Abstract
Arcanobacterium phocisimile, a newly described species with the type strain A. phocisimile 2698T isolated from a vaginal swab of a harbour seal and four additional A. phocisimile strains also isolated from four harbour seals could reliably be identified by phenotypic properties, by matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS), and by sequencing the genomic targets 16S rDNA and 16S-23S rDNA intergenic spacer region and the genes rpoB and gap. The A. phocisimile strains investigated in the present study were isolated together with several other bacterial species indicating that the pathogenic importance of A. phocisimile remains unclear. However, the detection of peptidic spectra by MALDI-TOF MS and the presented phenotypic and genotypic approach might help to identify A. phocisimile in future.
1. Introduction
Genus Arcanobacterium comprises the species Arcanobacterium haemolyticum, Arcanobacterium hippocoleae, Arcanobacterium pluranimalium, and Arcanobacterium phocae [1]. More recently A. canis and A. phocisimile were described as novel species of this genus [2, 3]. Arcanobacterium pyogenes together with Arcanobacterium bernardiae, Arcanobacterium bonasi, and Arcanobacterium bialowiezense was reclassified to the newly described species Trueperella [1]. The original description of A. phocisimile was based on physiological and biochemical characteristics, chemotaxonomic analysis, and 16S rDNA sequencing results of two strains isolated with several other bacterial species from a vaginal swab and an anal swab of two free living harbour seals of the German North Sea [3].
In the present study both initially described A. phocisimile strains and three additional strains obtained from three harbour seals were identified and further characterized phenotypically by MALDI-TOF MS analysis and genotypically by amplification and sequencing of various molecular targets.
2. Materials and Methods
2.1. Bacterial Strains
The A. phocisimile strains used in the present study included the previously described type strains A. phocisimile 2698T (LMG 27073T; CCM 8430T) and A. phocisimile 4112 [3]. Additionally investigated A. phocisimile 3047 was isolated (post mortem) together with Bacillus spp., Enterococcus spp., Erysipelothrix rhusiopathiae, and α-haemolytic streptococci in the year 2005 from the lung of a female harbour seal with bronchopneumonia and perforation of stomach. The harbour seal was found dead in Rantum, Sylt of the German North Sea. Also investigated A. phocisimile 4113 was recovered together with Pasteurella spp. and α-haemolytic streptococci from an anal swab and A. phocisimile 4125 together with Pseudomonas spp. and α-haemolytic streptococci also from an anal swab of two apparently healthy female harbour seals, respectively. A. phocisimile 4113 and A. phocisimile 4125 were isolated in 2007 during a monitoring program of free living harbour seals of the German North Sea.
2.2. Phenotypic and Genotypic Identification
All three newly investigated A. phocisimile strains were initially characterized phenotypically and by 16S rDNA sequencing [3, 4]. Both A. phocisimile strains previously mentioned in the species description [3] and the three A. phocisimile strains of the present study were further analysed by MALDI-TOF MS [5] and genotypically by amplification and sequencing of the previously described molecular target 16S-23S rDNA intergenic spacer region (ISR) and the genes rpoB and gap [4, 6, 7]. The primer sequences and the thermocycler programs are given in Table 1.
Table 1.
Oligonucleotide primer sequences and PCR conditions of the target genes used in the present study.
