Abstract
Purpose
This study aimed to explore the molecular mechanisms associated with bisphosphonate (BP)-related osteonecrosis of the jaw (ONJ) in patients with multiple myeloma (MM).
Methods
The gene expression profile GSE7116 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) from eleven patients with ONJ resulting from MM treated with BPs (ONJBPs) and ten MM patients without ONJ treated with BPs (MMBPs) were analyzed. Gene ontology (GO) and pathway enrichment analyses of DEGs were performed, followed by functional annotation and protein–protein interaction network construction. Finally, sub-network modules were constructed and analyzed.
Results
A total of 166 up- and 473 down-regulated DEGs were identified. The up-regulated DEGs were enriched in pathways related to cancer, and the down-regulated DEGs were enriched in pathways related to the immune system. Moreover, the GO terms enriched by the up-regulated DEGs were associated with misfolded proteins, and the down-regulated DEGs were associated with immune responses. After functional annotation, 16 transcription factors were identified, including X-box binding protein 1 (XBP1). In protein–protein interaction network analysis, tumor necrosis factor (TNF) and interleukin 1, beta (IL1B) had higher connectivity degrees. Among the constructed sub-network modules, module 1 was the best one, and DEAD (Asp-Glu-Ala-Asp) box helicase 5 (DDX5) was a hub gene. The DEGs in module 1 were mainly enriched in GO terms related to RNA splicing.
Conclusion
DEGs of ONJ were mainly enriched in pathways related to the immune system and RNA splicing. DEGs such as TNF, ILB1, DDX5, and XBP1 may be the potential targets of ONJ treatment.
Keywords: osteonecrosis of the jaw, multiple myeloma, bisphosphonates, differentially expressed genes, functional enrichment analysis
Introduction
Multiple myeloma (MM), a neoplasm of plasma cells, is a presently incurable hematological malignancy affecting one to five per 100,000 individuals each year worldwide.1,2 Frequently, there is invasion of the adjacent bone, which destroys skeletal structures and results in bone pain and fractures.3 Bisphosphonates (BPs) are synthetic analogs of naturally occurring pyrophosphates, which are capable of localizing to the bone and inhibiting osteoclastic function.4 BPs have been approved by other studies for the treatment of MM and solid tumors.5,6 However, a complication, avascular osteonecrosis of the jaw (ONJ), has recently been described as associated with their use.4,7
Osteonecrosis refers to the death of bone as a result of impaired blood supply.8 ONJ results from bone exposure in the oral cavity, with subsequent necrosis often following dental procedures or traumatic injuries.9 ONJ has been described in association with the use of zoledronic acid and pamidronate in various malignancies.4,10 Currently, the exact mechanism of this BP-mediated osteonecrosis is not certain, and it is likely multifactorial. Age, smoking, obesity, cancer diagnosis, and poor oral health have been shown to be predisposing factors for ONJ.11 Previous studies suggest that genetic factors could also be involved in BP-mediated ONJ risk.12,13 For example, Such et al published a study showing that the presence of one or two minor alleles of the cytochrome P450, subfamily 2C, polypeptide 8 gene was an independent prognostic marker associated with development of ONJ in MM patients treated with BPs.12 Nicoletti et al reported that RNA binding motif, single-stranded interacting protein 3, is significantly associated with BP-related ONJ.13 Progress in understanding the mechanisms of BP-mediated ONJ in patients with MM will contribute to solutions for this complication. However, our present knowledge is insufficient.
In the present study, we applied a biological informatics approach to analyze the gene expression profiles of GSE7116, and we performed functional analysis for differentially expressed genes (DEGs) between patients with ONJ resulting from MM treated with BPs (ONJBPs) and MM patients treated with BPs without ONJ (MMBPs). Additionally, functional enrichment and functional annotations were analyzed, and a protein–protein interaction (PPI) network was constructed. We aimed to provide a systematic perspective toward understanding underlying mechanisms and toward discovering new therapeutic targets for the treatment of ONJ.
Methods
Affymetrix microarray data
The raw CEL array data of GSE7116 were downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database based on the platform of the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which was deposited by Raje et al.14 The database contains 26 samples, including eleven samples of ONJBP, ten samples of MMBP, and five samples from normal donors. In this paper, the ONJBP and MMBP samples were used for the development of the Affymetrix microarray data.
