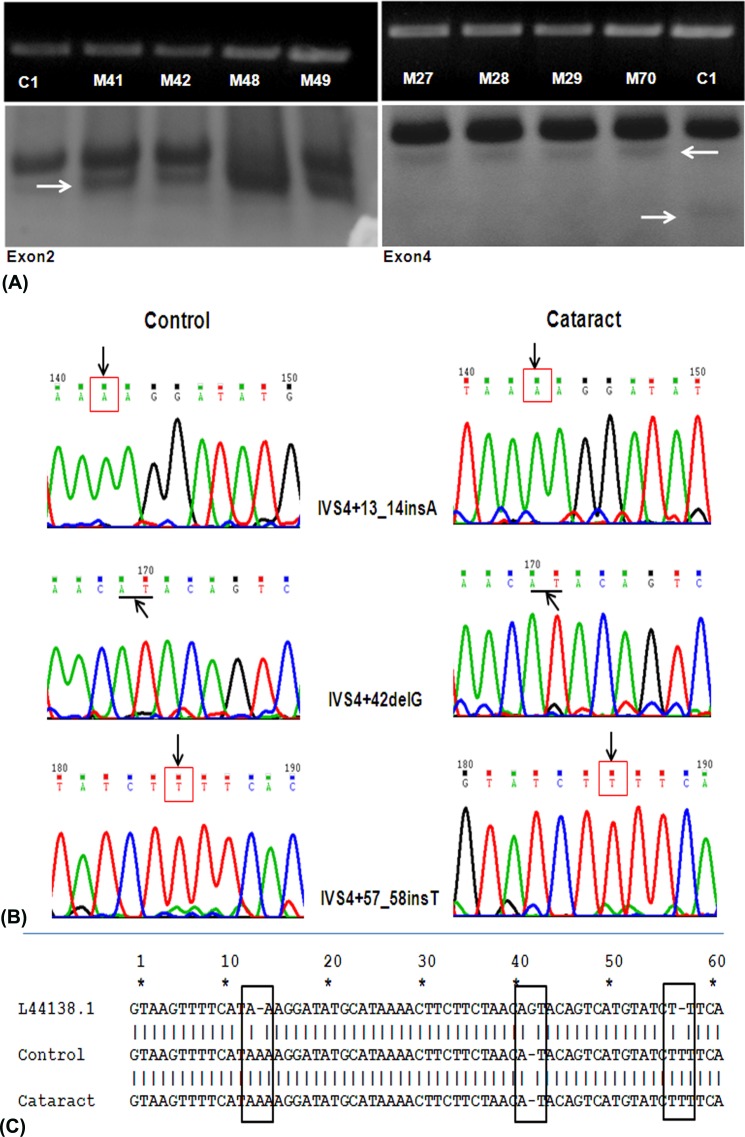
SSCP analysis was carried out for all 5 exons of SOD1 gene in both control LE (C) (n = 7) and cataract LE (M) (n = 20). Mobility shifts were found in exon2 (cases, n = 12 [60%]) and exon4 (control, n = 4 [57%]; cases, n = 12 [60%]) amplicons (Fig. 2A). DNA sequencing and subsequent blast analysis revealed three INDEL variations (IVS4+13_14insA, IVS4+42delG, and IVS4+57_58insT) in the 3′ region of the exon-intron4 boundary (Figs. 2B, 2C).
Figure 2.
(A) Polymerase chain reaction amplicons of exon2 and exon4 of SOD1 gene resolved in a 2% agarose gel (upper panel) and SSCP analysis (lower panel). C, control samples; M, cataract samples; arrows indicate mobility shifts. (B) Sequence chromatogram of exon4 of SOD1 gene from control and cataractous LE DNA samples. Arrows point to the site of nucleotide variation (insertion variations are indicated by boxes and deletion variations are indicated by underline). (C) BLAST analysis of exon4 of controls and cataractous samples against the GenBank sequence (L44318). Numbers denote the intronic nucleotide position; boxes represent the site of the nucleotide polymorphisms.