| Oligonucleotide primers | Sequence | Program∗ | Expected size of PCR product (bp) | References |
|---|---|---|---|---|
| (1) 16S rDNA UNI-L (2) 16S rDNA UNI-R (amplification primer) |
5′-AGAGTTTGATCATGGCTCAG-3′ 5′-GTGTGACGGGCGGTGTGTAC-3′ |
1 | 1,403 | [4] |
| (3) 16S rDNA-533F (4) 16S rDNA-907R (sequencing primer) |
5′-GTGCCAGCMGCCGCGGTAA-3′ 5′-CCGTCAATTCMTTTGAGTTT-3′ |
— | — | [4] |
| (5) Gap-F (6) Gap-R (gap primer) |
5′-TCGAAGTTGTTGCAGTTAACGA-3′ 5′-CCATTCGTTGTCGTACCAAG-3′ |
2 | 830 | [4] |
| (7) ISR-23S-F (8) ISR-23S-R (intergenic spacer region primer) |
5′-CCTAGCCTGGTGGTTGGGTAG-3′ 5′-GTGCGGGTAACCAGAAATAACTCTG-3′ |
3 | 345 | [6] |
| (9) C2700F (10) C3130R (rpoB primer) |
5′-CGWATGAACATYGGBCAGGT-3′ 5′-TCCATYTCRCCRAARCGCTG-3′ |
4 | 406 | [7] |
*PCR program 1: x1 (95°C, 600 sec), x30 (95°C, 30 sec, 58°C, 60 sec, 72°C, 60 sec), and x1 (72°C, 420 sec). 2: x1 (94°C, 180 sec), x30 (94°C, 30 sec, 50°C, 40 sec, 72°C, 60 sec), and x1 (72°C, 300 sec). 3: x1 (95°C, 240 sec), x30 (95°C, 8 sec, 66°C, 10 sec, 72°C, 10 sec), x1 (72°C, 420 sec). 4: x1 (95°C, 600 sec), x35 (94°C, 30 sec, 50°C, 30 sec, 72°C, 120 sec), and x1 (72°C, 600 sec).
3. Results and Discussion
All three strains newly characterized in the present study could reliably be identified as A. phocisimile by phenotypic properties and by 16S rDNA sequencing. The phenotypic properties appeared to be almost identical to both previously characterized A. phocisimile strains (Table 2). However, a positive pyrazinamidase reaction of A. phocisimile seems to be the only reliable biochemical property for differentiation of A. phocisimile from pyrazinamidase negative A. phocae.
Table 2.
Phenotypical properties of three A. phocisimile strains investigated in the present study and A. phocisimile 4112 and A. phocisimile 2698T described previously.
| Phenotypic properties | 3047 | 4113 | 4125 | 4112** | 2698T ** |
|---|---|---|---|---|---|
| Hemolysis on sheep blood agar | + | + | + | + | + |
| Hemolysis on rabbit blood agar | + | + | + | + | + |
| CAMP-like reaction with:∗ | |||||
| Staphylococcus aureus β-hemolysin | − | − | − | − | − |
| Streptococcus agalactiae | + | + | + | + | + |
| Rhodococcus equi | + | + | + | + | + |
| Arcanobacterium haemolyticum | − | − | − | − | − |
| Reverse CAMP reaction | + | + | + | + | + |
| Nitrate reduction | −1 | −1 | −1 | −1 | −1 |
| Pyrazinamidase | (+)1 | (+)1 | +1 | +1 | +1 |
| Pyrrolidonyl arylamidase | −1 | −1 | (+)1 | −1 | −1 |
| Alkaline phosphatase | −1 | −1 | −1 | (+)1 | (+)1 |
| β-Glucuronidase (β-GUR) | −1,3 | −1,3 | −1,3 | −1,3 | −1,3 |
| β-Galactosidase (β-GAL) | +1,3 | +1,3 | +1,3 | +1,3 | +1,3 |
| α-Glucosidase (α-GLU) | +1,2,3 | +1,2,3 | +1,2,3 | +1,2,3 | +1,2,3 |
| β-Glucosidase (β-GLU) | −2 | −2 | −2 | −2 | −2 |
| N-Acetyl-β-glucosaminidase (β-NAG) | −1, +3 | −1, +3 | −1, +3 | −1, +3 | −1, +3 |
| Esculin (β-glucosidase) | −1 | −1 | −1 | −1 | −1 |
| Urease | −1 | −1 | +1 | −1 | −1 |
| Gelatine | −1 | −1 | −1 | −1 | −1 |
| Fermentation of: | |||||
| D-Glucose | +1 | +1 | +1 | +1 | +1 |
| D-Ribose | −1 | +1 | +1 | +1 | +1 |
| D-Xylose | −1 | −1 | −1 | −1 | −1 |
| D-Mannitol | −1 | −1 | −1 | −1 | −1 |
| D-Maltose | +1 | +1 | +1 | +1 | +1 |
| D-Lactose | +1 | +1 | +1 | +1 | +1 |
| D-Saccharose | +1 | +1 | +1 | +1 | +1 |
| Glycogen | +1 | +1 | +1 | +1 | +1 |
| α-Mannosidase | +2 | +2 | +2 | +2 | +2 |
| Catalase | + | + | + | + | + |
| Serolysis on Loeffler agar | − | − | − | − | − |
| Caseinase | − | − | − | − | − |
| Starch hydrolysis | + | + | + | + | + |
The reactions are shown as follows: *synergistic CAMP-like reaction with indicator strains; **results mostly obtained from Hijazin et al., 2013 [3]; +: positive reaction; (+): weak positive reaction; −: negative reaction; 1: Api Coryne test system (Biomerieux, Nürtingen, Germany); 2: tablets containing substrates (Rosco Diagnostica A/S, Taastrup, Denmark); 3: 4-methylumbelliferyl conjugated substrates (Sigma, Steinheim, Germany).