Data preprocessing and differential expression analysis
The original array data were converted into expression measures, and we performed background correction, quartile data normalization, and probe summarization with the robust multi-array average (RMA)15 algorithm in the R Affy16 package.
Paired t-tests based on the multitest package in R were used to identify genes that were significantly differentially expressed between the ONJBP and MMBP samples. The log2-fold change (log2FC) was calculated. Only |log2FC| >0.58 and false discovery rate (FDR) <0.05 results were considered as the cutoff value for DEG screening.
Gene ontology and pathway enrichment analyses
Gene ontology (GO)17 is a tool for the unification of biology that is used to collect a structured, defined, controlled vocabulary for a large scale system of gene annotation. The Kyoto Encyclopedia of Genes and Genomes (KEGG)18 knowledge database is a collection of online databases dealing with genomes, enzymatic pathways, and biological chemicals. REACTOME19 is a curated, peer-reviewed pathway database of human biological processes. The Database for Annotation, Visualization and Integrated Discovery (DAVID),20 has been developed as a comprehensive set of functional annotation tools for relating the functional terms with gene lists using a clustering algorithm. To analyze DEGs at the functional level, we performed GO, KEGG, and REACTOME pathway enrichment analyses using the DAVID online tool to obtain the enriched biological process, molecular functions (MFs), cellular component (CC) terms, and pathways. A P-value <0.01 was set as the threshold value.
Functional annotation of DEGs
Based on the data-driven transcription factors (TFs), the DEGs were selected and annotated to determine whether these genes had functions related to transcriptional regulation. The Tumor Suppressor Gene (TSGene) database21 integrates tumor suppressor genes (TSGs) with large-scale experimental evidence to provide a comprehensive resource for the further investigation of TSGs and their molecular mechanisms in cancer. The Tumor-associated Gene (TAG) database22 is used to save new tumor-associated genes (TAGs) that play roles in carcinogenesis. In the current study, we extracted all the known oncogenes and TSGs from the TSGene and TAG databases.
PPI network construction and sub-network identification
The Search Tool for the Retrieval of Interacting Genes (STRING) database23 is a pre-computed global resource, which has been designed to evaluate PPI information. In a PPI network, each node stands for a gene, and the edges represent the interactions between nodes. Degree indicates the number of edges linked to a given node, and nodes with high degree are defined as hub genes that possess important biological functions. In the present research, the STRING online tool was applied to analyze the PPI network of DEGs, and only those experimentally validated interactions with a combined degree >0.7 were selected as significant.
Cytoscape24 is a general bioinformatics package used for visualizing biological network and integrating data. Clustering with overlapping neighborhood expansion (ClusterONE)25 is used to discover densely connected and possibly overlapping regions within a PPI network. In the current study, ClusterONE, based on Cytoscape, was used to identify sub-networks in the PPI network. Then, GO functional and pathway enrichment analyses were performed with a P-value <0.01.
Results
Identification of DEGs
For the dataset GSE7116, a total of 1,920 transcripts were obtained after data preprocessing. Among them, 449 were up-regulated transcripts, which corresponded to 166 DEGs and 1,471 down-regulated transcripts corresponding to 473 DEGs.
GO and pathway enrichment analyses
After GO functional enrichment analysis, the DEGs were enriched in three categories including biological process, MF, and CC. The top three GO terms of the three categories are shown in Table 1. The up-regulated DEGs mainly participated in the GO terms related to misfolded proteins, while the down-regulated DEGs mainly participated in the GO terms related to the immune system.
Table 1.
