Abstract
IMPORTANCE
The normal absorptive function and structural maintenance of the intestinal mucosa depend on a constant process of proliferation of enterocytic stem cells followed by progressive differentiation toward a mature phenotype. The mechanisms that govern enterocytic differentiation in the mucosa of the small intestine are poorly understood.
OBJECTIVE
To determine whether schlafen 3 (but not other schlafen proteins) act in vivo and whether its effects are limited to the small intestine. We have previously demonstrated in nonmalignant rat intestinal IEC-6 cells that schlafen 3 levels correlate with the expression of various differentiation markers in vitro in response to differentiation stimuli.
DESIGN
Randomized controlled experiment.
SETTING
Animal science laboratory.
PARTICIPANTS
Male Sprague-Dawley rats 8 to 13 weeks old.
MAINOUTCOMES AND MEASURES
Messenger RNA (mRNA) from jejunal and colonic mucosa was isolated, and transcript levels of schlafen proteins 1, 2, 3, 4, 5, 13, and 14; sucrase isomaltase (SI); dipeptidyl peptidase 4 (Dpp4); glucose transporter type 2 (Glut2); and villin were measured by quantitative reverse transcriptase–polymerase chain reaction. We tested parallel variations in protein levels by Western blotting and Dpp4 enzyme activity.
RESULTS
The transcript level of schlafen 3 (Slfn3) correlated with the levels of the differentiation markers SI, Dpp4, Glut2, and villin. However, the expression of schlafen proteins 1, 2, 4, 5, 13, and 14 did not correlate with the expression of the differentiation markers. The mucosal mRNA levels of Slfn3, SI, Glut2, and Dpp4 were all substantially higher in the rat jejunum than in colonic mucosa by a mean (SE) factor of 51.0 (13.2) for 6 rats (P < .05), 599 (99) for 8 rats (P < .01), 12.5 (5.5) for 8 rats (P < .01), and 14.0 (3.9) for 8 rats (P < .01), respectively. In IEC-6 cells, infection with adenovirus-expressing GFP-tagged Slfn3 significantly increased Slfn3 expression and Dpp4-specific activity compared with GFP-expressing virus (in 6 rats; P < .05).
CONCLUSIONS AND RELEVANCE
Taken together with our previous in vitro observations, the results suggest that small intestinal enterocytic epithelial differentiation in rats may be regulated by Slfn3 in vivo, as in vitro, and that these effects may be specific to the small intestinal enterocytic phenotype as opposed to that of the mature colonocyte. Slfn3 human orthologs may be targeted to stimulate intestinal differentiation in patients with short bowel syndrome
Numerous intracellular signals have been shown to influence small intestinal epithelial differentiation in response to certain stimuli. These include the Notch pathway,1 epidermal growth factor, transforming growth factor α (TGF-α), and TGF-β.2 Understanding the signals by which enterocytic differentiation is governed is important because abnormalities of enterocytic differentiation are a key part of mucosal atrophy after long-term fasting or starvation.3,4 Furthermore, enterocytic differentiation may represent an important and novel pharmacologic target to promote small bowel mucosal function in short gut syndrome.5
Schlafen 3 (Slfn3) may be of particular interest. In rat IEC-6 enterocytes in vitro, Slfn3 appears to be necessary for enterocytic differentiation in response to an array of diverse stimuli, including, at the least, TGF-β, repetitive deformation, and sodium butyrate.6 The schlafen family of proteins regulates a range of biological processes, including differentiation, tumorigenesis, and apoptosis in hematopoetic and epithelial cells.7–9 Slfn3 is a member of this family and is expressed in the intestinal mucosa, liver, and lungs.10 In vitro, the expression of Slfn3 in rat IEC-6 intestinal cells correlates with the terminally differentiated state of enterocytes because Slfn3 suppression reduces cell differentiation.6
Colonic cell lines such as Caco-2 are frequently used to model small intestinal epithelial biology in vitro,11,12 and “differentiation” becomes a moving target depending on the cell line, environment, stimuli, and end points under study. Although the normal colonic epithelium in vivo is certainly elaborately differentiated, normal colonocytes differ markedly from small bowel enterocytes in many ways, including the activity of glutamine synthetase13 and the sensitivity of alkaline phosphatase14 to mutagens15 and its resistance to apoptosis.16 We hypothesized that Slfn3 expression would differ between normal rat small intestinal mucosa and normal rat colonic mucosa, that these differences would correlate with differences in the expression of conventional markers of enterocytic differentiation, and that the pattern of intestinal expression of Slfn3 would differ from that of other schlafen proteins. To test these hypotheses, we compared Slfn3 expression with variations in the transcript and protein level of villin, sucrase iso-maltase (SI), glucose transporter type 2 (Glut2), and dipeptidyl peptidase 4 (Dpp4), as well as with differences in the specific activity of the Dpp4 enzyme. In further studies, we directly assessed the effects of specific overexpression of Slfn3 in IEC-6 cells using an adenoviral expression system.