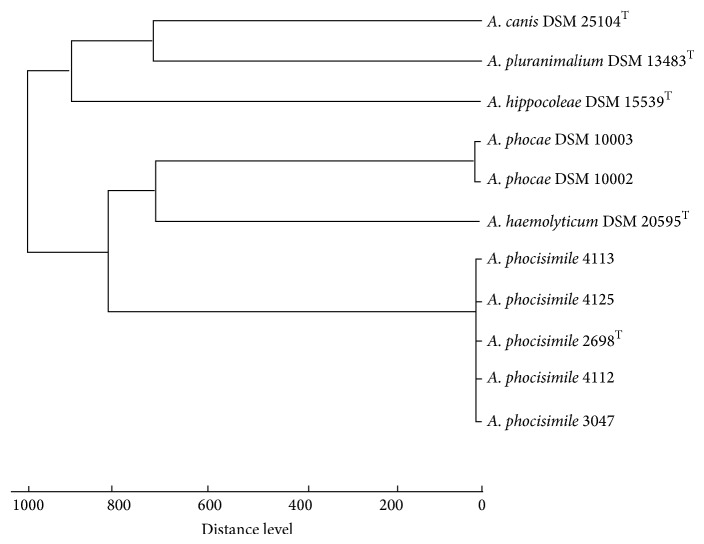
As shown by numerous authors MALDI-TOF MS is a powerful tool for species characterization of a broad spectrum of gram-positive and gram-negative bacteria [8–10]. This technique had previously been successfully used for rapid and reliable identification of bacteria of genera Arcanobacterium and Trueperella [5, 11]. The MALDI-TOF MS analysis of the present study revealed that by using the current Bruker data base, all five strains of this hitherto unknown species could not be identified to species level. However, using the MALDI Biotyper 3.1 software package the log (score) values of A. phocisimile 4112, A. phocisimile 3047, A. phocisimile 4113, and A. phocisimile 4125 matched against A. phocisimile 2698T with log (score) values between 2.69 and 2.74 indicating that all five strains belong to this newly described species. Inclusion of A. phocisimile in the Bruker reference database will allow for the identification of this new species in future. A dendrogram analysis of the MALDI-TOF MS results is presented in Figure 1.
Figure 1.
MALDI-TOF MS spectra from A. phocisimile 4113, A. phocisimile 4125, type strain A. phocisimile 2698T, A. phocisimile 4112, A. phocisimile 3047, and all other species of genus Arcanobacterium.
The genotypic classification by 16S rDNA sequencing revealed that the three novel A. phocisimile strains of the present study yielded 100% identity to both A. phocisimile strains described previously [3], also including the type strain A. phocisimile 2698T (Figure 2).
Figure 2.
