Abstract
In this study, the acid–base properties of the adenine cation radical are investigated by means of experiment and theory. Adenine cation radical (A•+) is produced by one-electron oxidation of dAdo and of the stacked DNA-oligomer (dA)6 by Cl2•− in aqueous glass (7.5 M LiCl in H2O and in D2O) and investigated by ESR spectroscopy. Theoretical calculations and deuterium substitution at C8–H and N6–H in dAdo aid in our assignments of structure. We find the pKa value of A•+ in this system to be ca. 8 at 150 K in seeming contradiction to the accepted value of ≤ 1 at ambient temperature. However, upon thermal annealing to ≥160 K, complete deprotonation of A•+ occurs in dAdo in these glassy systems even at pH ca. 3. A•+ found in (dA)6 at 150 K also deprotonates on thermal annealing. The stability of A•+ at 150 K in these systems is attributed to charge delocalization between stacked bases. Theoretical calculations at various levels (DFT B3LYP/6-31G*, MPWB95, and HF-MP2) predict binding energies for the adenine stacked dimer cation radical of 12 to 16 kcal/mol. Further DFT B3LYP/6-31G* calculations predict that, in aqueous solution, monomeric A•+ should deprotonate spontaneously (a predicted pKa of ca. −0.3 for A•+). However, the charge resonance stabilized dimer AA•+ is predicted to result in a significant barrier to deprotonation and a calculated pKa of ca. 7 for the AA•+ dimer which is 7 pH units higher than the monomer. These theoretical and experimental results suggest that A•+ isolated in solution and A•+ in adenine stacks have highly differing acid–base properties resulting from the stabilization induced by hole delocalization within adenine stacks.
1. Introduction
Electron and hole transfer through DNA is of significance to an increasing number of areas of research including radiation damage, DNA damage and repair processes, DNA photonics, and electronics.1,2 Of the variety of processes that affect excess electron and hole transfer through DNA, proton transfer is perhaps the most important.1–7 Intrabase-pair proton transfer processes have been proposed to slow or stop hole and excess electron transfer through the helix and thereby act as a crucial restriction on such spin and charge transfer.3–7 Such proton transfer processes will strongly depend on the pKa of the base ion-radical involved.
The pKa of a cation radical is invariably found to be lower than its parent compound. For example, the pKa of guanine (N1–H) is reported as 9.6, while the corresponding pKa of the guanine cation radical (G•+) in H2O at ambient temperature is ca. 3.9.3,4,8 The site of deprotonation in G•+ had been somewhat controversial as it is reported to be N1 in solution in pulse radiolysis studies and at the amine group (N2–H) in solid state.9a Our recent ESR studies employing isotopically (15N and deuterium) substituted dGuo confirm that the site of deprotonation in G•+ is at N1 in aqueous solution, and theoretical calculations9a confirm shifting of that locus of deprotonation of G•+ from N2 in a low dielectric solid9b to N1 in an aqueous environment.9a
One-electron oxidized adenine (adenine cation radical (A•+) and its deprotonated form (A(–H)•)) has also been well studied (i) employing pulse radiolysis and flash photolysis in aqueous solution,3,4,8,10,11 (ii) in γ-irradiated aqueous (D2O) glassy systems at low temperature,12 (iii) in X-ray irradiated single crystals,7,13–16 and in (iv) theoretical treatments.17–19,20a,b,21 Steenken and co-workers produced A•+ from dAdo in aqueous solution employing 248 nm laser photolysis (20 ns pulses), and from their measurements between pH’s 0 and 6, they concluded that the pKa of A•+ is e1.3,4 This value is much lower than the pKa of the corresponding N6–H atom (≥14 4) in neutral dAdo. Thus, ΔpKa(A/A•+) is much larger than the corresponding ΔpKa(G/G•+). However, in pulse radiolysis studies of dinucleoside phosphates ApG and GpA, rapid migration of the hole from one-electron oxidized adenine to G was proposed and the rate of this hole migration was shown to decrease by a factor of ca. 3 from pH 2.3 to 7.22 From these results, it was suggested that the pKa of A•+ (≤1) might be “too low” for these dinucleoside phosphate systems (see footnote 25 in ref 22). In view of the work to be presented here, these earlier results are the first results suggesting that base stacking may affect the pKa of A•+. In a much earlier NMR spectral study (360 MHz) of one-electron-oxidized adenine produced via laser CIDNP in 5′-AMP at various pH’s, the pKa of A•+ in aqueous (D2O) solution is reported as 4 ± 0.2.23 Theoretical calculations predict the pKa of A•+ as ca. 220a and 3.2.20b It is interesting to note that adenine has been suggested to play an important role of a hole carrier in long A–T stretches,24–27 and such hole transfer can occur over long A stacks if G does not interrupt the A sequence.28 From the very low pKa value reported by Steenken (≤1) of A•+, it would seem likely that A•+ would deprotonate rapidly and this deprotonation would stop the hole from being transferred further in these adenine tracts. This apparently does not occur, and one plausible explanation of this long-range hole transfer in these A-tracts could be that the rate of hole transfer in these adenine tracts is faster than the rate of its deprotonation. Recently, the rate of hole transfer in such adenine tracts was experimentally determined (kHT = 108 to 1010 s−1),25 and this value is slower than the expected rate of deprotonation base cation radicals.3,4 From this one would incorrectly predict deprotonation would occur during hole transfer. Recent work by Barton and co-workers suggests that the hole within these adenine tracts is delocalized over several adenine bases.27 Delocalization of the hole in A-tracts would likely stabilize the hole toward deprotonation, although no theoretical or experimental studies have treated this issue. From the above discussion it is clear that the nature of A•+ and its acid base properties are still open questions in DNA systems.
In this work, we present experimental and theoretical work that elucidates the acid–base properties of A•+. Following our previous works,1,9,12 we have produced A•+ by one-electron oxidation of dAdo by Cl2•− in an aqueous glass (7.5 M LiCl in H2O and in D2O at various pH’s and concentrations). A•+ was also formed via one-electron oxidation of the DNA-oligomer (dA)6 by Cl2•− in an aqueous glass (7.5 M LiCl in H2O). We have employed deuterium substitution in dAdo (at C8–H and N6–H) aided with theoretical calculations to determine the hyperfine coupling constants and to confirm our assignments of structure (Figure 1). Theoretical calculations are also employed to gain an understanding of the factors that increase the stability of A•+, and these calculations predict that base stacking can stabilize the cation radical state and can have profound effects on its prototropic equilibria. The experimental and theoretical results reported in this work suggest that, in constrast to G•+, the properties of A•+ in the isolated monomeric state can not explain the behavior of A•+ in DNA where stacking dramatically changes the properties of this cation radical.
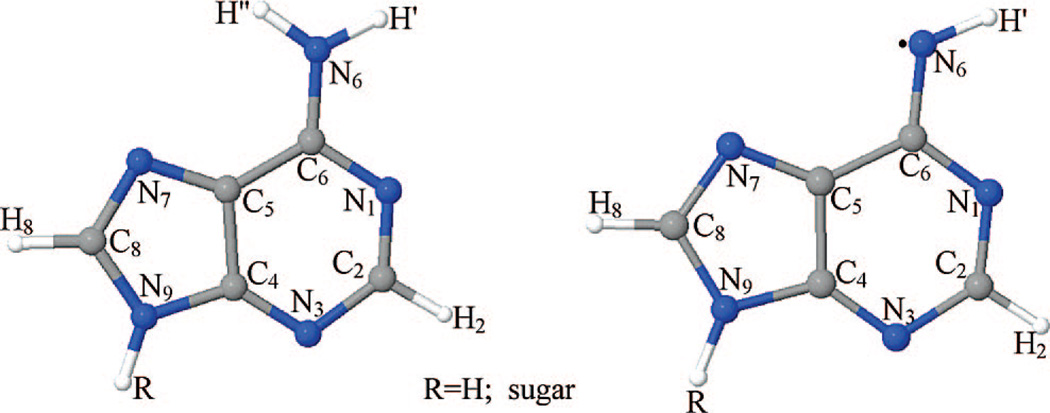
Figure 1.
Schematic diagram showing the atom numbering scheme of the adenine ring in the deoxyadenosine cation radical (A•+) and the syn conformation (with respect to N-atom N1 in the ring) of the deprotonated cation radical (A(–H)•) is shown.
2. Materials and Methods
2.1. Compounds
Lithium chloride (99% anhydrous, SigmaUltra) and 2′-deoxyadenosine (dAdo) monohydrate were purchased from Sigma (St. Louis, MO). We had obtained potassium persulfate (crystal) from Mallinckrodt, Inc. (Paris, KY). Deuteration at C-8 in the adenine moiety was carried out following the procedure of Huang et al.29 using triethylamine (TEA) (Fischer Scientific, NJ) and D2O (Deuterium oxide, 99.9 atom % D) (Aldrich, Sigma-Aldrich, Inc., St. Louis, MO). The degree of deuteration (≥98%) was confirmed by 1D NMR signal integration in DMSO-D6/99.9 atom % D (Wilmad LabGlass, Buena, NJ). The DNA-oligomer (dA)6 used in this study was obtained from IDT-Synthegen with standard desalting (Coralville, IA).
