Abstract
The breadth of genomic diversity found among organisms in nature allows populations to adapt to diverse environments1,2. However, genomic diversity is difficult to generate in the laboratory and new phenotypes do not easily arise on practical timescales3. Although in vitro and directed evolution methods4–9 have created genetic variants with usefully altered phenotypes, these methods are limited to laborious and serial manipulation of single genes and are not used for parallel and continuous directed evolution of gene networks or genomes. Here, we describe multiplex automated genome engineering (MAGE) for large-scale programming and evolution of cells. MAGE simultaneously targets many locations on the chromosome for modification in a single cell or across a population of cells, thus producing combinatorial genomic diversity. Because the process is cyclical and scalable, we constructed prototype devices that automate the MAGE technology to facilitate rapid and continuous generation of a diverse set of genetic changes (mismatches, insertions, deletions). We applied MAGE to optimize the 1-deoxy-d-xylulose-5-phosphate (DXP) biosynthesis pathway in Escherichia coli to overproduce the industrially important isoprenoid lycopene. Twenty-four genetic components in the DXP pathway were modified simultaneously using a complex pool of synthetic DNA, creating over 4.3 billion combinatorial genomic variants per day. We isolated variants with more than fivefold increase in lycopene production within 3 days, a significant improvement over existing metabolic engineering techniques. Our multiplex approach embraces engineering in the context of evolution by expediting the design and evolution of organisms with new and improved properties.
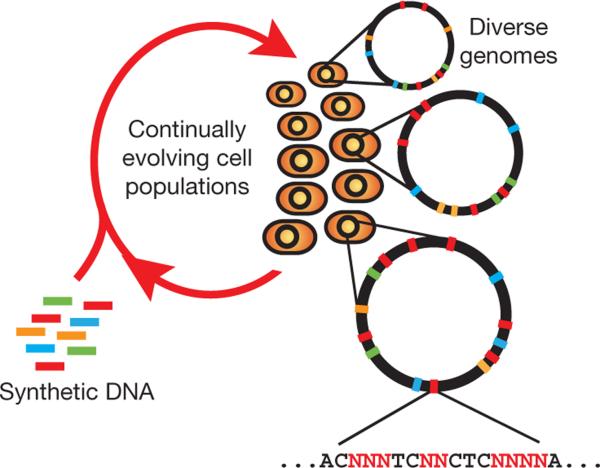
With the advent of next-generation fluorescent DNA sequencing10, our ability to sequence genomes has greatly outpaced our ability to modify genomes. Existing cloning-based technologies are confined to serial and inefficient introduction of single DNA constructs into cells, requiring laborious and outdated genetic engineering techniques. Whereas in vivo methods such as recombination-based genetic engineering (recombineering) have enabled efficient modification of single genetic targets using single-stranded DNA (ssDNA)11–14, no such attempts have been made to modify genomes on a large and parallel scale. MAGE provides a highly efficient, inexpensive and automated solution to simultaneously modify many genomic locations (for example, genes, regulatory regions) across different length scales, from the nucleotide to the genome level (Fig. 1).
Figure 1. Multiplex automated genome engineering enables the rapid and continuous generation of sequence diversity at many targeted chromosomal locations across a large population of cells through the repeated introduction of synthetic DNA.
Each cell contains a different set of mutations, producing a heterogeneous population of rich diversity (denoted by distinct chromosomes in different cells). Degenerate oligo pools that target specific genomic positions enable the generation of a diverse set of sequences at each chromosomal location.
Efficiency of the MAGE process was characterized using a modified E. coli strain (EcNR2). Mediated by the bacteriophage λ-Red ssDNA-binding protein β, allelic replacement is achieved in EcNR2 by directing ssDNA or oligonucleotides (oligos) to the lagging strand of the replication fork during DNA replication14. We optimized a number of parameters (see Supplementary Information, Supplementary Fig. 2 and Supplementary Table 1) to maximize efficiency of oligo-mediated allelic replacement. To generate sequence diversity in any region of the chromosome by allelic replacement, a pool of targeting oligos is repeatedly introduced into a cell. Under optimized conditions, we can successfully introduce new genetic modifications in >30% of the cell population (Supplementary Fig. 2d) every 2–2.5 h.