Alternate-Splice Transcript Analysis
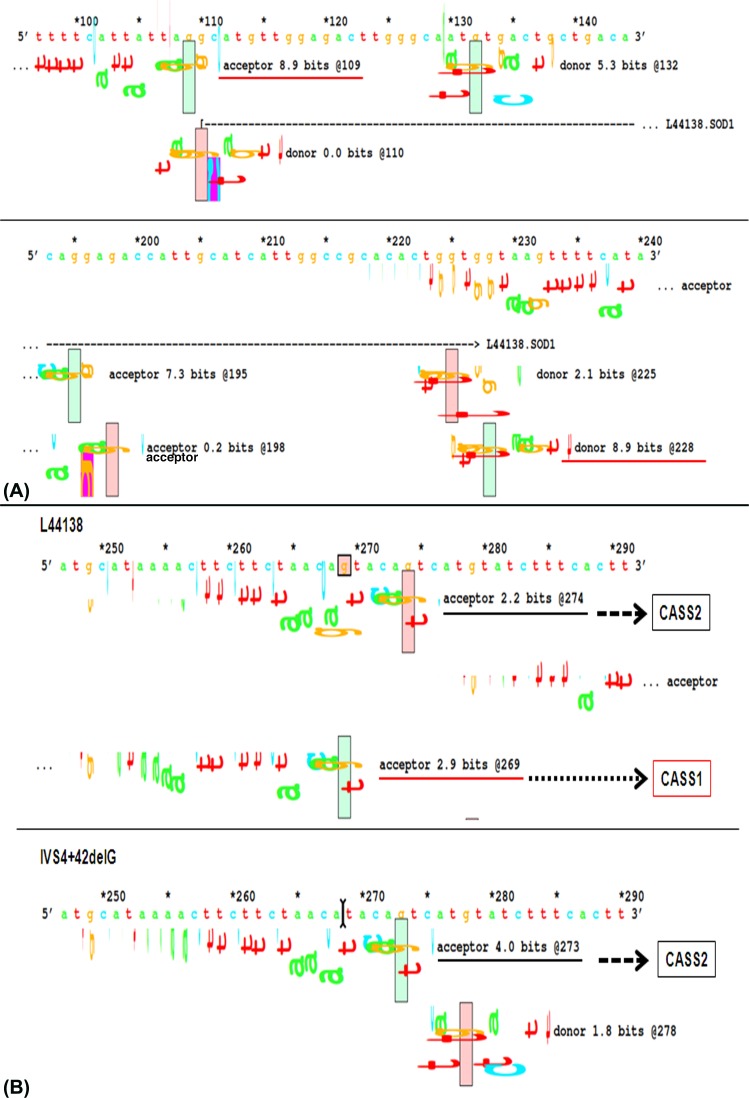
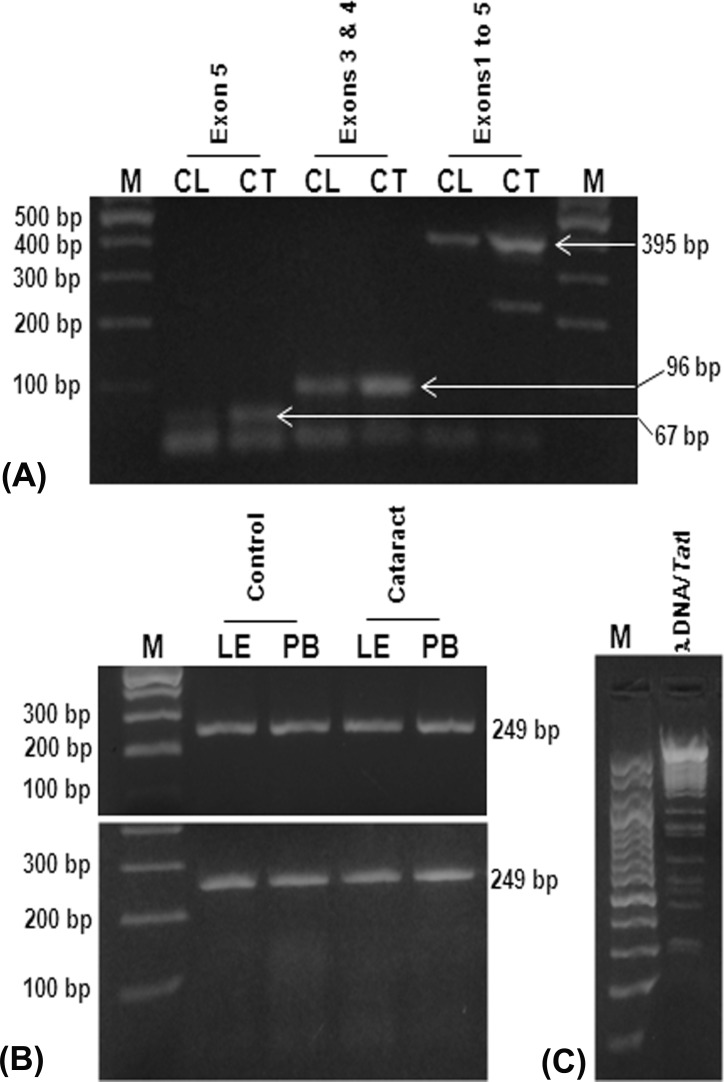
Splice-site nucleotide substitutions can be analyzed by comparing the individual information contents (Ri, bits) of the normal and variant splice-junction sequences.26,27,29,30 Theoretically, splice sites need to have Ri values of at least 0.0 bits to exist either as acceptors or as donors, but, empirically, it has been found that sites below 2.4 bits are not generally functional, and sites above 2.4 bits are strong enough to act as either donors or acceptors.26,27,29,30 We presumed that the observed INDEL variations in exon4 of cataractous samples could have influenced the strength of native splice sites and abolished their function. Splice-junction analysis of the normal (GenBank accession number L44138.1) and variant exon4 sequence showed no influence of INDELs on the strength of native-acceptor and native-donor splice sites, each having strength of 8.9 bits, which is strong enough to be involved in normal splicing events (Fig. 3A). Though we did not find any changes in the strength of native splice sites because of all three observed INDEL variations, the IVS4+42delG variation was found to have some effect on two other weak cryptic acceptor splice sites (CASS1 and CASS2). This particular SNP wiped off the CASS1 and enhanced the strength of the CASS2 from 2.2 to 4.0 bits (Fig. 3B). We believed that the increase in strength of CASS2 could make it able to be involved in alternate splicing events, whereby the resulting transcript might lack the complete exon4 coding region and retain part of proximal intron4 upstream to exon5 region. Next, we performed RT-PCR using primers that could amplify the entire coding region of SOD1 mRNA transcript (exon1 through exon5) to check the presence of alternatively spliced transcripts in cataractous condition. But, RT-PCR showed amplicons with 395-bp size in both clear (n = 3) and cataractous samples (n = 36), which indicates the absence of alternatively spliced SOD1 mRNA transcripts (Fig. 4A).