Gene ontology (GO) functional enrichment analysis for the up- and down-regulated differentially expressed genes (DEGs) (top three pathways with lower P-values)
| Category | Term | Description | Counts | P-value | Gene symbol |
|---|---|---|---|---|---|
| Up-regulated DEGs | |||||
| Biological process | GO:0071218 | Cellular response to misfolded protein | 2 | 4.43E-04 | ATXN3, UBE2W |
| Biological process | GO:0051788 | Response to misfolded protein | 2 | 9.23E-04 | ATXN3, UBE2W |
| Biological process | GO:0006515 | Misfolded or incompletely synthesized protein catabolic process | 2 | 1.57E-03 | ATXN3, UBE2W |
| MF | GO:0030125 | Clathrin vesicle coat | 2 | 7.52E-03 | EGFR, ZAK |
| MF | GO:0030131 | Clathrin adaptor complex | 2 | 1.65E-02 | EGFR, ZAK |
| MF | GO:0030119 | Adaptor-related protein-type membrane coat adaptor complex | 2 | 2.58E-02 | EGFR, ZAK |
| CC | GO:0004709 | Mitogen-activated protein kinase kinase kinase activity | 3 | 3.26E-04 | AP1G2, EGFR |
| CC | GO:0004702 | Receptor signaling protein serine/threonine kinase activity | 3 | 4.83E-03 | AP1G2, EGFR |
| CC | GO:0005057 | Receptor signaling protein activity | 4 | 5.64E-03 | AP1G2, EGFR |
| Down-regulated DEGs | |||||
| Biological process | GO:0002376 | Immune system process | 114 | 6.00E-15 | IL1B, TNF |
| Biological process | GO:0006955 | Immune response | 84 | 1.67E-14 | IL1B, TNF |
| Biological process | GO:0006950 | Response to stress | 150 | 5.66E-14 | IL1B, TNF |
| MF | GO:0044424 | Intracellular part | 369 | 7.77E-13 | IL1B, FUS |
| MF | GO:0005622 | Intracellular | 371 | 1.59E-12 | FUS, DDX5 |
| MF | GO:0044444 | Cytoplasmic part | 239 | 1.30E-11 | FUS, DDX5 |
| CC | GO:0005515 | Protein binding | 266 | 1.49E-13 | FUS, DDX5 |
| CC | GO:0000166 | Nucleotide binding | 94 | 2.33E-06 | FUS, DDX5 |
| CC | GO:1901265 | Nucleoside phosphate binding | 94 | 2.38E-06 | FOS, JUN |
Notes: Category refers to the GO functional categories; Counts refers to the number of enriched DEGs.
Abbreviations: MF, molecular function; CC, cellular component.
The significantly enriched KEGG and REACTOME pathways of the up-regulated DEGs and down-regulated DEGs are shown in Table 2. The up-regulated DEGs were enriched in 13 KEGG pathways, such as bladder cancer, non-small cell lung cancer, and six REACTOME pathways, such as constitutive PI3K/AKT signaling in cancer. The down-regulated DEGs were enriched in 72 KEGG pathways, such as Leishmaniasis and T cell receptor signaling pathways, and 230 REACTOME pathways, such as pathways related to the immune system and the adaptive immune system.
Table 2.
Pathway enrichment analysis for the up- and down-regulated differentially expressed genes (DEGs) (top three pathways with lower P-values)
| Category | Term | Description | Counts | P-value | Gene symbol |
|---|---|---|---|---|---|
| Up-regulated DEGs | |||||
| KEGG | 5219 | Bladder cancer | 3 | 1.90E-03 | E2F3, EGFR |
| KEGG | 5223 | Non-small cell lung cancer | 3 | 3.91E-03 | E2F3, EGFR |
| KEGG | 5214 | Glioma | 3 | 6.59E-03 | E2F3, EGFR |
| REACTOME | 425397 | Transport of vitamins, nucleosides, and related molecules | 2 | 1.32E-02 | SLC28A3, SLCO3A1 |
| REACTOME | 190236 | Signaling by fibroblast growth factor receptor | 3 | 5.19E-02 | EGFR, IRS1 |
| REACTOME | 382551 | Transmembrane transport of small molecules | 6 | 6.40E-02 | AQP11, ATP10A |
| Down-regulated DEGs | |||||
| KEGG | 5140 | Leishmaniasis | 14 | 1.85E-07 | IL1B, TNF |
| KEGG | 4621 | Nucleotide-binding oligomerization domain-like receptor signaling pathway | 12 | 7.06E-07 | IL1B, TNF |
| KEGG | 4660 | T cell receptor signaling pathway | 16 | 1.20E-06 | FOS, TNF |
| REACTOME | 168256 | Immune system | 68 | 2.64E-08 | IL1B, DHX9 |
| REACTOME | 202403 | T-cell receptor signaling | 11 | 1.89E-06 | CD3E, GRAP2 |
| REACTOME | 1280218 | Adaptive immune system | 41 | 2.69E-06 | MAP3K8, CD3E |
Notes: Category refers to the pathway functional categories; Counts refers to the number of enriched DEGs.