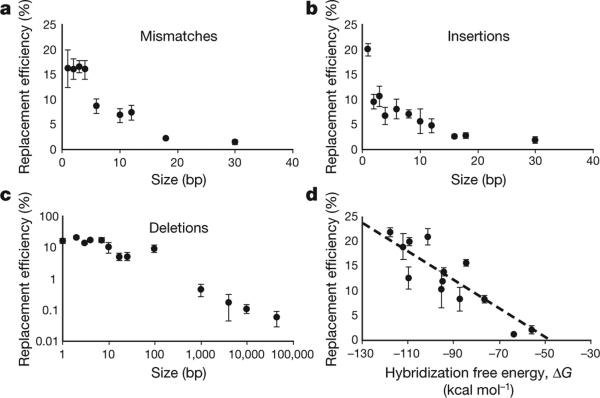
Oligo-mediated allelic replacement is capable of introducing a variety of genetic modifications at high efficiency. The efficiency of generating a mismatch or insertion modification is correlated to the amount of homologous sequence between the oligo and its chromosomal target (Fig. 2a, b); the efficiency of producing a deletion modification is correlated to the size of the deletion (Fig. 2c). Figure 2d shows that the predicted two-state hybridization free energy ΔG (ref. 15) between the oligo and target chromosomal sequence is a predictor of the allelic replacement efficiency. Thus, in a pool of oligos with degenerate sequences, oligos with more homology to the target will be incorporated in the chromosome at a higher frequency than those with less homology. This feature of MAGE enables tunable generation of divergent sequences along favourable evolutionary paths.
Figure 2. Characterization of allelic replacement efficiency as a function of the type and scale of genetic modifications.
a, Introducing mismatch mutations of up to 30 bp. b, Inserting exogenous sequences of up to 30 bp. c, Removing up to 45 kbp of chromosomal sequence using a single oligo. d, Correlation of replacement efficiency and two-state hybridization energy ΔG between the oligo and the targeted complement region in the genome. See Supplementary Fig. 1 for an illustration of oligo interaction with genomic targets and Supplementary Table 3 for a list of oligos and corresponding ΔG values. Dashed line is the linear regression correlation (y = −0.288x – 13.7, R2 = 0.799). All oligos used were 90 bp with two phosphorothioate bonds at the 3′ and 5′ ends. All error bars indicate ± s.d.; n = 3.
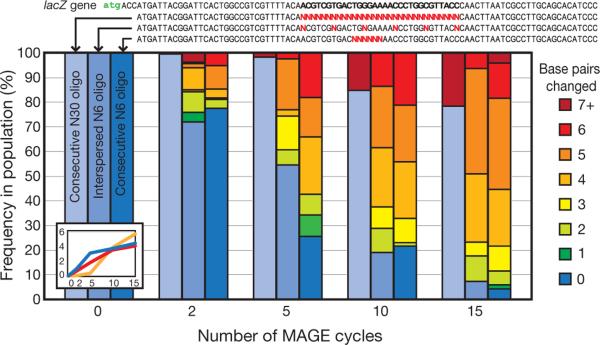
To determine the rate at which MAGE generates sequence diversity, we used three different 90-mer oligos to produce mismatch changes in a targeted region of the lacZ gene in three distinct cell populations. The cN6 and cN30 oligos contained 6 and 30 consecutive degenerate bases, respectively; the iN6 oligos contained 6 degenerate bases interspersed across a 30-bp region (Fig. 3). For these cell populations, the targeted lacZ region was sequenced in 96 random clonal isolates after MAGE cycles 2, 5, 10 and 15 that provided a snapshot of the genotypic variation in each population. Through successive cycles of MAGE, the chromosomal sequence of the lacZ region increasingly diverged away from wild type (Fig. 3). For cN6, after five MAGE cycles, we detected an average change of 3.1 bp per cell across the population (Fig. 3 inset, blue line), which equates to the generation of more than 4.3 × 109 bp of variation per day (3.1 bp changes per five cycles in 7 × 108 cells at 10 cycles per day). Within 15 cycles, cell populations containing all possible N6 genotype combinations were generated using either cN6 or iN6 oligos. Because the replacement efficiency for a 30-bp mismatching oligo is lower (1.5% from Fig. 2a), only 21.8% of the cN30 cell population had undergone allelic replacement after 15 cycles. We detected an average change of 5.6 bp per cell from the wild-type sequence across the whole cN30 population (Fig. 3 inset, orange line).
Figure 3. Sequence diversity generated across three separate cell populations as a function of the number of MAGE cycles.