Figure 3.
(A) Splice-junction analysis of native donor and acceptor splice sites; the strength of both acceptor and donor splice sites are indicated by 8.9 bits at the nucleotide positions 109 and 228, respectively; marked by underline. The strength of other possible cryptic splice sites was also shown. (B) Splice-junction analysis of IVS4+42delG polymorphism. This polymorphism wipes out the CASS1 at nucleotide position 269 (shown by dotted arrow) and enhances the strength of the CASS2 position 274 from 2.2 bits (upper panel) to 4.0 bits (shown by dashed arrows) at nucleotide position 273 (lower panel).
Figure 4.
(A) Real-time PCR analysis of different exonic regions of SOD1 transcripts in clear (CL) and cataract (CT) samples. M, 100 bp DNA ladder; lanes 2 and 3, amplicons corresponding to exon 5 (67 bp) region; lanes 4 and 5, amplicons corresponding to exons 3 and 4 (96 bp) region; lanes 5 and 6, amplicons corresponding to exons 1 to 5 (395 bp) region. (B) Polymerase chain reaction–RFLP analysis of IVS4+42delG polymorphism in DNA isolated from LE and peripheral blood (PB) samples. Neither DNA sample showed expected digestion fragments of 192 and 52 bp upon TatI restriction enzyme (lower panel), which showed intact exon4 fragment of 249 bp similar to undigested exon4 amplicons (upper panel), exon4/TatI restriction enzyme digestion (lower panel). (C) Assessment of functional activity of TatI restriction enzyme. M, 100 bp DNA ladder; lane 2, λ DNA upon digestion with TatI restriction enzyme at 65°C for 2 hours showed respective digestion pattern of the enzyme.
Genotyping of IVS4+42delG Polymorphism
IVS4+42delG polymorphism results in loss of the restriction site for Tat1 enzyme, which would otherwise yield two fragments of sizes 197 and 52 bp upon digestion in the native condition. Polymerase chain reaction–RFLP analysis of genomic DNA from LE (n = 27) and peripheral blood revealed the absence of restriction fragments in both cataract (n = 100) and noncataract control (n = 100) samples (Fig. 4B).