2.2. Sample Preparation
Following our work,9,12 solutions of 0.3 to 10 mg of dAdo, its C-8-deuterated derivative, and of (dA)6 (2 mg) were prepared in 1 mL of 7.5 M in H2O and in D2O in the presence of 3 to 6 mg of K2S2O8 as the electron scavenger. We performed several experiments with 15 M LiCl to assess the effect of LiCl on the prototropic equilibrium observed. pH’s of these solutions were adjusted by quick addition of adequate amounts (in µL) of 1 M NaOH or 1 M HCl in H2O or D2O under ice-cooled conditions. pH papers were used for pH measurements of these solutions. Due to this and also owing to the high ionic strength of these solutions, we consider these pH values as approximations.9,12 Deoxygenation of these solutions was carried out by bubbling thoroughly with nitrogen followed by drawing the solutions into 4 mm Suprasil quartz tubes (cat. no. 734-PQ-8, WILMAD Glass Co., Inc., Buena, NJ). The transparent glassy samples were then prepared by rapidly cooling the tubes containing these solutions to 77 K. The 7.5 to 15 M LiCl used in this work are not crystalline solids but glassy homogeneous supercooled liquids that on annealing soften to allow for molecular migration and solution phase chemistry. All these glassy samples were then stored in Teflon containers at 77 K in the dark.9,12
2.3. γ-Irradiation and Storage of Irradiated Samples
γ-Irradiation of the glassy samples of dAdo, of C-8-deuterated-dAdo (i.e., 8-D-dAdo), and of (dA)6 were carried out under liquid nitrogen using a model 109-GR9 irradiator, containing a shielded60Co source as per our earlier work9,12 with an absorbed dose of 2.5 kGy (for the nucleosides) and of 4.5 kGy (for the oligomer) at 77 K. All the irradiated samples were also stored at 77 K in the dark.
2.4. Formation of One-Electron Oxidized Adenine in dAdo and (dA)6 by Thermal Annealing of Samples
One-electron oxidized adenine species were produced by annealing γ-irradiated glassy samples in a variable temperature assembly (Air Products) in the dark via cooled nitrogen gas. The glassy samples were annealed for 12–20 min at 148 to 152 K where the glass softens sufficiently to allow for molecular migration which leads to the disappearance of Cl2•− with the concomitant formation of one-electron oxidized adenine (see Supporting Information Figure S1) as described in our earlier work.12 After γ-irradiation at 77 K and annealing, these glassy samples were immediately immersed and stored in liquid nitrogen (77 K). To observe deprotonation, samples were annealed further to slightly higher temperatures (ca. 160, 165 K) for 5–12 min and were stored immediately at 77 K in the dark for ESR studies. As in our previous works,1,9,12 formation of sugar radicals was not observed in these samples during annealing as long as they were kept in the dark at all times.
2.5. Electron Spin Resonance
For recording ESR spectra of these samples, we have used a Varian Century Series ESR spectrometer operating at 9.2 GHz with an E-4531 dual cavity, 9-in. magnet and with a 200 mW klystron.9,12 ESR spectra were recorded at 77 K and at the microwave power range 25–40 dB. We have used Fremy’s salt (with g = 2.0056, AN = 13.09 G) for field calibration (shown as reference marks in all figures with ESR spectra).9,12 As performed in our earlier works,9,12 a small singlet “spike” from irradiated quartz at g = 2.0006 was removed via subtraction from the recorded spectra before analyses.
2.6. Simulation of ESR Spectra
We have performed the anisotropic simulations of the experimental ESR spectra using SimFonia and WIN-EPR (Bruker) programs. The apparent g⊥ values of A•+ and A(–H)• were determined from the field position of the central component of their spectra (see Figures 2 and 3). Using these g⊥ values, we have obtained the values of g∥ by comparing the fits of the simulated spectra of A•+ and A(−H)• with the corresponding experimentally obtained spectra of A•+ and A(−H)• in 8-D-dAdo in D2O glass (see Figure 3). Theory predicts the major axis of the nitrogen coupling (Azz) is collinear with the intermediate proton couplings (Azz). This is expected since the pz-orbital on the C8 atom should be coaxial with the π-orbitals on the ring, and the optimized structures of A•+ and A(−H)• obtained via theoretical calculations (see Supporting Information Figure S7) are only slightly nonplanar. A•+ has 4 proton couplings (2 from the exocyclic N-atom, and 1 each from the C2 and C8 atoms), and A(–H)• has 3 proton couplings (1 with exocyclic N-atom and 1 each from C2 and C8). These proton coupling tensors all have the z-axis colinear, but the x and y axes are not. Hence, the calculated direction cosines, L (see Supporting Information Table T1), were employed to rotate the diagonalized A tensors to a common axis via (L−1A2L)1/2. The experimental spectra for A•+ obtained either from dAdo or from 8-D-dAdo (see Figures 2 and 3) do not have any observable resolution, and hence, we have used only the theoretically obtained values for its simulation. On the other hand, the experimental spectra of A(–H)• have observable resolutions both at the center and at the wings (see Figures 2 and 3) from the amine hydrogen couplings and the C-8 hydrogen coupling, as well as the nitrogen couplings. By selective deuteration, we observe changes in spectral components for A(−H)• which allows the estimation of the proton hyperfine couplings. In the C-8 deuterated A(−H)• in D2O only the poorly resolved nitrogen couplings are observed. Therefore, for A(−H)• we have adjusted the theoretically obtained HFCC values reported in the Table 1 to fit the experiment starting from the theoretically predicted values (see Table 1 footnote d for these adjusted values and the Supporting Information Table T1 for the original theoretical predictions for all cases considered).
Figure 2.
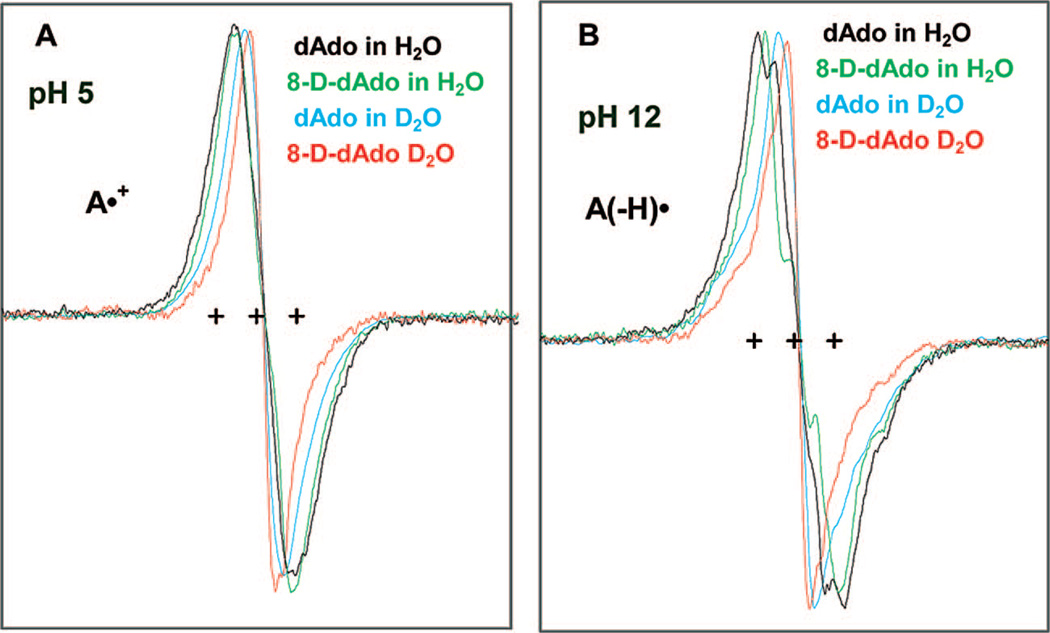
ESR spectra of one-electron oxidized adenine in dAdo or in 8-D-dAdo (3 mg/mL) produced by Cl2•− oxidation at 148 K (recorded at 77 K) in 7.5 M LiCl glass in H2O or in D2O in the presence of K2S2O8 at (A) native pH or pD (ca. 5) and at (B) pH or pD ca. 12. The three reference markers in this figure and in all the other figures of this report showing ESR spectra are Fremy’s salt resonances with the central marker is at g = 2.0056, and each of the three markers is separated from one another by 13.09 G.
Figure 3.
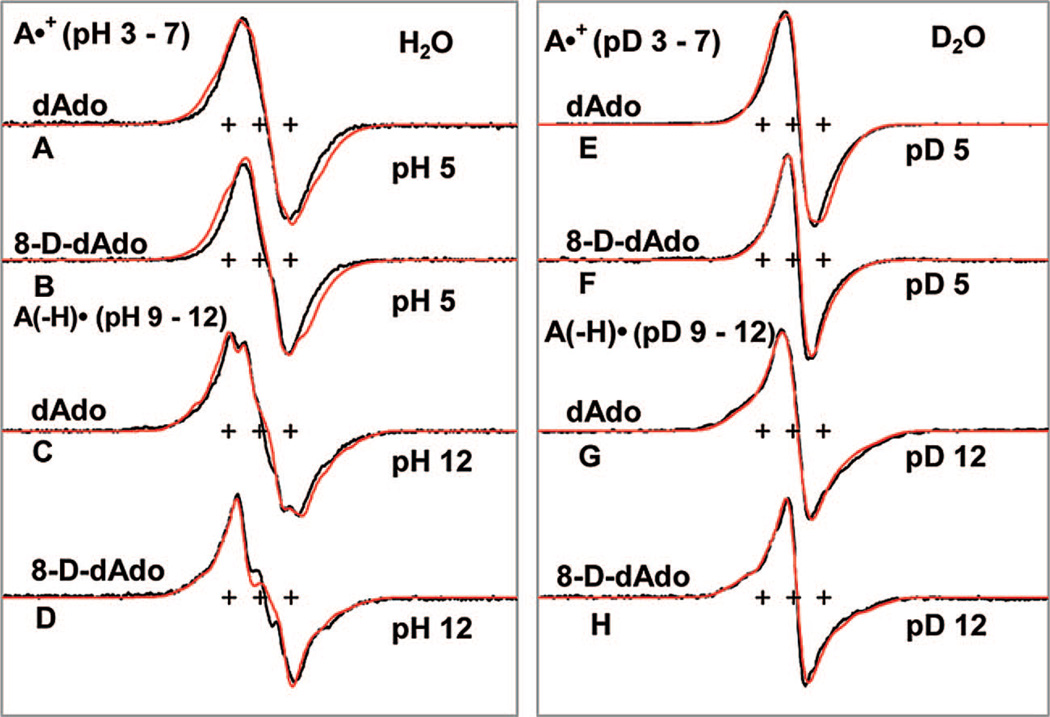
ESR spectra for one-electron oxidized dAdo or 8-D-dAdo (3 mg/mL) at various pH’s in 7.5 M LiCl glasses in H2O (A–D) (black) and in D2O (E–H) (black). The HFCC values and g-values used for the simulated spectra (red) are mentioned in Table 1. The spectra of A•+ and A(−H)• were recorded at 77 K after annealing to 148 and 152 K, respectively. These results show that, up to 150 K, a one-electron oxidized adenine moiety either in dAdo or in 8-D-dAdo remains as A•+ in the pH range 3–7 and as A(−H)• at and beyond pH 9 either in H2O or in D2O glasses.