Three 90-mer oligo pools were investigated: cN30, iN6 and cN6. cN30 contains oligos with 30 bp of consecutive degeneracy; iN6 contains oligos with 6 bp of degeneracy spaced every 5 bp; cN6 contains oligos with 6 bp of consecutive degeneracy. Frequency of strains in each population that contains 0 to 7+ bp of differences from the wild-type lacZ sequence are colour-coded. The inset shows average number of base pairs changed from wild type across the whole cell population as a function of the number of MAGE cycles using the three oligo pools cN30 (orange line), cN6 (blue line) and iN6 (red line).
The depth at which MAGE generates diversity is determined by a combination of three factors: (1) the degree of sequence variation desired at each locus; (2) the number of loci targeted; and (3) the number of MAGE cycles performed. When a single locus is targeted using a degenerate oligo pool, genetic diversity is generated across the population at that locus and is a function of the oligo pool complexity only. If more than one locus is targeted simultaneously, diversity is generated through the combinatorial arrangement of the different modified loci. The frequency at which each locus is modified can be computationally predicted through a binomial distribution (Supplementary Fig. 3). Although the cell population at any cycle may reflect only a subset of all variants theoretically possible, we can cumulatively generate more variants than the actual size of the cell population (~7 × 108 cells) through successive MAGE cycles. Thus, we can generate all variants regardless of population size through computational predictions and continuous cycling by MAGE.
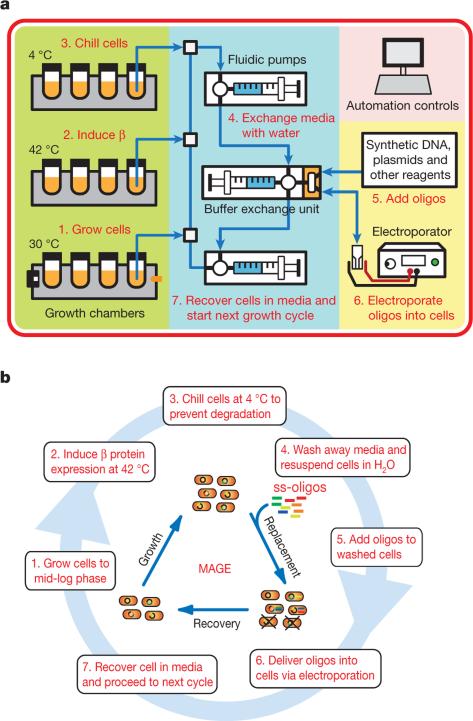
Given the cyclical and scalable nature of our approach, we constructed an integrated prototype device that automates the MAGE process to enable fast and reliable cellular programming. The device contains growth chambers to maintain healthy cell cultures and electroporation modules to repeatedly deliver DNA into the cells, thereby facilitating genome engineering and evolution (Fig. 4, Methods and Supplementary Information). Complex culturing conditions can be programmed into the device for growth of a diverse set of organisms and ecosystems.
Figure 4. MAGE automation.
a, Detailed schematic diagram of MAGE prototype including climate-regulated growth chambers with real-time cell density monitors (green), anti-fouling fluidics for transfer of cells between growth chambers and exchange of media and buffers (blue), and real-time generation of competent cells for transformation with synthetic DNA (yellow). Cultures are carried through different chambers at different temperature regimes (30 °C, 42 °C, 4 °C) depending on the necessary MAGE steps (that is, cell growth, heat-shock, cooling). Cells are made electrocompetent by concentration onto a filter membrane and resuspension with wash buffer. Oligos are delivered into cells by electroporation. b, Step-by-step diagram of MAGE cycling steps at a total run time of 2–2.5 h per cycle. Owing to high voltage (18 kV cm−1) electroporation, ~95% of cells are killed at each cycle. Hence, the electroporation event serves to both introduce oligos into cells and to dilute the cell population, cells are then recovered and grown to mid-log phase (7 × 108 cells ml−1) in liquid medium for the subsequent cycle.