Discussion
The aging human lens resides continuously in an OS atmosphere, which is believed to be one of the key factors in the gradual loss of lens transparency and subsequent cataract development.1,5–11 In our present study, we observed a much higher level of SOD activity in the LE than in the LN irrespective of the lens type. This indicates that a majority of defense enzymes against oxidative stress, especially SOD in lens, are derived from the lens epithelium.6,10 The lens epithelium also expresses all three isoforms of SOD.11 In our present study, we found lower levels of SOD1 and SOD2 activity in cataractous lenses irrespective of the type of cataracts. However, we could not estimate SOD3 activity as it was not possible to distinguish their activity from that of SOD1; these two enzymes are functionally similar in terms of their cofactor requirements and sensitivity to cyanide. Further, we observed that in cataractous lenses, SOD1 contributed only 60% to 70% of the total SOD activity as opposed to 90% in clear lenses.31 Although, in the present study, we found decreased or no change (in LE of cortical cataract) in the level of SOD2 activity in cataractous lenses, the level of activity was negligible in both clear and cataract lenses. Though we observed a higher level of SOD1 and no significant difference in SOD2 activity in cortical cataract activity than in nuclear and PSC, overall there was a significant decline in the levels of both SOD1 and SOD2 activity in different types of cataract. This suggests that a modulation in the level of SOD2 activity may not have any direct impact on the formation of different types of age-related cataract.
All SODs require trace elements as cofactors for their catalytic activity.32 Contradictory results have been published in relation to the level of trace elements in the cataract condition. A few studies have reported decreased levels of Zn12,33 and Mn,34 and a few have reported increased levels of Cu35–37 and Zn36,38 in senile cataractous lenses. In the present study, we found a higher level of Mn and Zn and no change in the level of Cu cofactors of SOD isoforms in cataractous lenses, especially in cortical cataract. Despite the presence of equimolar concentration of cofactors, the lens system failed to restore the optimal level of SOD activity in cataractous lenses. Therefore, we presumed factors other than the level of cofactors might be responsible for impairing SOD isoform activity. So, we performed mRNA expression analysis and found a significant downregulation of SOD1 and SOD3, but not of SOD2 in all types of cataract. Western blot analysis further confirmed the downregulation of SOD1 protein levels in cataractous lenses. But, we do not have any clue for the loss of immunoreactivity in Western blot for SOD2 and SOD3 in both clear and cataractous lenses. However, slow proteolysis, low new-protein synthesis, and extensive denaturation of proteins have been suggested as some of the reasons for loss of immunoreactivity.39 The present study suggests that the modulation of SODs, especially SOD1, has a very devastating effect on the antioxidative potential of the lenses. As a result, the system cannot counteract the increasing generation of O2.-, which could have resulted in lens damage and the subsequent loss of lens transparency.1,11 Recent studies have demonstrated that SOD1 is selectively expressed in lens tissue,40 and overexpression of the same was reported to prevent cataract formation in an in vitro whole lens model.41 As mRNA expression analysis, followed by Western blotting and enzyme activity experiments unequivocally revealed a significant decrease in the level of SOD1 mRNA and protein expression, and a subsequent decline in their activity, we believe that the impairment of SOD1 activity could be one of the risk factors in the cataractogenesis.