Methods
Animal Procedures and Tissue Harvest
All animal procedures were reviewed and approved by the Michigan State University Office of Radiation, Chemical, and Biological Safety and the Institutional Animal Care and Use Committee. Animal care was in accordance with the standards of the Public Health Service and the Association for Assessment and Accreditation of Laboratory Animal Care International. Male Sprague-Dawley rats 8 to 10 weeks old were used for all experiments. We harvested mucosa from progressively distal segments of the intestine. The jejunum, ileum 1, and ileum 2 small intestinal segments were harvested 10, 25, and 45 cm from the ligament of Treitz, respectively, whereas the segment that we labeled “ileum 3” was harvested 5 cm proximally to the cecum. The colonic samples were harvested 2 cm distal to the cecum.
Cell Culture
Nontransformed rat intestinal IEC-6 epithelial cells (American Type Culture Collection) were maintained at 37°C in 5% carbon dioxide, as previously described.4
Western Blot Analysis
Mucosal scrapings from target intestinal segments after harvest were immediately immersed in ice-cold lysis buffers (50mM Tris hydrochloride, 1mM ethylene diamine tetraace-tic acid, 1mM ethylene glycol tetraacetic acid buffer, 1% Triton X-100, 1% dichloroacetate, glycerol, 10mM NaPyroPO4, and 50mM sodium fluoride). Tissue was homogenized using Bullet Blender (Next Advance) and then centrifuged at 15 000g for 10 minutes at 4°C. Cultured IEC-6 cells were lysed in a lysis buffer, centrifuged at 15 000g for 10 minutes at 4°C, resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to Hybond ECL nitrocellulose membrane (Amersham Pharmacia Biotech), as previously described.17 Nonspecific binding sites were blocked for 1 hour at room temperature. Membranes were probed with antibodies to Slfn3, SI, Glut2, Dpp4, and villin (Santa Cruz Biotechnology) with appropriate secondary antibodies. Bands were visualized using the Odyssey imaging system (Licor) and analyzed with the Kodak Image Station 440CF. All exposures used for densitometric analysis were within the linear range.
RNA Isolation and Quantitative Reverse Transcriptase–Polymerase Chain Reaction
Total RNA was isolated from the cells using Tri-Reagent (Molecular Research Center, Inc) in accordance with the manufacturer’s instructions. The RNA concentration was measured spectrophotometrically at an optical density of 260 nm. The complementary DNA (cDNA) was prepared from RNA samples as described previously.18 The cDNA samples were analyzed by quantitative reverse transcriptase–polymerase chainreaction (qRT-PCR) using the BioRad MyiQ Real-Time PCR system and the BioRad SYBR Green supermix (BioRad Laboratories). Expression levels were determined from the threshold cycle (Ct) values using the method of 2−ΔΔCt and 18S expression as the reference control gene.18 Primers sequences used for qRT-PCR are listed in the eTable in the Supplement. The cycle conditions for the PCR were 1 cycle of 3 minutes at 95°C and 40 cycles of 30 seconds at 95°C, 30 seconds at the annealing temperature (57°C), and 30 seconds at 72°C.
Dpp4-Specific Activity Assay
Dpp4 activity was measured using the Dpp4-Glo assay (Pro-mega) according to the manufacturer’s protocol. The standard Dpp4 enzyme was purchased from Sigma. IEC-6 cells in a 6-well plate or in 10 mg of tissue were harvested in 0.5 mL of ice-cold phosphate-buffered saline and homogenized using Bullet Blender. The sample was centrifuged for 10 minutes at 10 000 rpm at 4°C. The supernatant was collected and stored at −80°C. Dpp4 activity was measured in 50 μL of asample diluted 30 times in phosphate-buffered saline.
Adenovirus Vector Construction and Cell Treatment
The Slfn3 cDNA of the rat Slfn3 gene was subcloned in-frame into pShuttle-CMV, linearized, and recombined into the plas-mid pAdTrack-CMV to create Ad-GFP-Slfn3. All viruses were found not to be replication-competent adenoviruses by use of PCR (E1 region amplification) and direct sequencing methods. Ad vector production and characterization were performed as described previously19 (80% confluent IEC-6 cells to 4000 vp/cell Ad-GFP [control] or Ad-GFP- Slfn3 for 48 hours).
Statistical Analysis
Values are group mean (SE) values of the nontransformed data. Prior to analysis, all data were checked to ensure that they fit a normal distribution and were corrected if necessary.4 Statistical analysis was performed using unpaired t tests or 1-way analysis of variance, as appropriate. Differences between mean values were considered to be statistically significant at P < .05.