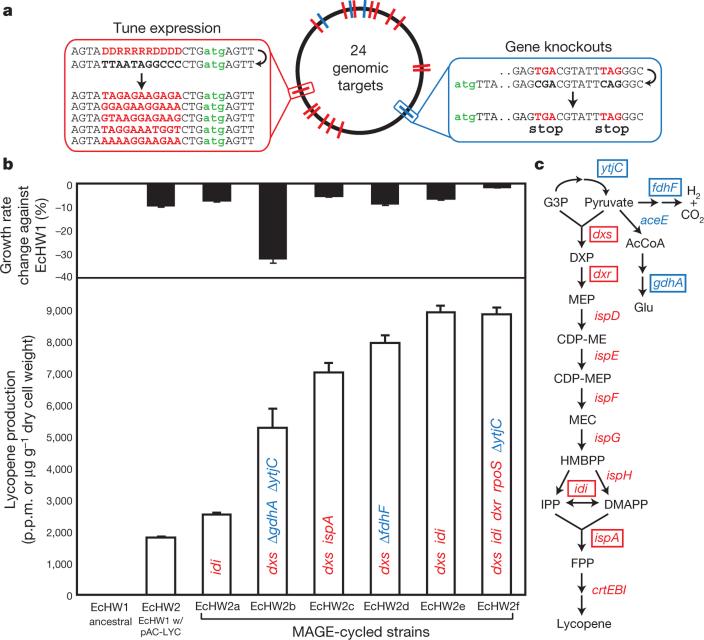
To demonstrate an application of the MAGE process, we optimized metabolic flux through the DXP biosynthesis pathway to overproduce the isoprenoid, lycopene, in an E. coli strain (EcHW2) that contained the pAC-LYC plasmid that is necessary for the final steps of lycopene production. Twenty endogenous genes (dxs, dxr, ispD, ispE, ispG, ispH, idi, ispA, appY, rpoS, crl, elbA, elbB, yjiD, purH, rnlA, yggT, ycgZ, ymgA, ariR) documented to increase lycopene yield16,17 were targeted to tune translation (Fig. 5a). Specifically, for each of the 20 genes, 90-mer oligos containing degenerate ribosome binding site (RBS) sequences (DDRRRRRDDDD; D = A, G, T; R = A, G) flanked by homologous regions on each side were used, with a total pool complexity of 4.7 × 105 (36 × 25 × 20). The replaced RBS regions were designed to be more similar to the canonical Shine–Dalgarno sequence (TAAGGAGGT)18, giving rise to enhanced translation efficiency. Additionally, four genes (ytjC, fdhF, aceE, gdhA) from secondary pathways19 were targeted for inactivation by oligos that introduced two nonsense mutations in the open reading frame, further improving flux through the DXP pathway. In contrast to prior strategies20–23 that were experimentally limited by the number of genetic components that could be manipulated at once, here we optimized 24 genes simultaneously to maximize lycopene production.
Figure 5. Optimization of the DXP biosynthesis pathway for lycopene production.
a, Genomic positions of 24 targeted genes with the RBS optimization strategy on the left (red) and gene knockout strategy on the right (blue). The gene knockout strategy involves the introduction of two nonsense mutations. All 90-mer oligos contain two phosphorothioated bases at the 3′ and 5′ termini. b, Black bars represent the growth rate of isolated variants (EcHW2a–f) relative to the ancestral EcHW1 strain. White bars represent lycopene production in p.p.m., which is normalized by dry cell weight in ancestral and mutant strains. Colour-coded labels in each white bar represent genetic modifications found by sequencing. All error bars indicate ± s.d.; n = 3. c, Modifications to the lycopene biosynthesis pathway of isolated variants EcHW2a–f with relevant genes highlighted by rectangular boxes. Blue labels represent knockout targets, red labels represent RBS tuning targets. AcCoA, acetyl-CoA; CDP-ME, 4-diphosphocytidyl-2-C-methyl-d-erythritol; CDP-MEP, 4-diphosphocytidyl-2C-methyl-d-erythritol-2-phosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; G3P, glyceraldehyde 3-phosphate; HMBPP, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate; IPP, isopentenyl diphosphate; MEP, 2-C-methyl-d-erythritol-4-phosphate; MEC, 2C-methyl-d-erythritol-2,4-cyclodiphosphate.