SOD1 is ubiquitously expressed and is composed of two equal subunits each containing a catalytic Cu ion and a stabilizing Zn ion.32 SOD1 encodes a 645-bp transcript that codes for a 154–amino acid protein product. SOD1 has always been a subject of research in several age-related and systemic diseases including cataract,10 amyotrophic lateral sclerosis (ALS),20 cancer,42 diabetes,43 and cardiovascular diseases.44 Several mutations20 and splice variants16,17 in the SOD1 gene have been reported to abolish its expression and enzymatic activity in amyotrophic lateral sclerosis,17,19,20 ocular diseases such as keratoconus,18,45 corneal diseases,46 and diabetic retinopathy.47,48
As several age-related diseases result from the accumulation of somatic mutations, we presumed that similar genetic alteration would also have occurred in LE, which would have impaired the tissue function. Since LE is important for the maintenance of lens homeostasis, we performed mutation screening in SOD1 from DNA isolated from the LE samples to understand the molecular mechanisms responsible for the impairment of SOD1 activity. A few samples, upon sequencing, revealed three INDEL variations (IVS4+13_14insA, IVS4+42delG, and IVS4+57_58insT) at 3′ exon-intron4 boundary rather than in the coding region. A similar observation was reported in ALS, in which mutation screening in coding regions of all five exons of SOD1 revealed no aberrations but revealed an intronic variation and up to 50% decreased activity.28 So far, three splice-site mutations have been identified in the fourth exon of the SOD1 gene in ALS patients48–51 and 7-bp deletion mutations in the second intron (IVS2+50del7 and c.16950delTAAACAG) in keratoconus patients.18,45 The INDEL variations that we observed in the present study, however, did not match with the previous studies. Intronic polymorphisms of any gene can cause splice-site aberrations and result in the deletion or insertion of amino acids as well as truncations. Such perturbations will significantly disrupt the structure and stability of the protein, and the result will be a major loss of function.28 As the observed INDEL variations in our study existed at the splice region, we believed they might have some effect on the alternate splicing mechanism. Therefore, we performed in silico splice-junction analysis using the Sequence Walker Program, which calculates the strength of each of the native and cryptic splice sites.29,30 Splice-junction analysis of IVS4+13_14insA and IVS4+57_58insT polymorphisms revealed that they do not have much impact on the splicing mechanism as they do not affect the strength of either the native or cryptic splice sites. However, IVS4+42delG polymorphism was found to enhance the strength of a cryptic acceptor splice site 2 (CASS2) from 2.2 to 4.0 bits, which is strong enough to generate a SOD1 transcript without exon4 during alternate splicing. We performed transcript profiling to check the presence of SOD1 transcripts lacking exon4 as a result of IVS4+42delG polymorphism. Of interest, our results did not find such transcripts in both clear and cataractous lenses. This finding indicates that the newly created CASS2, owing to IVS4+42delG polymorphism, does not have any direct influence on the SOD1 transcript splicing process. Further, we evaluated the prevalence of IVS4+42delG polymorphism in the cataractous and control populations and found a loss of restriction sites for TatI restriction enzyme in all cataractous and control samples. This indicates that the variation IVS4+42delG might be common in the Western Indian population. Hence, the mechanisms other than genetic mutations and/or polymorphisms might play a major role in the impairment of SOD1 expression and/or subsequent activity.
To summarize, in the present study, first, we attempted to check the activity level of SOD isoforms and found a decreased level of total SOD, SOD1, and SOD2 activity in cataractous condition. As the enzyme activity is attributed to the availability of cofactors, we decided to assess the level of Cu, Zn, and Mn and found higher levels of Zn and Mn and no change in the level of Cu. Since there is sufficient (as in Cu) or excess (as in Zn and Mn) quantity of cofactors in cataractous lenses, we concluded that the decrease in enzyme activity was not due to cofactors, and we thought that the decrease in activity could be a result of the decreased level of enzyme. So, we performed Western blotting for each enzyme isoform and found a decreased level of SOD1 protein in cataractous lenses, and no immunoreactivity for SOD2 and SOD3 in both clear and cataractous lenses. Further, we wanted to check whether this decreased level of protein expression was due to a decreased level of transcripts. Therefore, we performed quantitative real time (QRT)-PCR analysis and found downregulation of SOD1 but not SOD2 and SOD3 transcripts in cataractous lenses. Since SOD1 transcript alone was found to be decreased for all three parameters (activity, transcript, and protein levels), we thought that there could be some somatic or localized genetic modulation at the gene level that could have contributed to the decreased expression of SOD1 transcripts in lens epithelium. Hence, we screened for mutation in all exons of SOD1 gene from DNA isolated from lens epithelium and found three insertion/deletion (INDEL) variations in the splice sites. Since these variations were found at the splice sites, we thought to carry out in silico splice-junction analysis to examine whether these variations cause any changes in the native splice sites. One particular variation, IVS4+42delG, was found to increase the strength of CASS2 and to have the potential to be involved in alternate splicing, which could result in exon leaking. Hence, we amplified and sequenced (data not shown) the entire coding region of SOD1 and found a full-length transcript from exons 1 to 5 (395 bp) in both normal and cataract samples. We concluded that the decrease in transcript and protein levels was not due to alternate splicing of the SOD1 gene transcript. Parallel to this, we wanted to genotype this particular INDEL variation in both the normal and the cataractous population using PCR-RFLP. We analyzed the potential loss or gain of the restriction site resulting from this particular polymorphism using Restriction Mapper tool and found loss of site for TatI restriction enzyme in the variant gene. Results of genotyping showed the presence of the IVS4+42delG polymorphism in all tested samples of both normal and control populations. So, we concluded that this particular polymorphism might be common in the Western Indian population and may not have a major effect on modulating enzyme activity. Since it is purely a correlative study, solely observing changes in different factors that could potentially influence the enzyme activity, it was not possible to rule out that the decrease in SOD activity could be solely attributed to the decrease/loss of enzyme availability. Further, as correlation definitely does not demonstrate causation, we could not justify whether the decrease in SOD activity was a primary or secondary phenomenon to cataractogenesis.