Results
Slfn3 exhibits a distinct pattern of expression in rat intestinal mucosa. We compared the expression of various schlafen isoforms across the sampled segments of intestinal mucosa, studying genes for which the sequence was available in the National Center for Biotechnology Information database. The level of Slfn1 was significantly higher in the colon than in the various small intestinal segments (for 6 rats; Figure 1A; P < .05). Levels of Slfn2, Slfn5, and Slfn13 were significantly higher in the jejunum than in other segments (for 6 rats; Figure 1B, D, and E; P < .05). Only Slfn3 displayed significantly lower transcript levels in the colon than in all sampled small bowel segments (for 6 rats; Figure 1C; P < .05). The transcript levels of Slfn14 in the small intestine did not differ from those in the large intestine (Figure 1F), whereas Slfn4 expression was not detected in any segment of the intestine using either of 2 different sets of primers (data not shown).
Figure 1. Different Schlafen Proteins Exhibiting Different Patterns of Expression in the Rat Intestinal Mucosa.
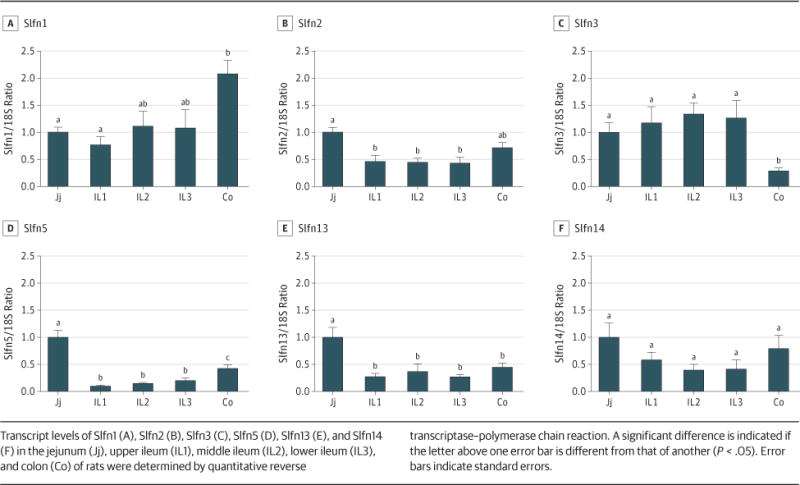
Transcript levels of Slfn1 (A), Slfn2 (B), Slfn3 (C), Slfn5 (D), Slfn13 (E), and Slfn14 (F) in the jejunum (Jj), upper ileum (IL1), middle ileum (IL2), lower ileum (IL3), and colon (Co) of rats were determined by quantitative reverse transcriptase–polymerase chain reaction. A significant difference is indicated if the letter above one error bar is different from that of another (P < .05). Error bars indicate standard errors.
The expression pattern of Slfn3 is similar to that of Dpp4, SI, and Glut2 among segments of the small intestine and colon. Like Slfn3, expression levels of villin, Dpp4, Glut2, and SI were significantly lower in the colon than in the jejunum or upper ileum (segments Il1 and Il2) (for 6 rats; Figure 2A–D; P < .05). The expression levels of SI and Glut2 were lower in the distal ileum (IL3) than in the upper ileum (IL1-2) or jejunum (for 6 rats; Figure 2C and D; P < .05).
Figure 2. Transcript Levels of Differentiation Markers, Which Vary Along the Length of the Rat intestinal Mucosa.
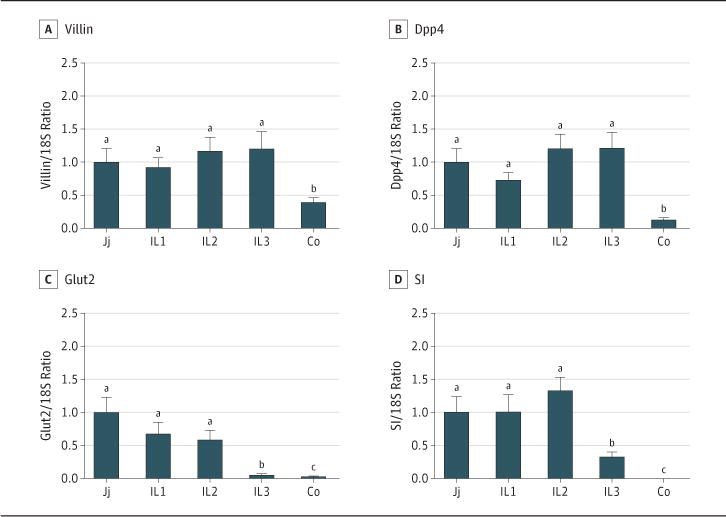
Transcript levels of villin (A), Dpp4 (B), Glut2 (C), and SI (D) were measured by quantitative reverse transcriptase–polymerase chain reaction in the mucosa of the jejunum (Jj), upper ileum (IL1), middle ileum (IL2), lower ileum (IL3), and colon (Co). Ordinate scales were matched to the ordinate scales in Figure 1 to facilitate interpretation. A significant difference is indicated if the letter above one error bar is different from that of another (P < .05). Error bars indicate standard errors.