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Functional annotation of DEGs
After researching the expression of TFs and TAGs, four TFs were up-regulated, including POU class 2 homeobox 2 and C-terminal binding protein 2, and 12 TFs were down-regulated, including X-box binding protein 1 (XBP1) and MYC-associated factor X. In addition, in the up-regulated DEGs, three were oncogenes, four were TSGs, and the functions of the other three DEGs were uncertain. In the down-regulated DEGs, 49 genes were TAGs. Among these DEGs, 13 were oncogenes, 21 were TSGs, and the functions of other 15 were uncertain.
PPI network construction
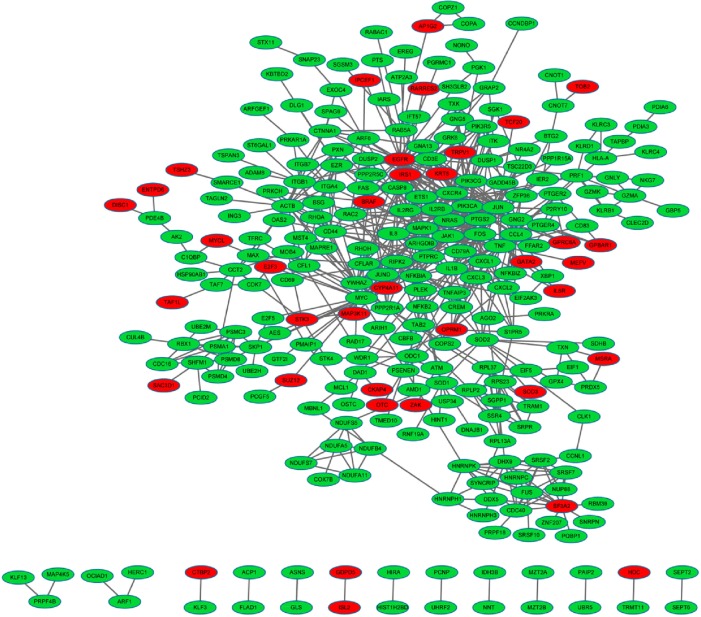
Based on the STRING database, PPI networks with a degree of more than 15 were then constructed using Cytoscape, and networks with 281 nodes and 643 edges were obtained (Figure 1). Fifteen nodes were selected as hub genes with a degree >15, including mitogen-activated protein kinase 1 (degree =30), tumor necrosis factor (TNF, degree =20), and interleukin 1, beta (IL1B, degree =17).
Figure 1.
The constructed protein–protein interaction network of differentially expressed genes (DEGs).
Notes: Red, up-regulated DEGs; green, down-regulated DEGs.
Sub-network identification and functional enrichment analysis
After clustering analysis of the PPI network using Cluster-ONE, we obtained 42 sub-network modules, among which module 1 was the best descriptor with 12 nodes and 40 edges (Figure 2). Heterogeneous nuclear ribonucleoprotein K (degree =10) and DEAD (Asp-Glu-Ala-Asp) box helicase 5 (DDX5, degree =4) had higher degrees in module 1.
Figure 2.
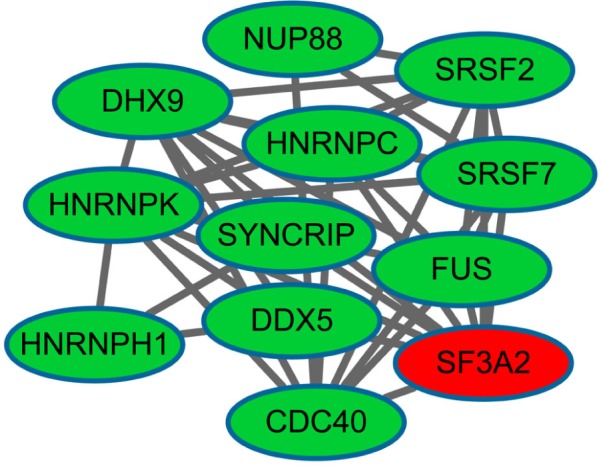
The constructed sub-network of differentially expressed genes (DEGs).