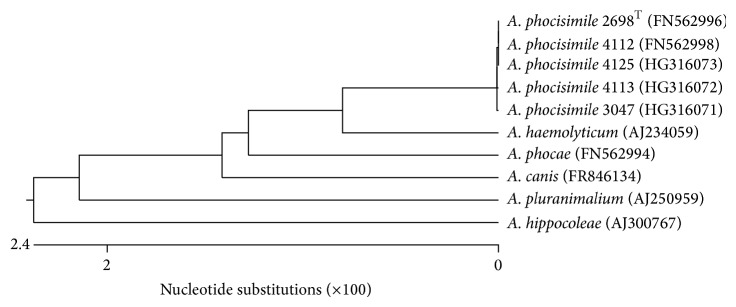
Dendrogram analysis of 16S rDNA sequences of the A. phocisimile strains of the present study and reference strains of genus Arcanobacterium obtained from NCBI GenBank.
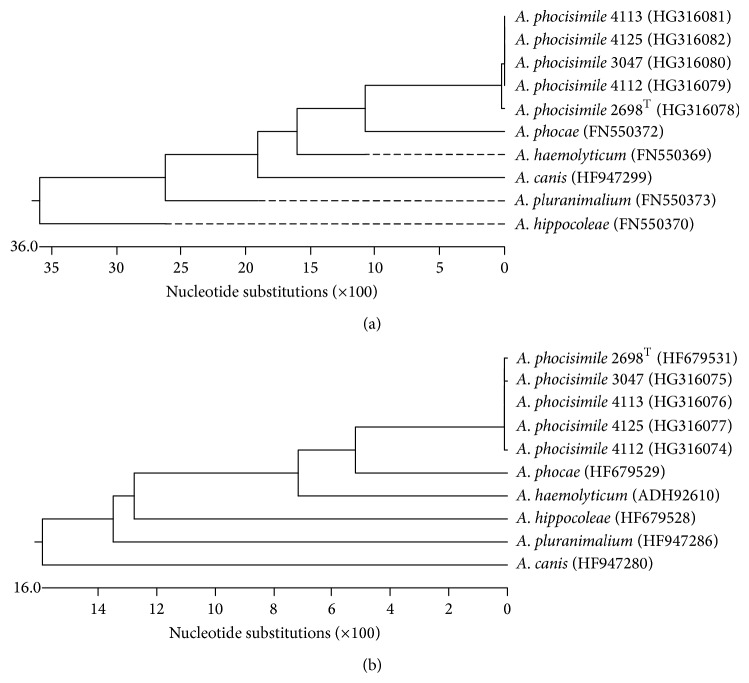
Comparable to previously described A. canis [11] all five A. phocisimile from the present study could additionally be classified by amplification and sequencing of ISR (FN563000, FN563002, HG316083, HG316084, and HG316085), gene rpoB (HG316078, HG316079, HG316080, HG316081, and HG316082), and gene gap (HF679531, HG316074, HG316075, HG316076, and HG316077) yielding for all three molecular targets an identity of ≥99.4%, ≥99.8%, and ≥99.8%, respectively, for all five strains among each other. A typical dendrogram using the sequencing results of the target genes rpoB and gap is shown in Figure 3.
Figure 3.
Dendrogram analysis of sequences of the genes rpoB (a) and gap (b) of the A. phocisimile strains of the present study and all other species of genus Arcanobacterium obtained from NCBI GenBank.
The results of the present study revealed that phenotypic properties, the determination of peptidic spectra by MALDI-TOF MS, and the various genotypic targets allow for a reliable identification of A. phocisimile and a further differentiation of A. phocisimile from closely related A. phocae which could also be isolated from marine mammals [12]. However, all A. phocisimile strains of the present study were isolated together with various other bacteria, partly from obviously healthy animals, indicating that the pathogenic importance of this species for marine mammals remains unclear.
4. Interpretive Summary
Arcanobacterium phocisimile type strain and four additional A. phocisimile strains isolated from harbour seals were identified phenotypically, by matrix assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS), by sequencing 16S rDNA, and, as novel molecular targets, by sequencing 16S-23S rDNA intergenic spacer region and the genes rpoB and gap indicating that MALDI TOF MS and the molecular targets might help to identify this novel species.