The protein levels of Slfn3 and the intestinal differentiation markers in the small intestine vary from those in the large intestine. We further measured protein levels of Slfn3, villin, Glut2, SI, and Dpp4 in the gut mucosa. As in our qRT-PCR studies, we found that the protein levels of Slfn3, villin, SI, and Dpp4 were all significantly lower in the colon than in the small intestine (for 5 rats; Figure 3A, B, and E; P < .05). Notably, the lower ileum (IL3) contained significantly less SI than did the other small intestinal segments (IL1-2 and J), but even the distal ileal mucosa had more SI than did the colon (for 5 rats; Figure 3D and E; P < .05).
Figure 3. Slfn3 and Differentiation Marker Protein Levels and Dpp4 Activity Mirroring Transcript Level in Rat Intestinal Mucosa.
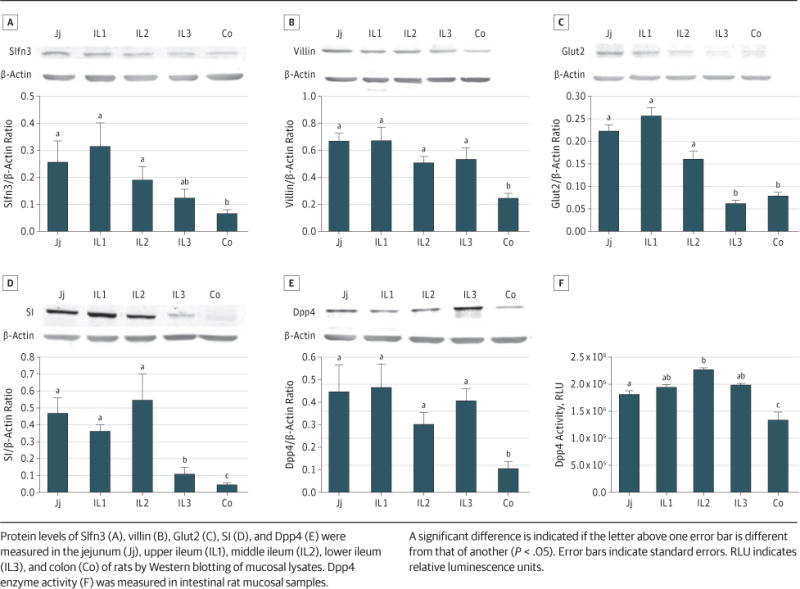
Protein levels of Slfn3 (A), villin (B), Glut2 (C), SI (D), and Dpp4 (E) were measured in the jejunum (Jj), upper ileum (IL1), middle ileum (IL2), lower ileum (IL3), and colon (Co) of rats by Western blotting of mucosal lysates. Dpp4 enzyme activity (F) was measured in intestinal rat mucosal samples. A significant difference is indicated if the letter above one error bar is different from that of another (P < .05). Error bars indicate standard errors. RLU indicates relative luminescence units.
Dpp4, a brush border enzyme that cleaves NH2-terminal peptides from polypeptides, is expressed in multiple cells and tissues. Dpp4 activity was higher in the middle ileum (IL2) than in the jejunum (for 5 rats; Figure 3F; P < .05), but, overall, Dpp4 activity was significantly higher in each segment of the small intestine than in the colon (for 5 rats; Figure 3F; P < .05).
Ad-GFP-Slfn3 transfection increased Slfn3 protein and messenger RNA (mRNA) levels and stimulated Dpp4-specific activity in IEC-6 cells. Because these results were only correlative, we sought to verify the direct effects of Slfn3 overexpression on a functional marker of intestinal epithelial differentiation. Ad-GFP-Slfn3 infection of IEC-6 cells substantially increased the level of Slfn3 transcript expression compared with GFP-treated mock transfectants (for 6 rats; Figure 4A; P < .05). Overexpression of Slfn3, compared with GFP-treated mock transfectants, in these IEC-6 cells significantly increased Dpp4-specific activity a mean (SE) factor of 1.98 (0.2) (for 3 rats; Figure 4B; P < .01).
Figure 4. Slfn3 Induction Modulating Dpp4 Activity In Vitro.