Table 1.
B3LYP/6-31G* Calculated Hyperfine Coupling Constants (HFCCs) and Spin Densities at Different Atoms in A•+ and A(−H)•
| hyperfine couplings (Gauss) ATot = Aiso + Aii |
||||||
|---|---|---|---|---|---|---|
| species | atom no. | atomic spin densities |
Aiso | Azz | Ayy | Axx |
| A•+ (monomer)a,b | N6 | 0.2499 | 4.59 | 7.67 | −3.8 | −3.88 |
| gzz = 2.0021 | N7 | 0.1036 | 1.83 | 3.45 | −1.67 | −1.78 |
| gyy = 2.0041 | N3 | 0.1525 | 2.60 | 4.55 | −2.25 | −2.297 |
| gxx = 2.0041 | C8–Hc | 0.1861 | −5.74 | −0.74 | 3.697 | −2.95 |
| line width (3.0, 2.5, 2.5 (D2O)) G, and 4.0, 4.0, 4.0 (H2O)) G |
N6–Hc N6–Hc |
- - |
−6.78 −6.69 |
−1.34 −1.49 |
6.38 5.85 |
−5.04 −4.36 |
| C2–Hc | 0.0893 | −3.21 | −0.75 | 2.51 | −1.77 | |
| A(−H)• (monomer)a,b,c,d,e,f | N6 | 0.6176 | 8.77 | 17.21 | −8.49 | −8.73 |
| gzz = 2.0021 | N1 | 0.1749 | 2.51 | 4.7 | −2.30 | −2.39 |
| gyy = 2.0045 | N3 | 0.2380 | 4.15 | 6.8 | −3.37 | −3.43 |
| gxx = 2.0045 | C8–Hc | 0.1431 | −4.29 | −0.32 | 2.55 | −1.74 |
| line widthd | ||||||
| N6–Hc | - | −13.97 | −10.36 | 13.47 | −2.38 | |
For A•+ (monomer) and A( −H)•, optimization and HFCC values were calculated using DFT (B3LYP) with a 6-31G** basis set in the gas phase. HFCC values obtained using 6–31G** and 6–31 G* basis sets were found to be very similar.
Direction cosines are given in the Table T1 in the Supporting Information.
The spin density of the atom with which the H-atom is bonded, e.g., spin density on the C-atom in C8–H.
For A(−H)•, the adjusted HFCC values (sum total of isotropic and anisotropic HFCC values) used to fit the experimental spectra in Figure 3 are N6 (21, 0.5, 0.5) G, N1 (5, 0.25, 0.25) G, N7 (9, 0.7, 0.7) G, C8–H (−5, −2.5, −7.5) G and N6–H (−20, 0.5, −11) G.
(4.5, 5.0, 6.0) G For A(−H)• in 8-D-dAdo in D2O glass, (5.5, 6.0, 5.8) G for A(−H)• in dAdo in D2O glass; (4.2, 4.2, 4.2) for A(−H)• in 8-D-dAdo in H2O glass and (6.5, 4.2, 4.2) is for A(−H)• in dAdo.
Calculations performed in the gas phase of A(−H)• (monomer) and AA(−H)• in the stacked conformation gave very similar HFCC values.
2.7. Theoretical (DFT) Calculations
The geometries of all species described below were fully optimized at the B3LYP/6–31G* level of theory using the Gaussian 03 suite of programs.30a We have found optimized ground-state geometries for (i) deoxyadenosine cation radical (A•+) and (ii) syn- and anti-conformations (with respect to the nitrogen atom, N1, in the ring) of the amine deprotonated species, A(−H)• (see Figure 1 for atom numbering of the adenine ring). The adenine dimer cation radical, i.e., (AB–AB)•+, was treated using the adenine base only. This dimer cation radical was considered in two conformations: (i) in a planar hydrogen-bonded conformation (AB–AB)p•+ and (ii) in the parallel stacked conformation (AB–AB)s•+. The starting geometry for optimization of the stacked conformation was generated by placing the two adenine rings symmetrically along the axis normal to the adenine ring plane and separated by 3.4 Å. Further, to consider the effect of solvent surrounding the deoxyadenosine cation radical (A•+), the geometry of A•+ was fully optimized in the presence of seven water molecules. In addition, the base stacked dimer cation radical ((AB–AB)s•+) was fully optimized in the presence of 18 water molecules. The polarized continuum model (PCM) using the integral equation formalism (IEF) was used to take into account the effect of the bulk solvent only for calculating the pKa of the adenine base cation radical in the monomer, AB•+, and in the stacked dimer (AB–AB)s•+ conformation (see Supporting Information Tables T2 and T3). We note that theoretical calculations of pKa values are very sensitive to the level of calculation; however, Thorp and co-workers20a and Goddard and co-workers20c have found the that B3LYP functional gives results which are in good agreement with experiment. The hyperfine coupling constants (HFCCs) of these adenine radicals were also calculated at the same level of theory and basis set. Recently, the use of the B3LYP functional and 6–31G* basis set was found to predict HFCCs in good agreement with experimental values in a variety of molecules.9a,31 Molecular structures were drawn using the JMOL molecular modeling program32 while spin densities were plotted using the GaussView33 molecular modeling software.
The basis set superposition error (BSSE) corrected stabilization or binding energy of the adenine cation radical dimer is calculated as follows:
| (1) |
where ΔE is the BSSE corrected stabilization or binding energy, Edimer is the total energy of the adenine dimer cation radical, Eneutral is the total energy of the neutral adenine (monomer), and Ecation radical is the total energy of the adenine cation radical (monomer), respectively. The BSSE correction was performed using the standard Boys–Bernardi counterpoise correction scheme30b as implemented in Gaussian03.30a To calculate the stabilization energy of the adenine dimer cation radical, we have also employed the MP2 and MPWB95 functional apart from B3LYP as implemented in Gaussian03.30a Unless otherwise stated, all the energy values mentioned in the text are BSSE corrected.
3. Results and Discussion
3.1. ESR Studies
3.1.1. ESR Spectra of One-Electron Oxidized Adenine at ca. pH 5 and at ca. pH 12 in H2O and in D2O glasses at 150 K
In Figure 2, we present the ESR spectra of one-electron oxidized dAdo and 8-D-dAdo at the native pH (ca. 5) and also at ca. pH 12 of 7.5 M LiCl either in H2O or in D2O at 148 K. At pH 5 the cation radical, A•+, is found, whereas, at pH 12, the deprotonated radical, A(−H)•, is found. The various spectra in Figure 2A and 2B show the results of deuterium substitution at various sites and help to assign hyperfine couplings. Exocyclic amine hydrogens in dAdo (or in Ado) are exchangeable in D2O, and deuterons show hyperfine couplings 1/6.5 of that of protons in the same environment.9a Thus hydrogen couplings that are observed in H2O are lost in D2O (see Figure 2). This, along with larger dipolar interactions from the solvent in H2O vs D2O, explains the increase in the spectral width in H2O compared to that in D2O. Isotopic substitution of the C8-H atom in dAdo by deuterium is found to narrow the line width by ca. 4–5 G either for A•+ (see Figure 2A) or for A(−H)• (see Figure 2B). The constant field position of the central component (apparent g⊥value) for each of the conditions in Figure 2A is consistent with A•+ maintaining its molecular and electronic structure on isotopic substitution. A similar argument holds good for A(−H)• found in H2O or in D2O for dAdo and for 8-D-Ado at pH 12 (see Figure 2B). Note that the ESR spectrum of A•+ shows a clear and observable difference in the field position of the central component (apparent g⊥ value) when compared to that in the spectrum of its deprotonated form A(−H)• (see Figure 3). The apparent g┴ value (2.0041) of A•+ is lower than the corresponding value (2.0045) of apparent g⊥ of A(−H)•, and these values have been used for simulation of ESR spectra of these radicals (see Table 1).
3.1.2. ESR Spectra of One-Electron Oxidized Adenine at Various pH Ranges, e.g., 3–7 and 9–12 in H2O and in D2O Glasses
The ESR spectra of A•+ formed after annealing at 148 K in the dark, via one-electron oxidation by Cl2•− in the glassy (7.5 M LiCl) samples of dAdo and 8-D-dAdo either in H2O or in D2O and at pH (pD) 5 are shown in Figure 3A, 3B and 3E, 3F along with spectral simulations in red. The spectra shown in Figure 3A, 3B and 3E, 3F remain identical in the pH or pD range 3–7. The spectral shape, total hyperfine splitting, and the field position of the central component of spectrum 3E match very nicely with that already reported for A•+ in the literature (Figure 7A in Sevilla et al.34a). Simulation (shown in red color) using the apparent g⊥ value (2.0041) and the theoretically calculated HFCC values (see Table 1) for A•+ in dAdo (monomer, gas phase) and also for A•+ in 8-D-dAdo are shown and have not been adjusted to fit the experiment. While the simulated spectra for A•+ in D2O glasses are reasonable fits with the corresponding experimentally obtained spectra (see Figure 3E and 3F), they are somewhat broader in the corresponding H2O spectra (see Figure 3A and 3B). Thus, from Figure 3E and 3F, we conclude that the narrowing of the line width upon C8-H deuteration is well predicted by theory. However, there is a lack of resolution of individual components which does not allow for further experimental assignment of couplings. We find that the simulations are broader than the experimental spectra (see Figure 3A and 3B). We suggest that, for A•+, the exocyclic N-H HFCCs (see Table 1) are predicted to be somewhat too large in H2O. Theoretical calculations for stacked AA•+ suggest that owing to stacking induced charge delocalization over both bases, the HFCC values of the exocyclic N-H decrease, compared to their corresponding values predicted for the monomer cation radical (vide infra, and see also Supporting Information Figure S2, and Table T1), and this may be a possible reason for the marginal fit.