In conclusion, the present study reveals a concomitant decrease in the levels of SOD1 transcript and protein expression, and enzyme activity in cataractous lenses. Although the decrease in the level of activity was suggested to be associated with suboptimal levels of cofactors, it could not be justified in the present study, as cataractous lenses showed optimal levels of Cu, Mn, and Zn. One of the intronic INDEL variations, IVS4+42delG, was found to have a strong impact on the splicing mechanism and could result in the generation of SOD1 transcript without exon4. However, the existence of such transcripts was not observed in the present study. PCR-RFLP analysis revealed the presence of this polymorphism in almost all subjects. Future research should focus on the factors that could influence the availability of cofactors for the SOD1 enzyme complex, and epigenetic regulation of SOD1 gene expression in age-related cataracts.
Acknowledgments
The authors thank Jim Ellis, PhD (US Department of Health and Human Services [DHHS]/National Institutes of Health [NIH]/Office of Director [OD]/Office of Research Services [ORS]/Division of Bioengineering and Physical Sciences [DBEPS]), of the National Institutes of Health, Bethesda, Maryland, for providing support in performing splice-junction analyses. The authors also thank C.S. Samaria Red Cross Eye Bank for providing human donor eyes, and all the participants in this study.
The authors alone are responsible for the content and writing of the paper.
Disclosure: S. Rajkumar, None; A.R. Vasavada, None; M.R. Praveen, None; R. Ananthan, None; G.B. Reddy, None; H. Tripathi, None; D.A. Ganatra, None; A.I. Arora, None; A.R. Patel, None
References
- 1. Spector A. Oxidative stress induced cataract: mechanism of action. FASEB J. 1995; 9: 1173– 1182. [PubMed] [Google Scholar]
- 2. Fridovich I. Superoxide anion radical (O2.-), superoxide dismutases, and related matters. J Biol Chem. 1997; 272: 18515– 18517. [DOI] [PubMed] [Google Scholar]
- 3. Spector A. The search for a solution to senile cataracts. Proctor Lecture. Invest Ophthalmol Vis Sci. 1984; 25: 130– 146. [PubMed] [Google Scholar]
- 4. World Health Organization. Blindness and Visual Disability; Part II of VII: major causes worldwide. Geneva, Switzerland: World Health Organization; 1997. Available at: http://www.who.int/inf-fs/en/fact143.html. Accessed August 15, 2004. [Google Scholar]
- 5. Reddy VN, Kasahara E, Hiraoka M, et al. Effects of variation in superoxide dismutases (SOD) on oxidative stress and apoptosis in lens epithelium. Exp Eye Res. 2004; 79: 859– 868. [DOI] [PubMed] [Google Scholar]
- 6. Bhuyan KC, Bhuyan DK. Superoxide dismutase of the eye: relative functions of superoxide dismutase and catalase in protecting the ocular lens from oxidative damage. Biochim Biophys Acta. 1978; 542: 28– 38. [DOI] [PubMed] [Google Scholar]
- 7. Fecondo JV, Augusteyn RC. Superoxide dismutase, catalase and glutathione peroxidase in the human lens. Exp Eye Res. 1983; 36: 15– 23. [DOI] [PubMed] [Google Scholar]
- 8. Ohrloff C, Hockwin O. Superoxide dismutase (SOD) in normal and cataractous human lenses. Graefes Arch Clin Exp Ophthalmol. 1984; 222: 79– 81. [DOI] [PubMed] [Google Scholar]
- 9. Scharf J, Dovrat A. Superoxide dismutase molecules in human cataractous lenses. Ophthalmol Res. 1986; 18: 332– 337. [DOI] [PubMed] [Google Scholar]
- 10. Fujiwara H, Takigawa Y, Suzuki T, et al. Superoxide dismutase activity in cataractous lenses. Jpn J Ophthalmol. 1992; 36: 273– 280. [PubMed] [Google Scholar]
- 11. Rajkumar S, Praveen MR, Gajjar D, et al. Activity of superoxide dismutase isoenzymes in lens epithelial cells derived from different types of age-related cataract. J Cataract Refract Surg. 2008; 34: 470– 474. [DOI] [PubMed] [Google Scholar]