Notes: Red, up-regulated DEGs; green, down-regulated DEGs.
After GO functional and KEGG pathway enrichment analyses of the DEGs in module 1, we found that these DEGs were mainly enriched in GO terms associated with RNA splicing and pathways of spliceosomes.
Discussion
Although BPs have been associated with ONJ, their role in its pathophysiology remains to be defined.26 Our current analysis of the gene expression profile revealed the underlying gene activity changes related to ONJ and enabled the identification of targets for management policy. In the present study, a total of 639 DEGs were identified as differing between ONJBP and MMBP samples through the gene expression profile of GSE7116. The down-regulated DEGs were enriched in pathways related to the immune system and biological process terms related to RNA splicing. In the PPI network and sub-network, the hub genes TNF, IL1B, and DDX5 had higher connectivity degrees. Additionally, TF of XBP1 was also found to be down-regulated. These results suggested that these genes and pathways might play important roles in the progression of BP-mediated ONJ.
In the present study, pathways associated with the immune system were enriched by several down-regulated DEGs, such as TNF and IL1B, which had higher connective degrees in the PPI network. Forster showed evidence that BP therapy negatively affected the function and survival of neutrophils, which were key players in the immune system.27 Another study reported that an immune defect may play a role in the pathophysiology of ONJ.28 Thus, the immune system possibly contributes to the pathogenesis of ONJBP. TNF and IL1B are potent bone-resorbing cytokines that may contribute to the development of the osteolytic bone disease observed in patients with MM.29 TNF encodes a multifunctional pro-inflammatory cytokine, and this cytokine is mainly secreted by macrophages. Importantly, TNF is involved in the regulation of a wide spectrum of biological processes, including cell proliferation, differentiation, and apoptosis.30 Kast reported that TNF was a necessary growth factor for the expansion and maintenance of MM cells.31 Tsimberidou et al also suggested that elevated TNF-α levels were associated with poor prognosis in patients with MM.32
The other cytokine, IL1B, is an important mediator of the inflammatory response, and it is involved in a variety of cellular activities, including cell proliferation, differentiation, and apoptosis.33 In MM, IL1B contributes to myeloma cell growth by an autocrine pathway.34 Recently, Tai et al reported that IL1B was important in the pathogenesis of MM.35 Additionally, a previous study found that concentrations of IL1B and TNF were significantly greater in patients with bone disease than in those without bone disease.36 These observations suggest that TNF and IL1B play important roles in the disruption of bone homeostasis in various tumors, and that anti-TNF/IL1B may be used as candidate agents to prevent MM-induced osteolysis. Interestingly, BPs were found to have inhibitory actions on cytokine secretion by macrophages; as a result, they have inhibitory effects on TNF and IL1B.37 In the present study, TNF and IL1B were down-regulated in the ONJBP samples, which was in accordance with the work done by Pennanen et al.37
In addition, BPs are pyrophosphate analogs with the addition of side chains, which bind to resorbing bone and are not metabolized. As a result, high concentrations will be maintained in bone for prolonged periods.38 Specially, TNF-α and IL-1 are key cytokines in the processes of bone resorption and osteoclast differentiation.38 Therefore, we speculated that the residual BPs might result in the accumulation of microdamage and then affect the mechanical integrity of the bone by targeting TNF and IL1B. Taken together, TNF and IL1B (and the related pathways in which they participate) may be candidate molecular markers associated with BP-mediated ONJ.