We have presented the ESR spectra of A(−H)• at pH or pD 12 in Figure 3C, 3D and 3G, 3H formed after annealing at 152 K, via one-electron oxidation by Cl2•− in the glassy (7.5 M LiCl) samples of dAdo and 8-D-dAdo either in H2O or in D2O along with the corresponding spectral simulations in red. For A(−H)•, identical ESR spectra are found at pH’s (pD’s) from 9 to 12. Simulations were performed employing apparent g┴ and g∥ values of 2.0045 and 20025 as well as HFCCs reported in Table 1 for A(−H)•. These simulations (shown in red color) match the experimental spectra (3C, 3D) and (3G, 3H) quite well. In these cases both the amine hydrogen couplings, the C-8 hydrogen coupling, and the nitrogen couplings are observable in the spectra. Hence, the corresponding calculated HFCC values reported in the Table 1 were adjusted to fit the experiment, and these are reported in footnote d of this table. In going from D2O to H2O glasses (compare Figure 3G vs 3C) we note the hyperfine couplings of the exchangeable N6–H proton coupling become clearly distinguishable. In addition, we note that the line width and spectral shape changes on deuteration at C8 (compare 3C with 3D and 3G with 3H) are well predicted by the simulations. The adjusted couplings reported in Table 1 for A(−H)• are expected to be good estimates of the major coupling values. We also note that theoretical calculations performed for A(−H)• (monomer) and AA(−H)• in the stacked conformation showed little effect of stacking and gave very similar HFCC values (see footnote h in Supporting Information Table T1).
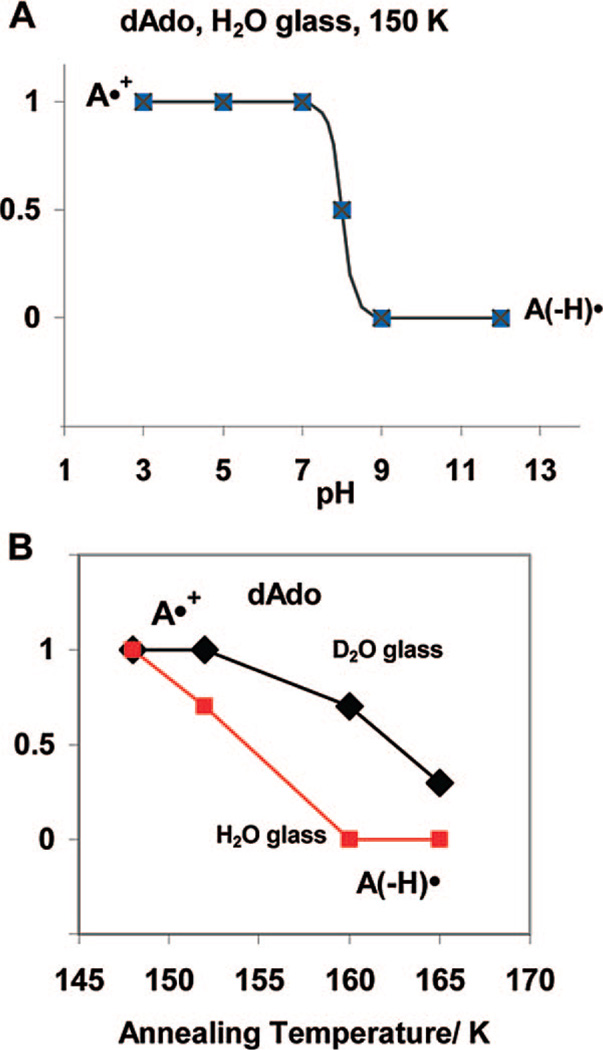
In the pH (or pD) range 3–7, the ESR spectrum for A•+ is found for dAdo and for 8-D-dAdo, whereas, in the pH (or pD) range 9–12, we find the ESR spectrum for A(−H)•. We have found at pH 8 at 150 K the ESR spectrum of A•+ and A(−H)• found in equal amounts. They are presumed to be in equilibrium at 150 K. This ESR spectrum and its analyses are now shown in Supporting Information Figure S3. This clearly indicates the pKa of A•+ in dAdo in the H2O glassy samples is ca. 8 (see Figure 4A) at 150 K. In D2O glassy samples at 155 K, we find the pKa of A•+ in dAdo is ca. 8.5. Moreover, we find no temperature dependence in the amounts of A•+ and A(–H)• at pH 7.5 from 77 to 150 K (see Supporting Information Figure S3).
Figure 4.
(A) The schematic diagram shows that A•+ is found for pH’s up to 7 and A(−H)• is found at pH’s of 9 and above at 150 K in 7.5 M LiCl H2O or D2O glasses. At pH 8, equal amounts of A•+ and A(−H)• are found; the pKa of A•+ is therefore ca. 8 at 150 K in these systems. In D2O glassy samples, the corresponding pKa of A•+ is ca. 8.5. (B) The prototropic equilibrium of A•+ is found at 150 K. However, annealing to a slightly higher temperature of ca. 160 K and above allows for molecular migration, and deprotonation is found at all pH’s investigated, i.e., 3 to 12, in H2O glasses. In D2O glasses, which soften at slightly higher temperatures, deprotonation of A•+ is nearly complete at 165 K.
3.1.3. Annealing Studies of A•+ Formed from dAdo
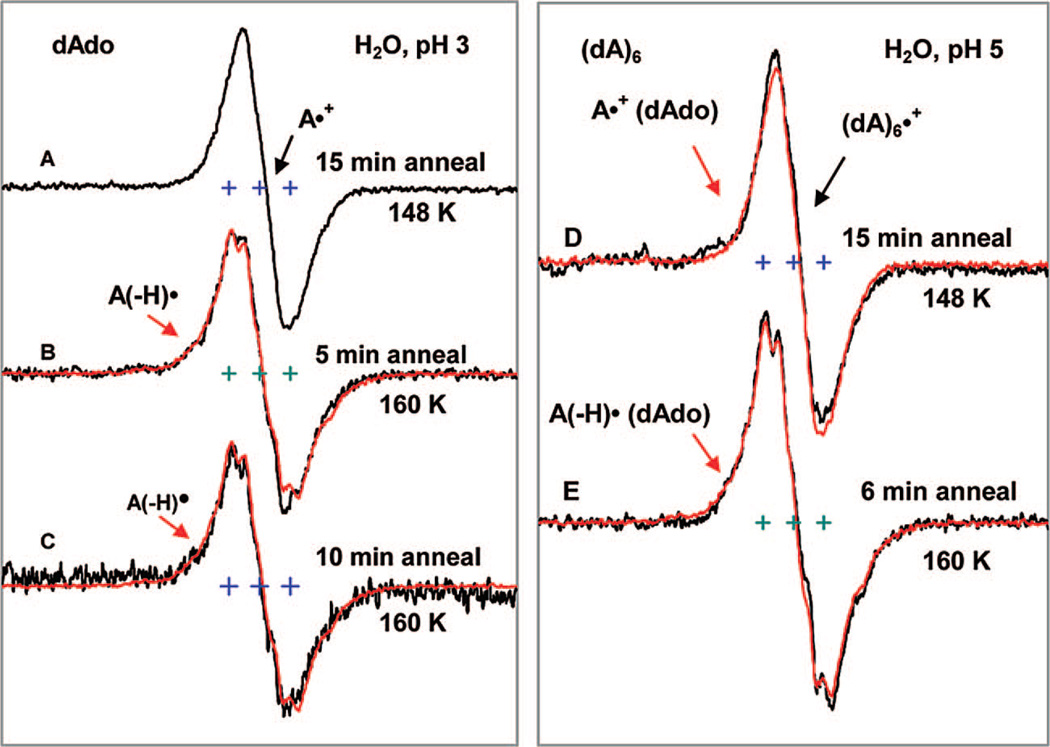
Spectra in Figure 5A to 5C show that, at pH 3, annealing A•+ in samples of 3 mg/mL dAdo in 7.5 M LiCl/H2O to 160 K results in complete deprotonation to form A(−H)•. Identical results are found at pH 5 (see Supporting Information Figure S4). No significant deprotonation of A•+ is observed after annealing at 148 K. However, after annealing to 152 K for 15 min, in the pH range 3–7, the ESR spectrum of one-electron oxidized dAdo in H2O glasses show about 25–30% deprotonation; i.e., the spectral composition is 70–75% A•+ and 25–30% A(−H)•.
Figure 5.
ESR spectra for one-electron oxidized dAdo (3 mg/mL) in pH 3 (A–C) and for one-electron oxidized DNA-oligomer (dA)6 (2 mg/mL) at the native pH (ca. 5) (D, E) in 7.5 M LiCl glass in H2O (black). Spectra A and D are the spectrum of corresponding A•+ recorded at 77 K after its production via annealing to 148 K in the dark. Spectra B, C at pH 3 and spectrum E at pH 5 show that, upon annealing at 160 K in the dark, A•+ is converted completely to A(−H)•. The authentic spectrum of A(−H)• (red) obtained by one-electron oxidation of dAdo at pH 12 in H2O glass is superimposed on spectra B, C, and E for comparison. All spectra were recorded at 77 K.