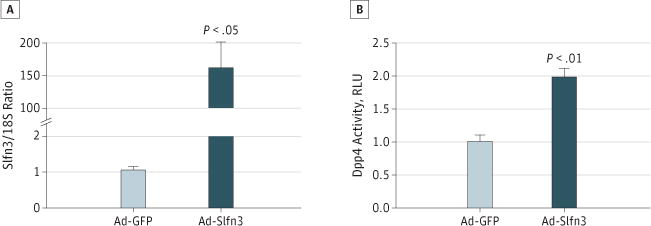
A, 80% confluent IEC-6 monolayers were infected with Ad-Slfn3 for 48 hours (in 6 rats). B, Ad-GFP-Slfn3 infection of IEC-6 cells increased Dpp4-specific activity in lysates from these IEC-6 monolayers (in 3 rats). Error bars indicate standard errors. RLU indicates relative luminescence units.
Discussion
Various stimuli and mechanisms have been proposed to influence intestinal differentiation in vitro and in vivo. However, relatively little is known about the differentiation between small and large intestinal epithelia. These studies suggest that Slfn3 is a potential specific regulator of small intestinal epithelial differentiation. Among the tested schlafen proteins, only Slfn3 expression correlated with the expression of differentiation markers along the longitudinal axis of the intestine. This was confirmed by the protein level and Dpp4 enzyme activity data. We further demonstrated in vitro that overexpression of Slfn3 increases Dpp4 enzymatic activity, an important marker of functional enterocytic differentiation.
Slfn3 belongs to a family of growth regulating genes with cytoplasmic and nuclear localization.20 This group contains an adenosine triphosphate–binding domain (AAA_4), a predicted transcriptional regulator area with a helix-turn-helix domain (COG2865), and a provisional common region (PHA02782). The adenosine triphosphate–binding region has RNA helicase-like activity and is involved in T-cell development and activation. According to their structural similarities, schlafen proteins are categorized into subgroups I (including Slfn1 and Slfn2), II (including Slfn3 and Slfn4), and III (including Slfn5, Slfn13, and Slfn14).20,21 Slfn1, Slfn2, and Slfn4 may be involved in the regulation of immune cell development and the differentiation of osteoclasts.7,9, 22 Slfn5 has been reported to be involved in the antitumor properties of IFN-α.23 The roles played by Slfn13 and Slfn14 have not yet been identified. Despite conserved regions, the schlafen proteins demonstrate distinct patterns of expression along the length of the intestine.
Enzymesand transportproteins of the brush border membrane are characteristic features of differentiated enterocytes. Our data suggest that Slfn3 may regulate the expression and activity of Dpp4, which is expressed in differentiated epithelia and endothelia of normal cells and cancer cells.24,25 The expression patterns ofSlfn3resemble those of mRNA and the other schlafen proteins and the specific activity of Dpp4 along the intestinal longitudinal axis, and Slfn3 overexpression increases Dpp4-specific activity in nonmalignant rat intestinal epithelial IEC-6 cells. We also reported previously that basal IEC-6 Dpp4 activity can be reduced by Slfn3 silencing.6 Slfn3 was induced over 162-fold, but Dpp4-specific activity was induced only 1.9-fold. The very large induction of Slfn3 is a function of the viral transfection system that was used, and Slfn3 is not easily titered. It is likely that this is much more than is required to induce a maximal Slfn3 effect. In addition, it is likely that other factors also influence Dpp4 transcription, such as hepatocyte nuclear factor 1α.26 Most importantly, however, such a doubling of Dpp4-specific activity, although modest compared with the much larger induction of Slfn3, is likely to be highly biologically significant. Dpp4 is an enzyme, so even small changes in its activity are likely to result in larger amplified changes in its reaction product. Dpp4 changes in vitro in response to such stimuli as butyrate or repetitive deformation by only 30% to 40%.27,28 In vivo, the Dpp4 protein level decreases by approximately 60% during defunctionalized atrophy of the intestinal mucosa.17 The 90% increase in Dpp4 activity observed here in response to Slfn3 overexpression seems of biological relevance in comparison. Dpp4 is expressed in the intestinal brush border of mature enterocytes, and its expression is controlled, in part, by diet.29 Atrophy of the intestinal mucosa reduces Dpp4 and SI protein levels.17 To demonstrate that the correlationbetweenSlfn3 and Dpp4 was not unique to a single enterocytic differentiation marker, we also studied the mRNA and SI, villin, and Glut2 protein levels. Villin is a calcium-regulated actin-binding protein of the microvillus core of the brush border, expressed in both crypts and villi of the small and large intestine.30 SI is a glucosidase enzyme expressed in the brush border of mature enterocytes and increasingly expressed during developmental maturation of the intestine.31 Levels of the fructose and glucose transporter Glut2 are regulated by components of diet such as fructose and fat.32 Expressions of both Glut2 and SI increase after polyamine-induced differentiation in vivo.33 In each case, we found higher levels of these markers in the small bowel than in the colon, along with higher levels of Slfn3.