- 12. Sulochana KN, Punitham R, Ramakrishnan S. Effect of cigarette smoking on cataract: antioxidant enzymes and constituent minerals in the lens and blood of humans. Indian J Pharmacol. 2002; 34: 428– 431. [Google Scholar]
- 13. Ozmen B, Ozmen D, Erkin E, et al. Lens superoxide dismutase and catalase activities in diabetic cataract. Clin Biochem. 2002; 35: 69– 72. [DOI] [PubMed] [Google Scholar]
- 14. Yan H, Harding JJ. Glycation-induced inactivation and loss of antigenicity of catalase and superoxide dismutase. Biochem J. 1997; 328: 599– 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y, Zhang L, Sun DL, et al. Genetic polymorphisms of superoxide dismutases, catalase, and glutathione peroxidase in age-related cataract. Mol Vis. 2011; 17: 2325– 2332. [PMC free article] [PubMed] [Google Scholar]
- 16. Hirano M, Hung W-Y, Cole N, et al. Multiple transcripts of the human Cu, Zn superoxide dismutase gene. Biochem Biophys Res Commun. 2000; 276: 52– 56. [DOI] [PubMed] [Google Scholar]
- 17. Kawata A, Kato S, Shimizu T, et al. Aberrant splicing of human Cu/Zn superoxide dismutase (SOD1) RNA transcripts. Neuroreport. 2000; 11: 2649– 2653. [DOI] [PubMed] [Google Scholar]
- 18. Udar N, Atilano SR, Nesburn AB, et al. SOD1: a candidate gene for keratoconus. Invest Ophthalmol Vis Sci. 2006; 47: 3345– 3351. [DOI] [PubMed] [Google Scholar]
- 19. Yulug IG, Katsanis N, deBelleroche J, et al. An improved protocol for the analysis of SOD1 gene mutations, and a new mutation in exon4. Hum Mol Genet. 1995; 4: 1101– 1104. [DOI] [PubMed] [Google Scholar]
- 20. Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993; 362: 59– 62. [DOI] [PubMed] [Google Scholar]
- 21. Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011; 60: 1304– 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chylack LT Jr, Wolfe JK, Singer DM, et al. The Lens Opacities Classification System III. The Longitudinal Study of Cataract Study Group. Arch Ophthalmol. 1993; 111: 831– 836. [DOI] [PubMed] [Google Scholar]
- 23. Pfaffl MW, Morgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002; 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orita M, Iwahana H, Kanazawa H, et al. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989; 86: 2766– 2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blum H, Beier H, Gross HJ. Improved silver staining of plant protein, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987; 8: 93– 99. [Google Scholar]
- 26. Rogan PK, Faux BM, Schneider TD. Information analysis of human splice site mutations. Hum Mutat. 1998; 12: 153– 171. [DOI] [PubMed] [Google Scholar]
- 27. Schneider TD. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997; 25: 4408– 4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Birve A, Neuwirth C, Weber M, et al. A novel SOD1 splice site mutation associated with familial ALS revealed by SOD activity analysis. Hum Mol Genet. 2010; 19: 4201– 4206. [DOI] [PubMed] [Google Scholar]
- 29. Schneider TD. Information content of individual genetic sequences. J Theor Biol. 1997; 189: 427– 441. [DOI] [PubMed] [Google Scholar]
- 30. Rogan PK, Schneider TD. Using information content and base frequencies to distinguish mutations from genetic polymorphisms in splice junction recognition sites. Hum Mutat. 1995; 6: 74– 76. [DOI] [PubMed] [Google Scholar]
- 31. Noor R, Mittal S, Iqbal J. Superoxide dismutase-applications and relevance to human diseases. Med Sci Monit. 2002; 8: RA210– 215. [PubMed] [Google Scholar]
- 32. McCord JM, Fridovich I. Superoxide dismutase: an enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969; 244: 6049– 6055. [PubMed] [Google Scholar]
