Abstract
In this paper we describe uPy, an extension module for the Python programming language that provides a uniform abstraction of the APIs of several 3D computer graphics programs called hosts, including: Blender, Maya, Cinema4D, and DejaVu. A plugin written with uPy is a unique piece of code that will run in all uPy-supported hosts. We demonstrate the creation of complex plug-ins for molecular/cellular modeling and visualization and discuss how uPy can more generally simplify programming for many types of projects (not solely science applications) intended for multi-host distribution. uPy is available at http://upy.scripps.edu
Keywords: Computer graphics, Python, software plug-in, Scientific data visualization, 3D Molecular visualization, Blender, Maya, Cinema4d
Introduction
Today's general-purpose 3D computer graphics software (3DCGS or hosts), such as Cinema4D, Maya, and Blender, are designed to model, simulate, animate and render everything from planets to people. At the same time, scientific and other domain-competent 3D software are used to model, simulate, visualize, and analyze data, typically from domain-specific entities; such entities include for example molecules (molecular graphics), internal organs (medical imagery), ecosystems and human-engineered structures. Many recent plugins adapt general 3DCGS to handle such domain-specific data; see for instance: BioBlender (http://bioblender.eu/), Blender for robotics (http://wiki.blender.org/index.php/Robotics:Contents), and mMaya1. But, specifically designing a 3DCGS plugin for each specific data type is time consuming and often leads to duplicated effort. Furthermore, these types of plug-ins and scripts are 3DCGS-dependent, because of the differences in 3DCSG's APIs. While there are similarities in the APIs, porting plugins to additional 3DCGS generates duplication of effort and code. While each 3DCGS offers low-level API access, typically through C++ or 3DCGS-specific scripting languages, over the past 10 years a growing number of 3DCGS, including: Autodesk's Maya, Maxon's Cinema4D, Houdini, Blender, Modo, RealFlow, and Rhino, are providing high-level API access through the Python programming language, thus creating the opportunity to develop a Python layer abstracting the 3DCSG's APIs and thus, presenting a unified API for plugin developers.
Over the past ten years, Python's has rapidly gained acceptance in both academia and industry and a large number of scientific tools have been developed in Python or made callable from Python (see for instance The Python Papers Monograph journal2). 3DCGS that embed a Python interpreter can leverage these third-party Python modules to cover a large range of scientific and domain-specific purposes. Nevertheless, integrating Python modules into a 3DCGS requires code that is specific to each 3DCGS's Python API which differs greatly both in syntax and design from one 3DCGS to the next. Since 3DCGS share many core functionalities and the domain-specific modules have a limited number of input/output data types, a single Python adaptor can provide a unified interface for plugging these modules into a variety of 3DCGS hosts at once. With such a connection made, domain-specific Python modules can access any host-specific Python wrapped API, and thus apply that host's core functionality to the data in order to efficiently/natively: (i) model and deform shapes, from spheres and boxes to highly complex polyhedra; (ii) create materials, light scenes and cameras; (iii) produce key-frame animations; (iv) generate realistic and other advanced renderings of scientific and domain-specific data; and (v) apply advanced physics simulations (rigid-body, soft-body, fluid, particle, hair) to the data. 3DCGS also provide their own Graphical User Interface (GUI) toolkits for developing GUIs for plug-ins. Thus, using the same approach, a single Python adaptor can also be used to customize the user interface (UI) of a 3DCGS host to create module-specific GUIs for every host with no code redundancy.
We developed a Python module called uPy (for ubiquitous Python), in order to provide a unified API for modeling geometry and designing user interfaces across a variety of 3DCGS. Scripts and plugins developed using this API will run in all the 3DCGS hosts supported by uPy, thus facilitating their maintenance, broadening their user-base, and promoting their reuse. uPy currently supports the following three popular 3DCGS as host applications: Blender2.49, 2.5× & 2.6×, MAXON Cinema4D12 & r13, and Autodesk Maya2011 & 2012. In addition, uPy supports our lab's freely distributed Python- and OpenGL-based 3D visualization engine DejaVu3.