One question could be, what is the effect of the LiCl matrix on the prototropic equilibrium of A•+? We have observed that (i) one-electron oxidized adenine in dAdo gave nearly identical spectra and pH dependence for A•+ and A(−H)• even at 15 M LiCl and, (ii) even at 15 M LiCl, A•+ is converted to A(−H)• upon annealing (see Supporting Information Figure S5). This suggests that the LiCl matrix does not have a large effect on the prototropic equilibrium of A•+ in dAdo.
As shown in Figure 4B, at 160 K, we find less conversion of A•+ to A(−H)• in D2O glasses in comparison to H2O glasses. For example, in D2O glasses ca. 30% of A•+ is converted to A(−H)• in dAdo on annealing to 160 K with near complete deprotonation at 165 K. This is attributed to the somewhat higher softening point of the D2O glass in comparison to the H2O glass.
3.1.4. Effect of Stacking on A•+ and A(−H)• Formed from dAdo in H2O Glasses
It is well established in the literature that, in aqueous solution at ambient temperature, dAdo dimerizes in a stacking geometry in the concentration range of millimolar and above.34b,c,e,f The thermodynamic parameters (ΔG°, ΔH°, ΔS°) of this dimerization have also been reported in the literature, and their values are −4.2 kJ mol−1, −27.2 kJ mol−1, and −75.2 J K−1 mol−1, respectively.34b,c Since concentration of dAdo in our system (3 mg/mL and above) falls in this range where dAdo is reported to associate and as the nature of our system (glassy at low temperature (ca. 148 to 165 K)) promotes stacking in dAdo, we propose that the adenine cation radical exists primarily in the stacked form (i.e., as a dimer) in our system. This explains its high pKa value (ca. 8) at 150 K. Progressive warming to higher temperatures allows for molecular migration and likely disrupts the dimer cation radical to the monomer cation radical with it is low pKa value, thereby leading to its immediate deprotonation. This would explain our observation that A•+ is completely converted to A(−H)• upon annealing to ca. 160 K in H2O glass (see Figure 4B).
We find that, with increasing concentration of dAdo, the extent of A•+ formed at 152 K increases (see Supporting Information Figure S6B). Thus, the stacking phenomenon tends to stabilize the cation radical state of the one-electron oxidized adenine moiety more with respect to its deprotonated state. We have also observed a shift of field position of the central component (apparent g⊥ value) of one-electron oxidized adenine formed by annealing at 152 K as a function of dAdo concentration (see Supporting Information Figure S6A). This change in the apparent g⊥ value is likely associated with increasing association of the adenines.
3.1.5. A•+ and A(−H) • Formed in the DNA-Oligomer (dA)6: Effect of Base Stacking
In this work we have used (dA)6 to probe the influence of stacking interaction on the ESR spectral characteristics of A•+ and A(−H)• with respect to the monomer (i.e., dAdo). CD and NMR studies show that oligomers (dA)n in aqueous solution at 273 K are in the stacked form.34d The dimer, (dA)2, is found to be 90% in the stacked form with greater degrees of stacking observed as n increases. Therefore, in our experimental conditions which promote stacking (frozen aqueous solution at 150 K), the oligomer (dA)6 should exist entirely in the stacked form. In Figure 5D, we present our results of (dA)6 at 148 K and after annealing to 160 K. The spectra of A•+ and A(−H)• formed in (dA)6 match closely with those for the monomer (compare spectrum 5D with 5A and spectrum 5E with 5C). This further strengthens our conclusion that the cation radical exists in the stacked form even in dAdo.
3.1.6. Annealing Studies of A•+ Formed the DNA-Oligomer (dA)6 in H2O Glasses
We find that A•+ in the DNA-oligomer (dA)6 forms A(−H)• upon annealing to 160 K in the dark at pH (ca. 5) in 7.5 M LiCl (H2O) glass (see spectra 5D and 5E). The behavior of (dA)6 is identical to that of dAdo on annealing to 160 K.
3.2. Theoretical Studies
3.2.1. Deoxyadenosine Cation Radical (A•+) Monomer Calculations
The B3LYP/6–31G(d) optimized structures of the deoxyadenosine radical cation (A•+) and deprotonated A•+ in syn- and anti-conformations (A(−H)•syn and A(−H)•anti) with respect to the N1 atom of A•+ are shown in sSupporting Information Figure S7. For A•+ we found that B3LYP/6–31G* calculated major hyperfine couplings (HFCCs) are localized at N6, N7, N3, C8, H′, H′′, and C2 atoms, respectively (see Table 1). In the gas phase, the A(−H)•syn conformation was more stable by ca. 1.04 kcal/mol than the corresponding A(−H)•anti conformation as predicted by the B3LYP/6–31G* method. The B3LYP/6–31G* calculated HFCCs of the A(−H)•syn conformation are localized on atoms N6, N1, N3, C8, and H″, respectively (see Table 1).
In Figure 6a and b, we present the optimized geometries of adenine cation radical dimer hydrogen bonded ((AB–AB)p•+) in C1 symmetry and in a stacked conformation [(AB–AB)s•+] in Cs symmetry (parallel orientation). Both calculations are for the gas phase.
Figure 6.
B3LYP/6-31G* optimized geometries of (a) the hydrogen bonded adenine cation radical dimer, (AB–AB)p•+, in C1 symmetry in the gas phase. (b) Adenine dimer cation radical [(AB–AB)s•+] in a stacked parallel conformation in the gas phase. The geometry was optimized in Cs symmetry. (c) Deoxyadenosine cation radical in the presence of seven water molecules (A•+ + 7H2O). The arrow on the elongated N6-H′ bond in this figure indicates that the elongation of this N6–H′ bond leads to deprotonation in the surrounding water shell (shown in e). (d) Adenine dimer cation radical in stacked conformation surrounded by 18 water molecules [(AB–AB)s•+ + 18H2O] (local minimum), and (e) hydronium ion formation from A•+ owing to deprotonation of the structure in (c).
The B3LYP/6–31G* optimized structure of the deoxyadenosine cation radical (A•+) in the presence of seven water molecules (A•+ + 7H2O) is shown in Figure 6c. To reduce the computational cost, we placed water molecules only near the adenine base. Based on the hydrogen-bond distances (see Figure 6c), it is evident that the hydrogen bonds between a water molecule and the H′ and H″ hydrogens of the NH2 group of the adenine base are quite strong. In the hydrated complex (A•+ + 7H2O), we found that the N6–H′ bond is elongated by ~0.1 Å as compared to the N6–H″ bond (see Figure 6c) or the corresponding N–H bonds in A•+ in the gas phase (see Supporting Information Figure S7). Leszczynski and co-workers recently studied the hydration of adenine in the presence of 12, 13, 14, and 16 water molecules using the B3LYP/6–31G(d) method.35 The locations of the water molecules in their study are nearly identical to our work (see Figure 6c). In the optimized structure, we found that the water molecules form hydrogen bonds with the N1, H′, H″, N3, H2, and N7 atoms of the adenine ring and lie in the range ~1.431 to 2.159 Å, respectively, and these values are very similar to the corresponding values of Leszczynski and co-workers.35
The B3LYP/6–31G* optimized structure of the adenine base cation radical dimer in the stacked conformation (AB–AB)s•+ is shown in Figure 6b. To simplify the computational complexity, we consider only the adenine base (AB) without the sugar moiety in the dimer calculation. Generally, it is found that the B3LYP method is not suitable for systems that are bound by weak π-π stacking interaction energies which are dispersion-like interactions.36 Since our stacked conformation (AB–AB)s also involves such interactions, first we have checked the reliability of the B3LYP method with the 6–31G* basis set and optimized the structures of the benzene dimer in the neutral state and also in the cation radical state as test cases. We note that the benzene cation radical ((C6H6)n•+, n = 2 to 6) has been studied by several groups using DFT and these clusters are stabilized owing to charge resonance interaction.37a–e In accordance with the existing literature, we found that the B3LYP method fails to predict π-stacking interactions for the neutral stacked conformation but predicts strong interactions for the benzene dimer cation owing to charge resonance interactions. Optimization using the B3LYP/6–31G* method shows that the benzene rings in the cation radical are separated by ca. 3.51 Å while in the neutral states the rings separate to a noninteracting distance of ca. 6 Å, respectively. We find that the BSSE corrected B3LYP/6–31G* calculated stabilization energy for the benzene cation radical dimer is −17.65 kcal/mol. The BSSE corrected MP2/6–31G*//B3LYP/6–31G* calculated (single point) stabilization energy for the benzene cation radical in the dimer conformation is found to be −17.56 kcal/mol. These values are in close agreement with the experimentally measured binding energy (17.6 kcal/mol) of the benzene radical cation dimer by Rusynial et al.38 using mass-selected, ion mobility drift-cell techniques. From these results it is clear that the π–π charge resonance interactions are well predicted in stacked π-system cation radical dimers but the far weaker π–π dispersion interactions in a neutral system are poorly treated as expected.