Conflict of Interests
The authors declare that they have no conflict of interests. The authors certify that they have no affiliation with or financial involvement in any organization or entity with a direct financial interest in the subject matter or materials discussed in the paper.
References
- 1.Yassin A. F., Hupfer H., Siering C., Schumann P. Comparative chemotaxonomic and phylogenetic studies on the genus Arcanobacterium Collins et al. 1982 emend. Lehnen et al. 2006: proposal for Trueperella gen. nov. and emended description of the genus Arcanobacterium . International Journal of Systematic and Evolutionary Microbiology. 2011;61(6):1265–1274. doi: 10.1099/ijs.0.020032-0. [DOI] [PubMed] [Google Scholar]
- 2.Hijazin M., Prenger-Berninghoff E., Sammra O., et al. Arcanobacterium canis sp. nov., isolated from otitis externa of a dog, and emended description of the genus Arcanobacterium Collins et al. 1983 emend. Yassin et al. 2011. International Journal of Systematic and Evolutionary Microbiology. 2012;62:2201–2205. doi: 10.1099/ijs.0.037150-0. [DOI] [PubMed] [Google Scholar]
- 3.Hijazin M., Sammra O., Ülbegi-Mohyla H., et al. Arcanobacterium phocisimile sp. nov., isolated from harbour seals. International Journal of Systematic and Evolutionary Microbiology. 2013;63:2019–2024. doi: 10.1099/ijs.0.045591-0. [DOI] [PubMed] [Google Scholar]
- 4.Sammra O., Balbutskaya A., Nagib S., et al. Properties of an Arcanobacterium haemolyticum strain isolated from a donkey. Berliner und Münchener Tierärztliche Wochenschrift. 2014;127:10–14. [PubMed] [Google Scholar]
- 5.Hijazin M., Hassan A. A., Alber J., et al. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for species identification of bacteria of genera Arcanobacterium and Trueperella . Veterinary Microbiology. 2012;157(1-2):243–245. doi: 10.1016/j.vetmic.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Hassan A. A., Ülbegi-Mohyla H., Kanbar T., et al. Molecular identification of Arcanobacterium bialowiezense and Arcanobacterium bonasi based on 16S-23S rRNA intergenic spacer region sequences. Veterinary Microbiology. 2008;130(3-4):410–414. doi: 10.1016/j.vetmic.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Ülbegi-Mohyla H., Hijazin M., Alber J., et al. Identification of Arcanobacterium pyogenes isolated by post mortem examinations of a bearded dragon and a gecko by phenotypic and genotypic properties. Journal of Veterinary Science. 2010;11(3):265–267. doi: 10.4142/jvs.2010.11.3.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seng P., Drancourt M., Gouriet F., et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clinical Infectious Diseases. 2009;49(4):543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 9.Murray P. R. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clinical Microbiology and Infection. 2010;16(11):1626–1630. doi: 10.1111/j.1469-0691.2010.03364.x. [DOI] [PubMed] [Google Scholar]
- 10.Bizzini A., Jaton K., Romo D., Bille J., Prod’hom G., Greub G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. Journal of Clinical Microbiology. 2011;49(2):693–696. doi: 10.1128/JCM.01463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sammra O., Balbutskaya A., Zhang S., et al. Further characteristics of Arcanobacterium canis, a novel species of veterinary importance. Veterinary Microbiology. 2013;167:619–622. doi: 10.1016/j.vetmic.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Ramos C. P., Foster G., Collins M. D. Phylogenetic analysis of the genus Actinomyces based on 16S rRNA gene sequences: description of Arcanobacterium phocae sp. nov, Arcanobacterium bernardiae comb. nov., and Arcanobacterium pyogenes comb. nov. International Journal of Systematic Bacteriology. 1997;47(1):46–53. doi: 10.1099/00207713-47-1-46. [DOI] [PubMed] [Google Scholar]