We stripped the mucosa from eachbowel segmentanddis-carded the muscular and serosal layers, and we confirmed this by a preliminary histological evaluation (data not shown). Nevertheless, it is possible that some of the mRNA and protein levels that we studied might have had a nonepithelial origin. Inflammatory cells, fibroblasts, endothelial cells, and other cell types are also found in the mucosa. Although we would not have expected substantial levels of the enterocytic differentiation markers to be present in these other cell types, it is certainly possible that the sampling of Slfn1 or Slfn2, for instance, could have been biased by an immune cell schlafen protein. However, Walsh et al10 have shown that Slfn3 immunoreactivity in the gut is strongly concentrated in the epithelial cells.
If Slfn3 is involved in enterocytic differentiation, what other signaling mechanisms might contribute to its effects? Enterocytic differentiation can involve multiple mechanisms. Notch signaling has been shown to regulate the fate of the cells of the mucosal crypt in differentiation after postmitotic events,1 whereas exogenous growth factors affect cell proliferation and differentiation along the rat gastrointestinal tract.34,35 Although the potential interaction between the Notch signal pathway and Slfn3 awaits further study, at least 1 growth factor, TGF-β, appears to stimulate Slfn3 expression.6 A previous in vitro study6 suggested that Slfn3 is necessary for the induction of enterocytic differentiation by repetitive deformation, TGF-β, and butyrate and that this effect might involve phosphatidylinositol 3-kinase (PI3K), focal adhesion kinase (FAK), or p38 signaling. Gauthier et al36 reported that suppression of FAK and PI3K signaling initiated a higher level of apoptosis in the jejunum compared with the colon in mice. The question as to whether FAK and PI3K signaling may be involved in the differential regulation of Slfn3 between the small and large intestines and the question as to what the exact roles of FAK and PI3K are in Slfn3 regulation of enterocyte differentiation remain to be answered.
Although both the small and the large intestine display active wingless-type MMTV integration site family (WNT) signaling, the pattern of its expression is dramatically different between human jejunum and the human colon.37 In vitro in the HCT-116 colon cancer line, Patel and colleagues38 suggested that Slfn3 may increase protein levels of TGF-β, p27, and E-cadherin but reduce protein levels of CDK-2 and β-catenin. E-cadherin has been shown to regulate WNT-dependent transcription in colon cancer cells via binding to β-catenin.39 Thus, Slfn3 might be involved in TGF-β-induced and/or WNT-induced small intestinal differentiation as well. All these signals represent important avenues for further study.
In this work, we analyzed the expression of schlafen proteins in vivo in the intestinal mucosa along the longitudinal gut axis. We determined that the patterns of Slfn3 expression resemble those of other small intestinal epithelial differentiation makers and brush border enzymes in vivo and can control the expression of Dpp4 in vitro. Although much work remains to be done to delineate the mechanism by which Slfn3 acts, our study supports the hypothesis that Slfn3 plays an important role in intestinal differentiation. These data extend our understanding of enterocyte-specific signaling in the maintenance of the digestive and absorptive function of the small intestinal mucosa. Targeting Slfn3 and its downstream effector signals may ultimately facilitate mucosal preservation and improve the recovery of patients with intestinal atrophy, atresia, or massive small bowel resections.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported in part by the National Institutes Health (grants 1 R56 DK096137-01 and RO1 DK067257 to Dr Basson) and a Veterans Affairs Merit Research Award (to Dr Basson).
Footnotes
Author Contributions: Drs Kovalenko and Basson had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Both authors.
Acquisition of data: Kovalenko.
Analysis and interpretation of data: Both authors.
Drafting of the manuscript: Both authors.
Critical revision of the manuscript for important intellectual content: Both authors.
Statistical analysis: Both authors.
Obtained funding: Both authors.
Administrative, technical, or material support: Both authors.
Study supervision: Both authors.
Conflict of Interest Disclosures: None reported.
Supplemental content at jamasurgery.com
Previous Presentation: This study was presented in part at the 2013 Association of VA Surgeons Annual Meeting; April 22, 2013; Milwaukee, Wisconsin.