As many as 15 billion genetic variants (4.3 × 108 bp variations per cycle for 35 MAGE cycles) were generated. Screening of variants was done by isolating colonies that produced intense red pigmentation on Luria–Bertani agar plates. Variants were isolated from ~105 colonies screened after 5–35 cycles of MAGE (see Supplementary Information), some exhibiting as much as a fivefold increase in lycopene production relative to the EcHW2 ancestral strain (Fig. 5b). Under similar experimental conditions, our highest lycopene yield of ~9,000 p.p.m. (μg per g dry cell weight) is better than documented yields17,19. Sequencing of six variants (EcHW2a–EcHW2f) revealed RBS convergence towards consensus-like Shine–Dalgarno sequences in genes localized at the beginning and end of the biosynthesis pathway (dxs, dxr, idi, ispA) as well as various gene knockouts from secondary pathways (ΔytjC, ΔgdhA, ΔfdhF) (Fig. 5c and Supplementary Table 2).
Different tuning parameters of the lycopene pathway can be individually and combinatorially assessed in the isolated variants. For example, translation optimization of idi alone (EcHW2a) increased lycopene production by 40%. Whereas optimizing dxs and idi increased lycopene production by 390% (EcHW2e), additional optimization at rpoS and dxr, along with inactivation of ytjC, improved the growth rate of EcHW2f to that of the EcHW1 ancestor. Interestingly, rpoS is the alternative RNA polymerase subunit sigma factor σS, the master stress response regulator, and its upregulation can increase stress resistance to the accumulation of lycopene, a very hydrophobic molecule17. ytjC is an uncharacterized phosphoglyce-rate mutase enzyme and is thought to increase metabolic flux through the DXP pathway by increasing the accumulation of glyceraldehyde 3-phosphate intermediates19. Inactivation of glutamate dehydrogenase (gdhA) increases lycopene production, but causes a 32% decrease in growth rate of EcHW2b relative to wild type. Combinations of genetic modifications can also be assessed against each other. For example, an optimized dxs and idi strain (EcHW2e) produces 12% more lycopene than a strain with dxs and ΔfdhF (EcHW2d) and 27% more than a strain with dxs and ispA modifications (EcHW2c). The optimized DXP biosynthesis pathway presented here can be used to produce many other isoprenoid compounds of industrial and pharmaceutical relevance24.
The diversity (that is, the degree of sequence change per target) generated by MAGE is adjustable and the specificity of targeting is always high. Oligos with defined sequences produce well-defined modification, whereas oligos with degenerate sequences produce high-diversity modifications tailored for exploring a vast sequence space. In this study, we used well-defined oligos to inactivate protein-coding sequences and high-diversity degenerate oligos to modify and sample different RBS sequences. We have also used the MAGE platform to perform whole-genome recoding of E. coli to enhance the incorporation of non-natural amino acids into proteins25 and construct safer and multi-virus-resistant strains (F.J.I. et al., manuscript in preparation). MAGE is thus a complementary technology to de novo genome synthesis26, allowing the tuning of synthetic and natural genomes in vivo for various applications.
MAGE is also an accelerated evolution platform that permits the repeated introduction and maintenance of many neutral (or deleterious) mutations in the cell population. Although these mutations would normally disappear in the population via genetic drift or natural selection, MAGE accelerates the rate of their accumulation in any individual cell, thus increasing the likelihood of finding sets of mutations that may interact synergistically to produce a surprisingly beneficial phenotype. Using this technology, we could engineer or evolve cells with higher transfection efficiency (for example, harnessing natural competence systems27), increased allelic replacement efficiency (for example, expressing higher levels or mutants28 of the λ-Red β protein) and perform large-scale bacterial artificial chromosome engineering. The simple allelic replacement mechanism could make this method amenable for use in other organisms, given that other ssDNA-binding protein homologues are functional29. Currently, 30 US dollars of commercially synthesized oligos can introduce up to 27 bp of modification at full degeneracy for a single genomic target. To target many loci, obtaining oligos from programmable DNA microchips30 can significantly decrease the cost in comparison to traditional oligo synthesis. We envision that large-scale pipelines to program synthetic organisms and ecosystems10 will greatly benefit from integration of hardware, software and wetware to engineer and evolve microbial, plant and animal systems.