To calculate the stabilization energy of the adenine cation radical in the stacked conformation, we employed B3LYP as well as MPWB95 and MP2 methods that better account for dispersion interactions. In the present study, we considered two different arrangements of the adenine ring in the stacked conformations: (i) parallel conformation, in which two adenine rings are placed parallel to each other, and (ii) antiparallel conformation, in which one of the adenine ring plane was rotated 180° with respect to the other adenine ring plane. The geometries in these two stacked conformations were fully optimized in their neutral and cationic radical state using MP2, B3LYP, and MPWB95 methods and the 6–31G* basis set (see Supporting Information Figures S11 and S12). The BSSE corrected stabilization energies (in kcal/mol) along with the interbase distances (in angstroms) are presented in Table 2. From Table 2, we note that stabilization energies for the dimer cation radical lie in the range −11.5 to −16.3 kcal/mol. In addition, for the dimer cation, the antiparallel conformation was found to be only slightly more stable (~ −0.7 to 3 kcal/mol) than the parallel conformation (see Table 2). However, in the neutral state, the antiparallel conformation was found to be ~ −5 to 6 kcal/mol more stable than the parallel conformation (see Table 2). Thus, in comparison to the neutral system where the two rings are stabilized mainly due to dispersion, the radical cation systems are held together very strongly in the stacked conformation due to charge resonance interactions. In addition, the stabilization energy −12.08 kcal/mol calculated using the B3LYP method for the radical cation is close to the value −15.64 kcal/mol as calculated by the MP2 method (see Table 2). Further, this detailed analysis clearly shows the applicability of the B3LYP method to study this class of systems.
Table 2.
Stabilization Energies of Adenine Dimer Cation Radical in Various Stacked Formsa
| method (all full optimization) |
symmetry | interbase distance (Å) |
Eint (kcal/mol) (BSSE corrected) |
||
|---|---|---|---|---|---|
| parallelb | antiparallelb | parallelb | antiparallelb | ||
| Dimer Cation Radical | |||||
| B3LYP/6-31G* | Cs | 3.4– 3.9 | – | −12.08 | – |
| MP2/6-31G* | Cs | 2.9– 3.4 | 2.9– 3.2 | −15.64 | −16.32 |
| MPWB95/6-31G* | Cs | 3.2– 3.5 | 3.1– 3.4b | −11.54 | −14.76 |
| MP2/6-31G* | C1 (skew) | 2.9– 3.5 | −13.54 | ||
| Neutral Dimer | |||||
| MPWB95/6-31G* | Cs | 3.5– 3.6 | 3.2– 3.4 | 3.06 | −2.91 |
| MP2/6-31G* | Cs | 3.4– 3.5 | −0.91 | ||
| MP2/6-31G* | C1 | 3.1– 3.4 | −5.8 | ||
Gas phase calculations. Energies calculated using eq 1. All calculations are BSSE corrected.
The parallel conformation was generated by placing the two adenine rings symmetrically along the axis normal to the adenine ring plane. The starting geometry for antiparallel conformation was obtained by rotating one of the two adenine rings in the parallel conformation by 180° (see Supporting Information Figures S11 and S12).
From Table T1 (see Supporting Information), we found that, in the stacked conformation (see Figure 6b), the Mulliken spin densities are equally delocalized in both adenine rings (as evidenced by the HFCC values) and, in each ring, the spin densities are located mainly on atoms N6, N3, C8, H′, and H′′, respectively.
The B3LYP/6–31G* optimized structure of hydrogen-bonded (AB–AB)p•+ is shown in Figure 6a as well as in Figure S8 in the Supporting Information. This optimized structure ((AB–AB)p•+) was obtained when the stacked (AB–AB)s•+ conformation was optimized in the C1 symmetry. In the hydrogen-bonded dimer cation radical ((AB–AB)p•+), the two adenine rings are twisted and hydrogen bonded with each other having hydrogen bond distances 1.293 and 2.353 Å, respectively. The BSSE corrected B3LYP/6–31G* calculated binding energy of the hydrogen bonded dimer cation radical was found to be 18.98 kcal/mol. Recently, Nam et al.21 studied the adenine cation radical dimer using the B3LYP/6–31+G(d,p) method. They reported a planar hydrogen-bonded adenine dimer cation radical having a binding energy of 1.08 eV (24.91 kcal/mol, without BSSE correction). The difference in the binding (24.91 and 18.98 kcal/mol) results from slightly different structures and lack of BSSE correction. The calculated spin distribution in (AB–AB)p•+ is more localized on one of the adenine rings on atoms N6, N3, and C8 (see Table T1 in the Supporting Information).
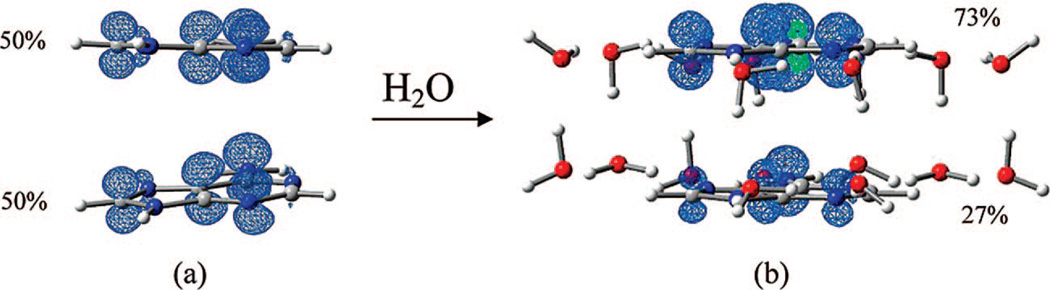
To examine the effect of the surrounding water molecules on the stacked conformation, we optimized the stacked adenine cation radical in the presence of 18 water molecules using the B3LYP/6–31G* method. The optimized local minimum structure of (AB–AB)s•+ + 18 H2O is shown in Figure 6d. We found that in the (AB–AB)s•+ + 18 H2O complex the two adenine rings are stacked within ca. 3.45 to 3.62 Å. These are smaller distances than those found for the gas-phase stacked system (see Figure 6b). We also found the Mulliken spin densities and the HFCCs in the (AB–AB)s•+ + 18 H2O complex favor one of the adenine rings (see Table T1 in the Supporting Information). The spin density distribution for stacked (AB–AB)s•+ in the gas phase and in the hydrated environment (AB–AB)s•+ + 18 H2O are shown in Figure 7a and b.
Figure 7.
B3LYP/6-31G* calculated spin-density distribution shown in percentages in (a) the base stacked adenine dimer cation radical [(AB–AB)s•+] in the gas phase and (b) the base stacked adenine dimer cation radical surrounded by 18 water molecules. On optimization, the water molecules adjust to favor spin on one of the adenine rings in the [(AB–AB)s•+ + 18H2O] complex.
From Figure 7, it is clearly evident that spin densities are more localized (73% vs 27%) on one of the adenine rings in the (AB–AB)s•+ + 18 H2O complex; however, it is fully delocalized in the gas phase (AB–AB)s•+. It appears that the surrounding water molecules adjust their structure on optimization to more strongly interact with one of the adenine rings and thereby tend to localize the spin density. The driving force for the localization stems from the stronger hydrogen bonds formed with the adenine ring with the greater charge (spin). Thus, a partially charge-separated state such as (A•+–A) in the hydrated environment is likely the minimum energy configuration. In order to test whether the spin densities shown in Figure 7 are artifacts of the B3LYP calculations, we also plotted the spin densities using Hartree-Fock (HF) as well as BHandHLYP methods and we have found very similar results (see Supporting Information Figure S10). In a recent study for [dApdA]•+ by Kumar and Sevilla,39a it has been observed that the spin densities are delocalized on both of the adenine rings; however, the spin densities were found to be localized on one of the adenine rings for [[dApdA(−H)•]. Similar results were found by Parinello and co-workers.39b We also note that the spin densities in the benzene dimer cation radical are well established to be delocalized on both rings equally by EPR studies and theory.37c EPR studies of the benzene dimer cation radical also shows that the hyperfine coupling constants for the benzene dimer cation are exactly one-half of those for the monomer as expected.37c This phenomenon is found for other aromatics such as naphthalene and anthracene.37f,g Thus, we conclude that this delocalization of charge in the aromatic dimer cation radical is not an artifact of the calculations and will apply to adenine as well.