- 33. Dawczynski J, Blum M, Winnefeld K, et al. Increased content of zinc and iron in human cataractous lenses. Biol Trace Elem Res. 2002; 90: 15– 23. [DOI] [PubMed] [Google Scholar]
- 34. Cekiç O, Bardak Y, Totan Y, et al. Nickel, chromium, manganese, iron and aluminum levels in human cataractous and normal lenses. Ophthalmic Res. 1999; 31: 332– 336. [DOI] [PubMed] [Google Scholar]
- 35. Rácz P, Erdöhelyi A. Cadmium, lead and copper concentrations in normal and senile cataractous human lenses. Ophthalmic Res. 1988; 20: 10– 13. [DOI] [PubMed] [Google Scholar]
- 36. Rasi V, Costantini S, Moramarco A, et al. Inorganic element concentrations in cataractous human lenses. Ann Ophthalmol. 1992; 24: 459– 464. [PubMed] [Google Scholar]
- 37. Cekic O. Copper, lead, cadmium and calcium in cataractous lenses. Ophthalmic Res. 1998; 30: 49– 53. [DOI] [PubMed] [Google Scholar]
- 38. Stanojević-Paović A, Hristić V, Cuperlović M, et al. Macro- and microelements in the cataractous eye lens. Ophthalmic Res. 1987; 19: 230– 234. [DOI] [PubMed] [Google Scholar]
- 39. Behndig A, Svensson B, Marklund SL, Karlsson K. Superoxide dismutase isoenzymes in the human eye. Invest Ophthalmol Vis Sci. 1998; 39: 471– 475. [PubMed] [Google Scholar]
- 40. Diehn JJ, Diehn M, Marmor MF, et al. Differential gene expression in anatomical compartments of the human eye. Genome Biol. 2005; 6: R74.1– 74.13. Available at: http://genomebiology.com/2005/6/9/R74. Accessed May 10, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin D, Barnett M, Grauer L, et al. Expression of superoxide dismutase in whole lens prevents cataract formation. Mol Vis. 2005; 11: 853– 858. [PubMed] [Google Scholar]
- 42. Oberley LW, Buettner GR. Role of superoxide dismutase in cancer: a review. Cancer Res. 1979; 39: 1141– 1149. [PubMed] [Google Scholar]
- 43. Flekac M, Skrha J, Hilgertova J, et al. Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med Genet. 2008; 9: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alameddine FM, Zafari AM. Genetic polymorphisms and oxidative stress in heart failure. Congest Heart Fail. 2002; 8: 157– 172. [DOI] [PubMed] [Google Scholar]
- 45. Bonis PD, Laborante A, Pizzicoli C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. 2011; 17: 2482– 2494. [PMC free article] [PubMed] [Google Scholar]
- 46. Behndig A, Karlsson K, Johansson BO, et al. Superoxide dismutase isoenzymes in the normal and diseased human cornea. Invest Ophthalmol Vis Sci. 2001; 42: 2293– 2296. [PubMed] [Google Scholar]
- 47. Berkowitz BA, Gradianu M, Bissig D, et al. Retinal ion regulation in a mouse model of diabetic retinopathy: natural history and the effect of Cu/Zn superoxide dismutase overexpression. Invest Ophthalmol Vis Sci. 2009; 50: 2351– 2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kernell A, Lundh BL, Marklund SL, et al. Superoxide dismutase in the anterior chamber and the vitreous of diabetic patients. Invest Ophthalmol Vis Sci. 1992; 33: 3131– 3135. [PubMed] [Google Scholar]
- 49. Valdmanis PN, Belzil VV, Lee J, et al. A mutation that creates a pseudoexon in SOD1 causes familial ALS. Ann Hum Genet. 2009; 73: 652– 657. [DOI] [PubMed] [Google Scholar]
- 50. Zu JS, Deng HX, Lo TP, et al. Exon 5 encoded domain is not required for the toxic function of mutant SOD1 but essential for the dismutase activity: identification and characterization of two new SOD1 mutations associated with familial amyotrophic lateral sclerosis. Neurogenetics. 1997; 1: 65– 71. [DOI] [PubMed] [Google Scholar]
- 51. Sapp PC, Rosen DR, Hosler BA, et al. Identification of three novel mutations in the gene for Cu/Zn superoxide dismutase in patients with familial amyotrophic lateral sclerosis. Neuromuscul Disord. 1995; 5: 353– 357. [DOI] [PubMed] [Google Scholar]