METHODS
Strains and culture conditions
The λ prophage was obtained from strain DY33031, modified to include the bla gene and introduced into wild-type MG1655 E. coli by P1 transduction at the bioA/bioB gene locus and selected on ampicillin to yield the strain EcNR1 (λ-Red+). Replacement of mutS with the chloramphenicol resistance gene (cmR cassette) in EcNR1 produced EcNR2 (mutS−, λ-Red+). EcNR2 was grown in low salt LB-min medium (10 g tryptone, 5 g yeast extract, 5 g NaCl in 1 l dH2O) for optimal electroporation efficiency. A premature stop codon was introduced into the cmR gene of EcNR2 with oligo cat_fwd_stop (Supplementary Table 3) to produce EcFI5, thus inactivating the cmR gene. An oligo (cat_fwd_restore) containing the wild-type sequence was used to restore the CmR phenotype. The pAC-LYC plasmid32 containing genes crtE, crtB and crtI was electroporated into EcNR1 to generate EcHW1, which produces lycopene at basal levels. Replacement of mutS with a kanamycin resistance gene in EcHW1 produced EcHW2.
Oligonucleotides and DNA sequencing
All oligonucleotides were obtained from Integrated DNA Technologies with standard purification. Oligonucleotides used in the MAGE process contained the following modifications: (1) 30–110 bp in length for optimization experiments; (2) different numbers of phosphorothioated bases; and (3) degenerate nucleotides as described elsewhere in this paper. Additional primers were purchased to amplify relevant genetic regions of the lycopene pathway to sequence strains that expressed high levels of lycopene. DNA sequencing to confirm allelic replacements was performed by Agencourt Bioscience.
LacZ and chloramphenicol replacement efficiency assays
Replacement efficiency was characterized by performing the allelic replacement protocol on EcNR2 cells using 90-mer oligos (Supplementary Table 3) that produced a premature stop codon in the chromosomal lacZ gene. In general, 250–500 cells were plated on LB-min containing 5-bromo-4-chloro-3-indoyl-β-d-galactoside and isopropyl-β-d-thiogalactoside (USB Biochemicals) agar plates. Efficiency of allelic replacement was calculated by taking the ratio of the number of white colonies to the total number of colonies on plates. A similar strategy was used in the cmR gene recovery experiments with strain EcFI5 where 30–110mer oligos were used to determine optimal oligo length for allelic replacement (Supplementary Fig. 2a). These oligos contained two phosphorothioate bonds at both the 5′ and 3′ termini. Cells were plated on LB-min-chloramphenicol and LB-min agar plates and grown overnight. Efficiency of allelic replacement was calculated by taking the ratio of the number of colonies on LB-minchloramphenicol plates to the number of colonies on LB-min plates.
MAGE automation device
Cells were grown in sterilized 10-ml glass vials placed in thermally conductive blocks. The growth chambers were climate-regulated through temperature controllers that actuate Peltier heating/cooling elements. Cultures acclimatized quickly (<15 s) in chambers held at different temperatures due to small volumes (for example, 3 ml). Aeration of the culture was achieved through agitation of the chambers at 300 r.p.m. using an orbital shaker. Real-time monitoring of growth rates was achieved by detecting changes in light transmittance across the chamber from light-emitting diodes emitting at 600 nm. Cultures were transferred between chambers through solenoid isolation valves using syringe pumps. Cells were made electrocompetent by a filtration system, which uses a syringe pump to concentrate cells onto a filter membrane (0.22 μm pore size) and resuspend them off the membrane with appropriate electroporation buffer (for example, dH2O). Single-stranded oligos (or PCR products) were electroporated into cells in a conductive cuvette with an electric pulse (18 kV cm−1). After each cycle, the entire system (chambers, syringes, filters) was washed with 70% ethanol followed by dH2O three times to reduce contamination and biofilm formation. All instruments were digitally controlled through software written in the LabView programming environment (National Instruments).
Cultures were initially inoculated into 3 ml of medium in the growth chamber. The device then executed repeatedly and continuously through the following steps: (1) grow cells at 30 °C to a pre-set density (that is, OD600 nm of 0.7); (2) induce cells for allelic replacement via 42 °C heat shock for 15 min; (3) chill cells at 4 °C to halt cellular metabolism; (4) wash cells through 15–20 iterations of filtration and resuspension with dH2O; (5) mix cell suspension and synthetic DNA; (6) deliver DNA into cells by electroporation; and (7) resuspend electro-porated cells with growth media.