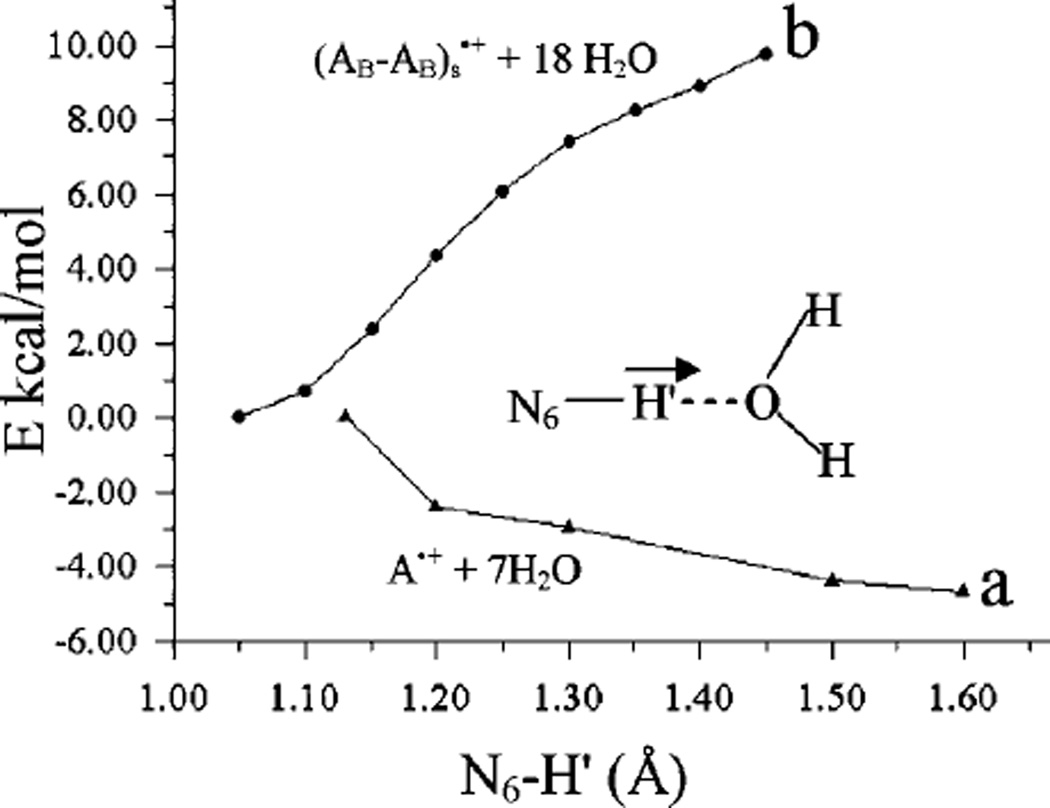
Using the B3LYP/6–31G* method with the PCM model for solvent interactions, we also calculated the pKa of the adenine cation radical (A•+) and the stacked adenine cation radical (AB–AB)s•+ and present the calculation details in Tables T2 and T3 in the Supporting Information. The theoretical calculation of pKa of a molecule is very sensitive, and a small change in the calculated free energy (G) can change the pKa values by several units (orders of magnitude in Ka). In our case, we calculated the pKa using the free energy of H+ in the gas phase as −6.28 kcal/mol based on the Sackur–Tetrode equation.20a Deprotonation from the NH2 group of AB•+ was considered, and the calculated pKa values of AB•+ in anti- and syn-conformations are −0.26 and −1.04 (see Supporting Information Figure S9) while the corresponding values for (AB–AB)s•+ are 7.02 and 6.44, respectively. Based on the calculated pKa values, the monomer, AB•+, is found to be more acidic than the dimer, (AB–AB)s•+, by ca. 7 pKa units. In this context, we calculated the potential energy surface (PES) for the deprotonation of the NH2 group considering the optimized structures of A•+ + 7 H2O and (AB–AB)s•+ + 18 H2O shown in Figure 8. In calculating the PES, the N6–H′ bond was elongated from its equilibrium bond distance in 0.1 Å steps. At each fixed N6–H′ distance the geometry was fully optimized. For A•+ + 7 H2O, we found that A•+ deprotonates in the presence of water without a barrier; however, in the stacked conformation a substantial uphill process is involved in stretching the N6–H′ bond up to 1.45 Å. We conclude that the pKa of the A•+ monomer is quite low while in the stacked system A•+ has a far higher pKa value. When water molecules are considered explicitly, we find partial localization of the spin; however, the charge resonance interactions in the stacked system still significantly inhibit the deprotonation reaction. The present study shows that the deprotonation reaction from the A•+ monomer is facile in the presence of a proton acceptor (such as, the first hydration shell), while in the stacked system this deprotonation has a significant uphill barrier.
Figure 8.
Potential energy surface (PES) of N6–H′ bond dissociation of the (a) A•+ + 7H2O cation radical and (b) (AB–AB)s•+ + 18H2O cation radical in stacked conformation. The distances and energies are given in angstroms (Å) and kcal/mol, respectively. See Figure 6 for structures.
4. Conclusions
The salient findings drawn from the present study are as follows:
Identification of A•+ and A(−H)• in model systems: In this study, employing ESR spectroscopy and theoretical calculations, the adenine cation radical (A•+) and its deprotonated radical (A(−H)•) have been identified in glassy systems of dAdo and (dA)6. The literature suggests that both the monomer dAdo and the DNA-oligomer (dA)6 should have the adenine bases in a stacked configuration in aqueous solutions at low temperatures.34b,c,e,f Our results suggest that this stacking stabilizes A•+ by partial hole delocalization to adjacent adenines. We note that, in spite of base stacking, an isolated A•+ is found in irradiated adenosine •HCl hemihydrate single crystals16 and A(−H)• is found in adenosine single crystals.13 However, in each case the initial hole has undergone a deprotonation reaction from the parent structure which allows for its localization on a single base. Hole localization on the deprotonated base in base stacks has been reported in our previously reported calculations for adenine dinucleoside phosphate systems as well.39a These calculations show that, in the A(−H)• stack, spin is localized only on the A(−H) portion of the dinucleoside phosphate as expected; however, the spin is delocalized in the AA stack in the cation radical of the dinucleoside phosphate.39a In aqueous solutions of Ado and dAdo at ambient temperatures and low concentrations where no stacking occurs,34b,c pulse radiolysis experiments find only A(−H)• as expected from the low pKa of monomeric A•+.3,4,8,10,11
pKa values and dimer cation stabilization (theory and experiment): Experimentally we find that A•+ in dAdo in our aqueous system has the pKa ca. 8 at 150 K, which deprotonates upon thermal annealing at 160 K (see Figures 4 and 5). However, theoretical calculations for the pKa (B3LYP/6–31G*) predict the pKa of A•+ is ca. −0.3 and also predicts the pKa of the AA•+ dimer is far higher, ca. 7. Theoretical calculations also predict that base stacking of the cation radical over the neutral adenine results in a high binding energy (12 to 16 kcal/mol depending on level of calculation; see Table 2). Thus, the stacked dimer cation radical system is predicted to be quite stable, and this stabilization comes through the delocalization of the charge over the two adenine bases. We calculate a negligible barrier to deprotonation in the monomer cation radical and a large barrier to deprotonation in the stacked systems (Table 3). These results are consistent with the calculated pKa’s of ca. −0.3 and ca. 7. It is interesting to compare these values with that of the parent adenine (pKa ca. 144). We see the pKa drops by ca. 14 units for a full charge. The drop of ca. 7 pKa units when the cation charge is shared over two adenines is therefore expected.
Nature of charge delocalization in adenine stacks: We note that the predicted nature of charge delocalization in stacked adenine (see Figure 6b) is quite different that for guanine stacks in which the charge is predicted to localize mainly on one of the guanine bases.39a,40 Calculations predict a very small nuclear relaxation energy (NRE) of ca. 0.10 eV on formation of the adenine cation radical in the AT base pair.41 As a consequence, the adenine cation radical has nearly the same structure as it has in the neutral state. However, the corresponding NRE on guanine cation radical formation in the GC base pair is far larger (0.34 eV).41,42 Thus, adenine stacks in dsDNA are naturally situated to delocalize charge (hole), whereas guanine stacks undergo reorganization and localize the hole. Our calculations show that the effect of the first hydration layer of solvent (18 waters) on the hole delocalization in the adenine dimmer cation radical in a stacked geometry allows for polarization and partial localization of the hole to favor one adenine (see Figure 7b); yet even so, the stacked base moieties of the adenine dimer cation radical would still share charge and remain temporarily stable to deprotonation. Of course, in solution, some hindrance to this delocalization of charge (hole) would be expected from the solvent-induced polarization and the resultant partial localization. The implications of these results suggest that, in DNA, stacking will stabilize the adenine cation toward deprotonation and that solvent dynamics and associated polarization of charge will have a strong effect on the movement of the hole through adenine stacks as has been suggested previously.27,42,43
Table 3.
Summary of the Experimental and Theoretically Calculated Properties of A•+ and of A2•+
Supplementary Material
Acknowledgment
This work was supported by the NIH NCI under Grant No. R01CA045424. A.K. and M.D.S. are thankful to the Arctic Region Supercomputing Center (ARSC) for a generous grant of CPU time and facilities. A.K. is personally thankful to the ARSC staff for their constant cooperation and timely help. Computational studies were also supported by a computational facilities grant NSF CHE-0722689.