It has been well documented that RNA splicing plays a biologically important function.39 A previous study showed that RNA splicing occurs frequently in human cancer cells.40 In our study, biological process terms related to RNA splicing were enriched by several down-regulated DEGs, such as DDX5. Moreover, DDX5 was also a hub gene in the sub-network. The DDX5 protein is a member of the DEAD-box (DDX) family of RNA helicases.41 DDX5 is involved in the alternative regulation of pre-mRNA splicing.42 It is now well established that DDX5 has the ability to stimulate cell proliferation and to prevent apoptosis.43 At present, studies have reported that DDX5 is overexpressed in human colon cancers and breast cancer, which strongly suggests that DDX5 promotes tumorigenesis.44,45 Importantly, Zhan et al demonstrated that DDX5 was up-regulated in MM in relation to normal bone marrow.46 As a result, we speculate that the down-regulation of DDX5 in ONJBP may be caused by BPs. DDX5 and the biological process terms related to RNA splicing may be key factors in ONJ.
We also found that the TF of XBP1 was down-regulated in the present study. This gene product is a basic-region leucine zipper protein that is also identified as a cellular TF, which binds to an enhancer in the promoter of the T cell leukemia virus type 1 promoter.47 Inside the vascularized solid tumors, cells undergo endoplasmic reticulum stress, but they can survive in such adverse microenvironments by an adaptive mechanism called the unfolded protein response (UPR).48 XBP1 is a critical transcriptional activator of the UPR, and it is responsible for regulating the functions of genes in cell survival.48 Recent studies have implicated XBP1 overexpression in human carcinogenesis and tumor growth under hypoxic conditions; specifically, elevated XBP1 mRNA levels have been detected in hepatocellular carcinomas and breast tumors.49,50 Additionally, the abundant expression of XBP1 has been detected in human MM cells.51 A gene expression profiling study has also confirmed the specific expression of XBP1 in MM.46 Therefore, compounds that target XBP1 may show therapeutic activity for the treatment of some cancers. In our study, the expression levels of XBP1 were found to decline in ONJBP samples, which suggests that XBP1 may play an important role in the progression of BP-related ONJ in patients with MM. Although there are few studies of the association between XBP1 and ONJ, we can speculate that XBP1 may be an important TF related to BP-mediated ONJ.
Bioinformatics technologies have the potential to identify and validate candidate agents for serious diseases; however, the present study has a few limitations. The sample size for microarray analysis was small, which may cause a high rate of false positive results. Moreover, the current study lacked experimental verification. Further genetic and experimental studies with larger sample sizes are still needed to confirm our observations.
Conclusion
In conclusion, our data provide a comprehensive set of bio-informatics analyses of DEGs that may be involved in BP-mediated ONJ. The findings in the current study may contribute to our understanding of the underlying molecular mechanisms of BP-mediated ONJ. DEGs such as TNF, IL1B, and DDX5, pathways associated with the immune system, and biological process terms related to RNA splicing, may have the potential to be used as targets for ONJ diagnosis and treatment. The identification of genetic factors in the management of this complication may result in safer long-term use of BPs in MM.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant number 81372835) and the Jilin University First Hospital Junior Scientist Grant (number JDYY42013021).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 2.Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008;111:2516–2520. doi: 10.1182/blood-2007-10-116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kyle R, Rajkumar S. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2008;23:3–9. doi: 10.1038/leu.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggiero SL, Mehrotra B, Rosenberg TJ, Engroff SL. Osteonecrosis of the jaws associated with the use of bisphosphonates: a review of 63 cases. J Oral Maxillofac Surg. 2004;62:527–534. doi: 10.1016/j.joms.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Stewart AF. Clinical practice: hypercalcemia associated with cancer. N Engl J Med. 2005;352:373–379. doi: 10.1056/NEJMcp042806. [DOI] [PubMed] [Google Scholar]