References
- 1.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 2005;19(14):1686–1691. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw D, Gohil K, Basson MD. Intestinal mucosal atrophy and adaptation. World J Gastroenterol. 2012;18(44):6357–6375. doi: 10.3748/wjg.v18.i44.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Teitelbaum DH. Epidermal growth factor/TNF-α transactivation modulates epithelial cell proliferation and apoptosis in a mouse model of parenteral nutrition. Am J Physiol Gastrointest Liver Physiol. 2012;302(2):G236–G249. doi: 10.1152/ajpgi.00142.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovalenko PL, Kunovska L, Chen J, Gallo KA, Basson MD. Loss of MLK3 signaling impedes ulcer healing by modulating MAPK signaling in mouse intestinal mucosa. Am J Physiol Gastrointest Liver Physiol. 2012;303(8):G951–G960. doi: 10.1152/ajpgi.00158.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne TA, Wilmore DW, Iyer K, et al. Growth hormone, glutamine, and an optimal diet reduces parenteral nutrition in patients with short bowel syndrome: a prospective, randomized, placebo-controlled, double-blind clinical trial. Ann Surg. 2005;242(5):655–661. doi: 10.1097/01.sla.0000186479.53295.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan L, Yu Y, Sanders MA, Majumdar AP, Basson MD. Schlafen 3 induction by cyclic strain regulates intestinal epithelial differentiation. Am J Physiol Gastrointest Liver Physiol. 2010;298(6):G994–G1003. doi: 10.1152/ajpgi.00517.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz DA, Katayama CD, Hedrick SM. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity. 1998;9(5):657–668. doi: 10.1016/s1074-7613(00)80663-9. [DOI] [PubMed] [Google Scholar]
- 8.Oh PS, Patel VB, Sanders MA, et al. Schlafen-3 decreases cancer stem cell marker expression and autocrine/juxtacrine signaling in FOLFOX-resistant colon cancer cells. Am J Physiol Gastrointest Liver Physiol. 2011;301(2):G347–G355. doi: 10.1152/ajpgi.00403.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Zuylen WJ, Garceau V, Idris A, et al. Macrophage activation and differentiation signals regulate schlafen-4 gene expression: evidence for Schlafen-4 as a modulator of myelopoiesis. PLoS One. 2011;6(1):e15723. doi: 10.1371/journal.pone.0015723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh MF, Hermann R, Sun K, Basson MD. Schlafen 3 changes during rat intestinal maturation. Am J Surg. 2012;204(5):598–601. doi: 10.1016/j.amjsurg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnabl KL, Field C, Clandinin MT. Ganglioside composition of differentiated Caco-2 cells resembles human colostrum and neonatal rat intestine. Br J Nutr. 2009;101(5):694–700. doi: 10.1017/S0007114508048289. [DOI] [PubMed] [Google Scholar]
- 12.Beau I, Berger A, Servin AL. Rotavirus impairs the biosynthesis of brush-border-associated dipeptidyl peptidase IV in human enterocyte-like Caco-2/TC7 cells. Cell Microbiol. 2007;9(3):779–789. doi: 10.1111/j.1462-5822.2006.00827.x. [DOI] [PubMed] [Google Scholar]
- 13.Andriamihaja M, Davila AM, Eklou-Lawson M, et al. Colon luminal content and epithelial cell morphology are markedly modified in rats fed with a high-protein diet. Am J Physiol Gastrointest Liver Physiol. 2010;299(5):G1030–G1037. doi: 10.1152/ajpgi.00149.2010. [DOI] [PubMed] [Google Scholar]
- 14.Loret S, Rusu D, El Moualij B, et al. Preliminary characterization of jejunocyte and colonocyte cell lines isolated by enzymatic digestion from adult and young cattle. Res Vet Sci. 2009;87(1):123–132. doi: 10.1016/j.rvsc.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Park HS, Goodlad RA, Wright NA. Crypt fission in the small intestine and colon: a mechanism for the emergence of G6PD locus-mutated crypts after treatment with mutagens. Am J Pathol. 1995;147(5):1416–1427. [PMC free article] [PubMed] [Google Scholar]
- 16.Habtezion A, Toivola DM, Asghar MN, et al. Absence of keratin 8 confers a paradoxical microflora-dependent resistance to apoptosis in the colon. Proc Natl Acad Sci U S A. 2011;108(4):1445–1450. doi: 10.1073/pnas.1010833108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovalenko PL, Basson MD. Changes in morphology and function in small intestinal mucosa after Roux-en-Y surgery in a rat model. J Surg Res. 2012;177(1):63–69. doi: 10.1016/j.jss.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovalenko PL, Zhang Z, Cui M, Clinton SK, Fleet JC. 1,25 dihydroxyvitamin D-mediated orchestration of anticancer, transcript-level effects in the immortalized, non-transformed prostate epithelial cell line, RWPE1. BMC Genomics. 2010;11:26. doi: 10.1186/1471-2164-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan L, Kovalenko PL, Patel SS, Seregin S, Amalfitano A, Basson MD. Induction of intestinal epithelial differentiation by intraluminal delivery of an adenoviral vector coding for Schlafen 3. Gastroenterology. 2012;142(5):S724. [Google Scholar]