Footnotes
Supporting Information Available: Figure S1 showing step-wise formation of A•+ in a glassy sample. Figure S2 the ESR spectrum for A•+ 8-D-dAdo with the simulated spectra of A•+. Figure S3 gives the spectra and analyses of one-electron oxidized dAdo at pH’s 7.5 and 8. Figure S4 shows formation of A(–H)• on annealing A•+. Figure S5 compares results in 7.5 M LiCl glasses with those found in 15 M LiCl glasses. Figure S6 shows the sensitivity of the g value and A•+ stabilization with dAdo concentration. Figures S7 and S8 show the B3LYP/6-31G* optimized geometries of the various species considered in this work. Figure S9 shows the calculated pKa values in an adenine base cation radical in syn and anti conformations. Figure S10 shows the HF-MP2/6-31G*, BHandHYP/6-31G*, and HF/6-31G* calculated spin density distributions in the base stacked adenine dimer cation radical. Figures S11 and S12 show the MP2/6-31G* optimized geometries of a stacked adenine dimer cation radical and of a stacked neutral adenine dimer. Table T1 gives the hyperfine coupling constant (HFCC) values and spin densities at various atoms of A•+ and of A(−H)• in different conformations. Tables T2 and T3 present the details of pKa calculations. Geom-1 to 12 represent the optimized geometries of the various species considered in this study. Finally, a full ref 30a is given. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.(a) Becker D, Adhikary A, Sevilla MD. In: Charge Migration in DNA Physics, Chemistry and Biology Perspectives. Chakraborty T, editor. Berlin, Heidelberg: Springer-Verlag; 2007. pp. 139–175. [Google Scholar]; (b) Wang Q, Fiebig T. In: Charge Migration in DNA Physics, Chemistry and Biology Perspectives. Chakraborty T, editor. Berlin, Heidelberg: Springer-Verlag; 2007. pp. 221–248. [Google Scholar]
- 2.Wagenknecht H-A. In: Charge Transfer in DNA: From Mechanism to Application. Wagenknecht H-A, editor. Weiheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. pp. 1–26. [Google Scholar]
- 3.Steenken S. Chem. Rev. 1989;89:503–520. [Google Scholar]
- 4.(a) Steenken S. Free Radical Res. Commun. 1992;16:349–379. doi: 10.3109/10715769209049187. [DOI] [PubMed] [Google Scholar]; (b) Steenken S. Biol. Chem. 1997;378:1293–1297. [PubMed] [Google Scholar]
- 5.Colson A-O, Besler B, Close DM, Sevilla MD. J. Phys. Chem. 1992;96:661–668. [Google Scholar]
- 6.Bertran J, Oliva A, Rodriguez-Santiago L, Sodupe M. J. Am. Chem. Soc. 1998;120:8159–8167. [Google Scholar]
- 7.Nelson WH, Sagstuen E, Hole EO, Close DM. Radiat. Res. 1998;149:75–86. [PubMed] [Google Scholar]
- 8.Candeias LP, Steenken S. J. Am. Chem. Soc. 1989;111:1094–1099. [Google Scholar]
- 9.(a) Adhikary A, Kumar A, Becker D, Sevilla MD. J. Phys. Chem. B. 2006;110:24171–24180. doi: 10.1021/jp064361y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hole EO, Nelson WH, Sagstuen E, Close DM. Radiat. Res. 1992;129:119–138. [PubMed] [Google Scholar]
- 10.O’Neill P, Davies SE. Int. J. Radiat. Biol. 1987;52:577–587. doi: 10.1080/09553008714552071. [DOI] [PubMed] [Google Scholar]
- 11.Candeias LP, Steenken S. J. Am. Chem. Soc. 1992;114:699–704. [Google Scholar]
- 12.Adhikary A, Becker D, Collins S, Koppen J, Sevilla MD. Nucleic Acids Res. 2006;34:1501–1511. doi: 10.1093/nar/gkl026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Close DM, Nelson WH. Radiat. Res. 1989;117:367–378. [PubMed] [Google Scholar]
- 14.Close DM, Nelson WH, Sagstuen E, Hole EO. Radiat. Res. 1994;137:300–309. [PubMed] [Google Scholar]
- 15.Kar L, Bernhard WA. Radiat. Res. 1983;93:232–253. [Google Scholar]
- 16.Nelson WH, Sagstuen E, Hole EO, Close DM. Radiat. Res. 1992;131:272–284. [PubMed] [Google Scholar]
- 17.Chen Y, Close DM. Struct. Chem. 2002;13:203–209. [Google Scholar]
- 18.Wetmore SD, Boyd RJ, Eriksson LA. J. Phys. Chem. B. 1998;102:10602–10614. [Google Scholar]
- 19.Hwang CT, Stumpf CL, Yu Y-Q, Kenttämaa HI. Int. J. Mass Spectrom. 1999;182/83:253–259. [Google Scholar]
- 20.(a) Baik M-H, Silverman JS, Yang IV, Ropp PA, Szalai VS, Thorp HH. J. Phys. Chem. B. 2001;105:6437–6444. [Google Scholar]; (b) Chen X, Syrstad EA, Nguyen MT, Gerbaux P, Tureček F. J. Phys. Chem. A. 2004;108:9283–9293. doi: 10.1021/jp0529725. [DOI] [PubMed] [Google Scholar]; (c) Jang YH, Goddard WA, III, Noyes KT, Sowers LC, Hwang S, Chung DS. Chem. Res. Toxicol. 2002;15:1023–1035. doi: 10.1021/tx010146r. [DOI] [PubMed] [Google Scholar]
- 21.Nam SH, Park SH, Ryu S, Song JK, Park SM. Chem. Phys. Lett. 2008;450:236–242. [Google Scholar]
- 22.Candeias LP, Steenken S. J. Am. Chem. Soc. 1993;115:2437–2440. [Google Scholar]
- 23.Scheek RM, Stob S, Schleich T, Alma NCM, Hilbers CW, Kaptain R. J. Am. Chem. Soc. 1981;103:5930–5932. [Google Scholar]
- 24.Giese B, Amaudrut J, Köhler A-K, Spormann M, Wessely S. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 25.(a) Kawai K, Majima T. In: Charge Transfer in DNA: From Mechanism to Application. Wagenknecht H-A, editor. Weiheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. pp. 117–151. [Google Scholar]; (b) Lewis FD, Daublain P, Zhang L, Cohen B, Vura-Weis J, Wasielewski MR, Shafirovich V, Wang Q, Raytchev M, Fiebig T. J. Phys. Chem. B. 2008;112:3838–3843. doi: 10.1021/jp710718p. [DOI] [PubMed] [Google Scholar]
- 26.Augustyn KE, Genereux JC, Barton JK. Angew. Chem., Int. Ed. 2007;46:5731–5733. doi: 10.1002/anie.200701522. [DOI] [PubMed] [Google Scholar]
- 27.Shao F, O’Neill MA, Barton JK. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17914–17919. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joy A, Ghosh AK, Schuster GB. J. Am. Chem. Soc. 2006;128:5346–5347. doi: 10.1021/ja058758b. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Yu P, LeProust E, Gao X. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frisch MJ, et al. Gaussian03, revision B.04. Pittsburgh, PA: Gaussian, Inc.; 2003. (for complete reference, see Supporting Information). Boys SF, Bernardi F. Mol. Phys. 1970;19:553..
- 31.Hermosilla L, Calle P, García de la Vega JM, Sieiro C. J. Phys. Chem. A. 2006;110:13600–13608. doi: 10.1021/jp064900z. [DOI] [PubMed] [Google Scholar]
- 32. http://jmol.sourceforge.net. Jmol development team, An Open-Science Project. 2004
- 33.GaussView. Pittsburgh, PA: Gaussian, Inc.; 2003. [Google Scholar]
- 34.(a) Sevilla MD, Becker D, Yan M, Summerfield SR. J. Phys. Chem. 1991;95:3409–3415. [Google Scholar]; (b) Ts’o POP. In: Fine Structure of Proteins and Nucleic Acids. Fasman GD, Tiemasheff SN, editors. New York: Marcel Dekker; 1970. p. 151. [Google Scholar]; (c) Broom AD, Schweizer MP, Ts’o POP. J. Am. Chem. Soc. 1967;89:3612–3622. [Google Scholar]; (d) Olsthoorn CSM, Bostelaar LJ, De Rooij JFM, Van Boom JH, Altona C. Eur. J. Biochem. 1981;115:309–321. doi: 10.1111/j.1432-1033.1981.tb05240.x. [DOI] [PubMed] [Google Scholar]; (e) Itahara T, Imaizumi K. J. Phys. Chem. B. 2007;111:2025–2032. doi: 10.1021/jp0664521. [DOI] [PubMed] [Google Scholar]; (f) Leonard NJ. Acc. Chem. Res. 1979;12:423–429. [Google Scholar]
- 35.Sukhanov OS, Shishkin OV, Gorb L, Podolyan Y, Leszczynski J. J. Phys. Chem. B. 2003;107:2846–2852. [Google Scholar]
- 36.Zhao Y, Truhlar DG. Acc. Chem. Res. 2008;41:157–167. doi: 10.1021/ar700111a. [DOI] [PubMed] [Google Scholar]
- 37.(a) Pieniazek PA, Krylov AI, Bradforth SE. J. Chem. Phys. 2007;127:044317. doi: 10.1063/1.2749506. [DOI] [PubMed] [Google Scholar]; (b) Rusyniak MJ, Ibrahim YM, Wright DL, Khanna SN, El-Shall MS. J. Am. Chem. Soc. 2003;125:12001–12013. doi: 10.1021/ja035504m. [DOI] [PubMed] [Google Scholar]; (c) Itagaki Y, Benetis NP, Kadam RM, Lund A. Phys. Chem. Chem. Phys. 2000;2:2683–2689. [Google Scholar]; (d) Todo M, Okamoto K, Seki S, Tagawa S. Chem. Phys. Lett. 2004;399:378–383. [Google Scholar]; (e) Okamoto K, Seki S, Tagawa S. J. Phys. Chem. A. 2006;110:8073–8080. doi: 10.1021/jp061527m. [DOI] [PubMed] [Google Scholar]; (f) Howarth OW, Frenkel GK. J. Am. Chem. Soc. 1966;88:4514–4515. [Google Scholar]; (g) Badger B, Brocklehurst B. Nature. 1968;219:263. [Google Scholar]
- 38.Rusyniak M, Ibrahim Y, Alsharaeh E, Meot-Ner (Mautner) M, El-Shall MS. J. Phys. Chem. A. 2003;107:7656–7666. [Google Scholar]
- 39.(a) Kumar A, Sevilla MD. J. Phys. Chem. B. 2006;110:24181–24188. doi: 10.1021/jp064524i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mantz YA, Gervasio FL, Laino T, Parinello M. J. Phys. Chem. A. 2007;111:105–112. doi: 10.1021/jp063080n. [DOI] [PubMed] [Google Scholar]
- 40.(a) Yoshioka Y, Kitagawa Y, Takano Y, Yamaguchi K, Nakamura T, Saito I. J. Am. Chem. Soc. 1999;121:8712–8719. [Google Scholar]; (b) Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. J. Am. Chem. Soc. 1998;120:12686–12687. [Google Scholar]
- 41.(a) Li X, Cai Z, Sevilla MD. J. Phys. Chem. A. 2002;106:9345–9351. doi: 10.1021/jp021669q. [DOI] [PubMed] [Google Scholar]; (b) Li X, Cai Z, Sevilla MD. J. Phys. Chem. B. 2001;105:10115–10123. [Google Scholar]; (c) Li XF, Sevilla MD. Adv. Quantum Chem. 2007;52:59–87. [Google Scholar]
- 42.(a) Conwell EM, Park JH, Choi HY. J. Phys. Chem. B. 2005;109:9760–9763. doi: 10.1021/jp044485f. [DOI] [PubMed] [Google Scholar]; (b) Conwell EM. Proc. Natl. Acad. Sci. U.S.A. 2005;102:8795–8799. doi: 10.1073/pnas.0501406102. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Conwell EM, McLaughlin PM, Bloch SM. J. Phys. Chem. B. 2008;112:2268–2272. doi: 10.1021/jp077344x. [DOI] [PubMed] [Google Scholar]
- 43.Henderson PT, Jones D, Hampikian G, Kan YZ, Schuster GB. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8353–8358. doi: 10.1073/pnas.96.15.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.