- 6.Cartenì G, Bordonaro R, Giotta F, et al. Efficacy and safety of zoledronic acid in patients with breast cancer metastatic to bone: a multicenter clinical trial. Oncologist. 2006;11:841–848. doi: 10.1634/theoncologist.11-7-841. [DOI] [PubMed] [Google Scholar]
- 7.Vannucchi AM, Ficarra G, Antonioli E, Bosi A. Osteonecrosis of the jaw associated with zoledronate therapy in a patient with multiple myeloma. Br J Haematol. 2005;128:738. doi: 10.1111/j.1365-2141.2005.05382.x. [DOI] [PubMed] [Google Scholar]
- 8.Bamias A, Kastritis E, Bamia C, et al. Osteonecrosis of the jaw in cancer after treatment with bisphosphonates: incidence and risk factors. J Clin Oncol. 2005;23:8580–8587. doi: 10.1200/JCO.2005.02.8670. [DOI] [PubMed] [Google Scholar]
- 9.Assael LA. New foundations in understanding osteonecrosis of the jaws. J Oral Maxillofac Surg. 2004;62:125–126. doi: 10.1016/j.joms.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Marx RE. Pamidronate (Aredia) and zoledronate (Zometa) induced avascular necrosis of the jaws: a growing epidemic. J Oral Maxillofac Surg. 2003;61:1115–1117. doi: 10.1016/s0278-2391(03)00720-1. [DOI] [PubMed] [Google Scholar]
- 11.Hoff AO, Toth B, Hu M, Hortobagyi GN, Gagel RF. Epidemiology and risk factors for osteonecrosis of the jaw in cancer patients. Ann N Y Acad Sci. 2011;1218:47–54. doi: 10.1111/j.1749-6632.2010.05771.x. [DOI] [PubMed] [Google Scholar]
- 12.Such E, Cervera J, Terpos E, et al. Cyp2c8 gene polymorphism and bisphosphonate-related osteonecrosis of the jaw in patients with multiple myeloma. Haematologica. 2011;96(10):1557–1559. doi: 10.3324/haematol.2011.042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicoletti P, Cartsos VM, Palaska PK, Shen Y, Floratos A, Zavras AI. Genomewide pharmacogenetics of bisphosphonate-induced osteonecrosis of the jaw: the role of RBMS3. Oncologist. 2012;17:279–287. doi: 10.1634/theoncologist.2011-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raje N, Woo S-B, Hande K, et al. Clinical, radiographic, and biochemical characterization of multiple myeloma patients with osteonecrosis of the jaw. Clin Cancer Res. 2008;14:2387–2395. doi: 10.1158/1078-0432.CCR-07-1430. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 16.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croft D, O’Kelly G, Wu G, et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29:420–427. doi: 10.1093/bioinformatics/bts715. [DOI] [PubMed] [Google Scholar]
- 23.Franceschini A, Szklarczyk D, Frankild S, et al. String v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashcroft J. Bisphosphonates and phossy-jaw: breathing new life into an old problem. Lancet Oncol. 2006;7:447–449. doi: 10.1016/S1470-2045(06)70704-5. [DOI] [PubMed] [Google Scholar]
- 27.Forster C. The Role of the Innate Immune System in Bisphosphonate-induced Osteonecrosis of the Jaw [dissertation] Toronto: University of Toronto; 2011. [Google Scholar]
- 28.Ruggiero SL, Drew SJ. Osteonecrosis of the jaws and bisphosphonate therapy. J Dent Res. 2007;86:1013–1021. doi: 10.1177/154405910708601101. [DOI] [PubMed] [Google Scholar]
- 29.Sati HI, Greaves M, Apperley JF, Russell RG, Croucher PI. Expression of interleukin-1beta and tumour necrosis factor-alpha in plasma cells from patients with multiple myeloma. Br J Haematol. 1999;104:350–357. doi: 10.1046/j.1365-2141.1999.01193.x. [DOI] [PubMed] [Google Scholar]
- 30.Gordon JR, Galli SJ. Mast cells as a source of both preformed and immunologically inducible TNF-alpha/cachectin. Nature. 1990;346:274–276. doi: 10.1038/346274a0. [DOI] [PubMed] [Google Scholar]