- 20.Neumann B, Zhao L, Murphy K, Gonda TJ. Subcellular localization of the Schlafen protein family. Biochem Biophys Res Commun. 2008;370(1):62–66. doi: 10.1016/j.bbrc.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 21.Geserick P, Kaiser F, Klemm U, Kaufmann SH, Zerrahn J. Modulation of T cell development and activation by novel members of the Schlafen (slfn) gene family harbouring an RNA helicase-like motif. Int Immunol. 2004;16(10):1535–1548. doi: 10.1093/intimm/dxh155. [DOI] [PubMed] [Google Scholar]
- 22.Lee NK, Choi HK, Yoo HJ, Shin J, Lee SY. RANKL-induced schlafen2 is a positive regulator of osteoclastogenesis. Cell Signal. 2008;20(12):2302–2308. doi: 10.1016/j.cellsig.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Katsoulidis E, Mavrommatis E, Woodard J, et al. Role of interferon alpha (IFNalpha)-inducible Schlafen-5 in regulation of anchorage-independent growth and invasion of malignant melanoma cells. J Biol Chem. 2010;285(51):40333–40341. doi: 10.1074/jbc.M110.151076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matteucci E, Giampietro O. Dipeptidyl peptidase-4 (CD26): knowing the function before inhibiting the enzyme. Curr Med Chem. 2009;16(23):2943–2951. doi: 10.2174/092986709788803114. [DOI] [PubMed] [Google Scholar]
- 25.Busek P, Malík R, Sedo A. Dipeptidyl peptidase IV activity and/or structure homologues (DASH) and their substrates in cancer. Int J Biochem Cell Biol. 2004;36(3):408–421. doi: 10.1016/s1357-2725(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 26.Gu N, Tsuda M, Matsunaga T, et al. Glucose regulation of dipeptidyl peptidase IV gene expression is mediated by hepatocyte nuclear factor-1alpha in epithelial intestinal cells. Clin Exp Pharmacol Physiol. 2008;35(12):1433–1439. doi: 10.1111/j.1440-1681.2008.05015.x. [DOI] [PubMed] [Google Scholar]
- 27.Han O, Li GD, Sumpio BE, Basson MD. Strain induces Caco-2 intestinal epithelial proliferation and differentiation via PKC and tyrosine kinase signals. Am J Physiol. 1998;275(3, pt 1):G534–G541. doi: 10.1152/ajpgi.1998.275.3.G534. [DOI] [PubMed] [Google Scholar]
- 28.Basson MD, Turowski GA, Rashid Z, Hong F, Madri JA. Regulation of human colonic cell line proliferation and phenotype by sodium butyrate. Dig Dis Sci. 1996;41(10):1989–1993. doi: 10.1007/BF02093601. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki H, Katayama T, Yamamoto K, Kumagai H. Transcriptional regulation of tyrosine phenol-lyase gene of Erwinia herbicola AJ2985. Biosci Biotechnol Biochem. 1995;59(12):2339–2341. doi: 10.1271/bbb.59.2339. [DOI] [PubMed] [Google Scholar]
- 30.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277(36):33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 31.Traber PG. Control of gene expression in intestinal epithelial cells. Philos Trans R Soc Lond B Biol Sci. 1998;353(1370):911–914. doi: 10.1098/rstb.1998.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kellett GL, Brot-Laroche E, Mace OJ, Leturque A. Sugar absorption in the intestine: the role of GLUT2. Annu Rev Nutr. 2008;28:35–54. doi: 10.1146/annurev.nutr.28.061807.155518. [DOI] [PubMed] [Google Scholar]
- 33.Wild GE, Searles LE, Koski KG, Drozdowski LA, Begum-Hasan J, Thomson AB. Oral polyamine administration modifies the ontogeny of hexose transporter gene expression in the postnatal rat intestine. Am J Physiol Gastrointest Liver Physiol. 2007;293(2):G453–G460. doi: 10.1152/ajpgi.00077.2006. [DOI] [PubMed] [Google Scholar]
- 34.Housley RM, Morris CF, Boyle W, et al. Keratinocyte growth factor induces proliferation of hepatocytes and epithelial cells throughout the rat gastrointestinal tract. J Clin Invest. 1994;94(5):1764–1777. doi: 10.1172/JCI117524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitchen PA, Goodlad RA, FitzGerald AJ, et al. Intestinal growth in parenterally-fed rats induced by the combined effects of glucagon-like peptide 2 and epidermal growth factor. JPEN J Parenter Enteral Nutr. 2005;29(4):248–254. doi: 10.1177/0148607105029004248. [DOI] [PubMed] [Google Scholar]
- 36.Gauthier R, Laprise P, Cardin E, et al. Differential sensitivity to apoptosis between the human small and large intestinal mucosae: linkage with segment-specific regulation of BCL-2 homologs and involvement of signaling pathways. J Cell Biochem. 2001;82(2):339–355. doi: 10.1002/jcb.1172. [DOI] [PubMed] [Google Scholar]
- 37.Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129(2):626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Patel VB, Yu Y, Das JK, Patel BB, Majumdar AP. Schlafen-3: a novel regulator of intestinal differentiation. Biochem Biophys Res Commun. 2009;388(4):752–756. doi: 10.1016/j.bbrc.2009.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