Colorimetric screen and assay for lycopene production
Cells from the cycled EcHW2 population were plated on LB-min-chloramphenicol agar plates and grown for 1 day at 30 °C and 2 days at room temperature to produce red colonies. The 24 gene targets were divided into three oligo pools, one containing 10 targets, a second containing 14 targets and a third containing all 24 targets, which were cycled through the MAGE process in three separate cultures. In total, 105 colonies with increased colour intensity by visual inspection were screened after cycles 5, 10, 15, 20, 25, 30 and 35. From 127 isolates, six strains (EcHW2a–f, with representation from each pool) were selected for direct DNA sequencing across all gene targets and quantitatively measured for lycopene production. For lycopene quantification, these isolated colonies were grown in LB-min medium for 24 h. Lycopene was extracted from 1 ml of cells as follows: centrifuged at 16,000g for 30 s, removal of supernatant media and resuspended with water, and recentrifuged at 16,000g for 30 s. Once the supernatant was removed, the cells were resuspended in 200 μl acetone and incubated in the dark for 15 min at 55 °C with intermittent vortexing. The mixture was centrifuged at 16,000g for 30 s and the supernatant lycopene solution was transferred to a clean tube for quantification. Absorbance at 470 nm of the extracted lycopene solution was measured using a spectrophotometer and calibrated against a known lycopene standard (Sigma-Aldrich, catalogue number L9879) to determine the lycopene content. Dry cell weight was determined by baking a washed cell pellet at 105 °C for 24 h in the dark. Lycopene yield of different EcHW2 derivatives (EcHW2a–f) was calculated by normalizing the amount of lycopene extracted to the dry cell weight.
Supplementary Material
Acknowledgements
We are grateful to J. Jacobson for his insights and advice throughout this work. We thank D. Court for his insights and sharing strain DY330, N. Reppas for advice and sharing strain EcNR2, F. X. Cunningham for sharing pAC-LYC, and B. H. Sterling for assistance in constructing the EcFI5 strain. We also thank M. Jewett, J. Aach, D. Bang, S. Kosuri and members of the Church laboratory for advice and discussions. We thank the NSF, DOE, DARPA, the Wyss Institute for Biologically Inspired Engineering and training fellowships from the NIH and NDSEG (H.H.W.) for supporting this research.
Footnotes
Author Contributions H.H.W., F.J.I. and G.M.C. conceived the study jointly with P.A.C.; H.H.W. and F.J.I. designed and performed experiments with assistance from P.A.C., Z.Z.S., G.X. and C.R.F.; H.H.W. and F.J.I. wrote the manuscript; G.M.C. supervised all aspects of the study.
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 2.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 3.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nature Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 4.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 5.Crameri A, Raillard S-A, Bermudez E, Stemmer WP. C. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature. 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 6.Joo H, Lin Z, Arnold FH. Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature. 1999;399:670–673. doi: 10.1038/21395. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YX, et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 8.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nature Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 9.Cadwell RC, Joyce GF. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 10.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 12.Costantino N, Court DL. Enhanced levels of l Red-mediated recombinants in mismatch repair mutants. Proc. Natl Acad. Sci. USA. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nature Protocols. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl Acad. Sci. USA. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin YS, Stephanopoulos G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab. Eng. 2007;9:337–347. doi: 10.1016/j.ymben.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Kang MJ, et al. Identification of genes affecting lycopene accumulation in Escherichia coli using a shot-gun method. Biotechnol. Bioeng. 2005;91:636–642. doi: 10.1002/bit.20539. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine – Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab. Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopeneoverproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nature Biotechnol. 2005;23:612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 21.Farmer WR, Liao JC. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol. Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- 22.Kim SW, Keasling JD. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001;72:408–415. doi: 10.1002/1097-0290(20000220)72:4<408::aid-bit1003>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 23.Yuan LZ, Rouviere PE, Larossa RA, Suh W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 2006;8:79–90. doi: 10.1016/j.ymben.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Khosla C, Keasling JD. Metabolic engineering for drug discovery and development. Nature Rev. Drug Discov. 2003;2:1019–1025. doi: 10.1038/nrd1256. [DOI] [PubMed] [Google Scholar]
- 25.Cropp TA, Schultz PG. An expanding genetic code. Trends Genet. 2004;20:625–630. doi: 10.1016/j.tig.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 27.Metzgar D, et al. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 2004;32:5780–5790. doi: 10.1093/nar/gkh881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama M, Ohara O. Improvement of recombination efficiency by mutation of Red proteins. Biotechniques. 2005;38:917–924. doi: 10.2144/05386RR02. [DOI] [PubMed] [Google Scholar]
- 29.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc. Natl Acad. Sci. USA. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 31.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham FX, Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.