- 31.Kast RE. Evidence of a mechanism by which etanercept increased TNF-alpha in multiple myeloma: new insights into the biology of TNF-alpha giving new treatment opportunities–the role of bupropion. Leuk Res. 2005;29:1459–1463. doi: 10.1016/j.leukres.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Tsimberidou AM, Waddelow T, Kantarjian HM, Albitar M, Giles FJ. Pilot study of recombinant human soluble tumor necrosis factor (TNF) receptor (p75) fusion protein (TNFR: Fc; Enbrel) in patients with refractory multiple myeloma: increase in plasma TNF alpha levels during treatment. Leuk Res. 2003;27:375–380. doi: 10.1016/s0145-2126(02)00082-6. [DOI] [PubMed] [Google Scholar]
- 33.Vangsted AJ, Klausen TW, Abildgaard N, et al. Single nucleotide polymorphisms in the promoter region of the IL1B gene influence outcome in multiple myeloma patients treated with high-dose chemotherapy independently of relapse treatment with thalidomide and bortezomib. Ann Hematol. 2011;90:1173–1181. doi: 10.1007/s00277-011-1194-3. [DOI] [PubMed] [Google Scholar]
- 34.Lacy MQ, Donovan KA, Heimbach JK, Ahmann GJ, Lust JA. Comparison of interleukin-1 beta expression by in situ hybridization in monoclonal gammopathy of undetermined significance and multiple myeloma. Blood. 1999;93:300–305. [PubMed] [Google Scholar]
- 35.Tai YT, Li XF, Breitkreutz I, et al. Role of B-cell-activating factor in adhesion and growth of human multiple myeloma cells in the bone marrow microenvironment. Cancer Res. 2006;66:6675–6682. doi: 10.1158/0008-5472.CAN-06-0190. [DOI] [PubMed] [Google Scholar]
- 36.Lichtenstein A, Berenson J, Norman D, Chang MP, Carlile A. Production of cytokines by bone marrow cells obtained from patients with multiple myeloma. Blood. 1989;74:1266–1273. [PubMed] [Google Scholar]
- 37.Pennanen N, Lapinjoki S, Urtti A, Mönkkönen J. Effect of liposomal and free bisphosphonates on the IL-1 beta, IL-6 and TNF alpha secretion from RAW 264 cells in vitro. Pharm Res. 1995;12:916–922. doi: 10.1023/a:1016281608773. [DOI] [PubMed] [Google Scholar]
- 38.Cao JJ. Effects of obesity on bone metabolism. J Orthop Surg Res. 2011;6:30. doi: 10.1186/1749-799X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Lo HS, Yang H, et al. Computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. Cancer Res. 2003;63:655–657. [PubMed] [Google Scholar]
- 40.Stimpfl M, Tong D, Fasching B, et al. Vascular endothelial growth factor splice variants and their prognostic value in breast and ovarian cancer. Clin Cancer Res. 2002;8:2253–2259. [PubMed] [Google Scholar]
- 41.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 42.Abdelhaleem M. RNA helicases: regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, Lin C, Zhao S, Wang H, Liu ZR. Phosphorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell proliferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem. 2007;282:16811–16819. doi: 10.1074/jbc.M610488200. [DOI] [PubMed] [Google Scholar]
- 44.Stucke VM, Gorses D, Hofmann F. DEAD-box RNA helicase p68 is not required for nuclear translocation of beta-catenin in colon cancer cells. Cell Cycle. 2008;7:830–832. doi: 10.4161/cc.7.6.5614. [DOI] [PubMed] [Google Scholar]
- 45.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) Am J Transl Res. 2010;2:223–234. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida H, Nadanaka S, Sato R, Mori K. XBP1 is critical to protect cells from endoplasmic reticulum stress: evidence from Site-2 protease-deficient Chinese hamster ovary cells. Cell Struct Funct. 2006;31:117–125. doi: 10.1247/csf.06016. [DOI] [PubMed] [Google Scholar]
- 48.Shajahan AN, Riggins RB, Clarke R. The role of X-box binding protein-1 in tumorigenicity. Drug News Perspect. 2009;22:241–246. doi: 10.1358/dnp.2009.22.5.1378631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee K, Tirasophon W, Shen X, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujimoto T, Onda M, Nagai H, Nagahata T, Ogawa K, Emi M. Upregulation and overexpression of human X-box binding protein 1 (hXBP-1) gene in primary breast cancers. Breast Cancer. 2003;10:301–306. doi: 10.1007/BF02967649. [DOI] [PubMed] [Google Scholar]
- 51.Munshi NC, Hideshima T, Carrasco D, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]