Other software projects sharing similar goals include: (i) the Enthought tool suite (http://www.enthought.com/), in which the TraitsUI package provides a unified API for creating user interfaces based on wxPython/PyQt, and the MayaVI package, which provides some 3D geometry modeling capabilities. TraitsUI, however, does not support the custom UI tools of the uPy host applications. (ii) The Blurdev project (http://code.google.com/p/blur-dev/) extends the Autodesk Softimage and 3dsMax software with a Python interpreter. Blurdev enables scripting in Python and the integration of 3rd party Python packages, such as Qt, for creating plugin interfaces in these host applications. However, Blurdev's approach is limited to the Windows Operating Systems. Finally (iii) the Cortex project (http://code.google.com/p/cortex-vfx/) is a low-level development framework of libraries reusable in different host software for abstracting the APIs of various rendering backends including: RenderMan, Maya, OpenGL, Houdini and Nuke. There is currently no unifying API for the UI of Cortex's rendering backends. Moreoever, building and installing Cortex is not trivial. Since, Cortex is a C++ library with associated Python bindings, Cortex is amenable to interoperate with uPy. Compared to these approaches, uPy is lightweight, operating system agnostic, and easy to install as it is written in pure Python (i.e. uPy requires no C or C++ extensions to compile). Furthermore, uPy could easily be extended to support the Enthought suite, Autodesk Softimage, and 3dsMax as additional host applications, thus enabling the interoperation of all of these other software tools.
uPy's unified API enables programmers and users to explore the advantages and drawbacks of each host's capabilities and to utilize applications of proprietary tools that may be unique to certain hosts. With its simple implementation and architecture, uPy has helped us develop new scientific visualization tools covering a broad range of fields, from molecular modeling to mescoscale cellular visualization (see below). While we are using uPy for scientific visualization, uPy is completely generic and can be used to extend 3DCSGs with functionality of any type and from any application domain.
Architecture
Founded on Python, we developed uPy as two independent modules. The first module is a modeling Helper class that enables access to all the basic modeling features of the host applications. The second module is a user interface (UI) Adaptor class that provides a unified API for accessing the host UI system. uPy also provides Tkinter and Qt backends allowing the UI of plugins written using uPy to be instantiated in any Python application.
Figure 1 illustrates the simplicity of the uPy architecture in which the independent modules (modeling Helper and UI Adaptor) comprise two Python files per host that wrap that host's overlapping functions. Extending uPy for a new host application that already embeds a Python interpreter requires only the creation of these two files. Each of the two files implements host-specific code for a particular set of functions, which abstracts common host functionalities, such as widget creation and geometry generation.
Figure 1. uPy unites scientific software with various hosting applications.
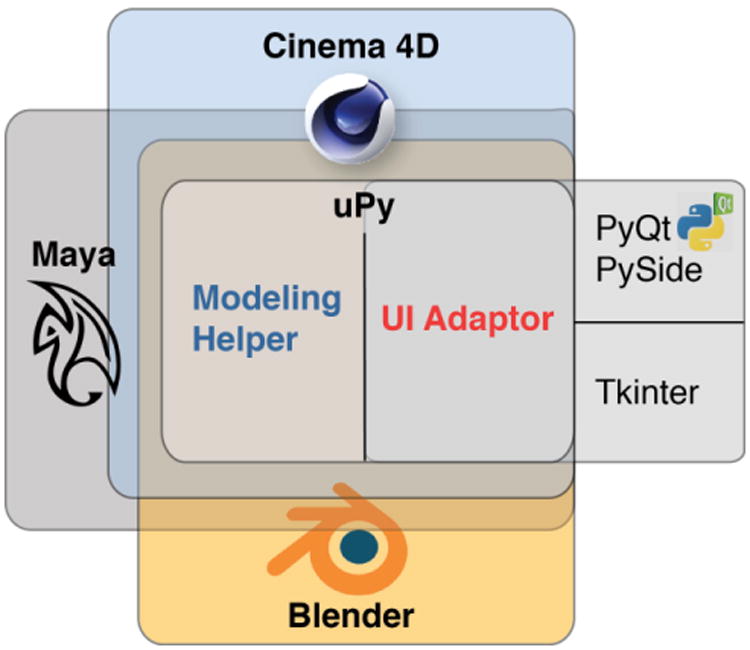
This diagram represents the feature coverage offered by uPy across the different Python wrapped hosts. The host-specific modeling and UI functions are exposed by a uPy Modeling Helper class and a uPy UI Adaptor class for each supported host.
Implementation
Although uPy can reduce redundant effort for most general programming pipelines, our lab focuses on the scientific domain of molecular and cellular biology, therefore we provide four examples of scientific interoperation and describe implementation within this context. Writing scientific visualization plug-ins for different applications requires code to: (i) generate the data, (ii) create, visualize, and modify the graphical representation of the data and (iii) create a GUI that exposes the plug-in's user-settable parameters.
While domain-specific standalone packages can generate the data independent of any host API, building a graphical representation and modifying it requires access to each supported host-specific API (see Figure 2a). To avoid redundancy, uPy's Modeling Helper class wraps the host-specific snippet of code (blue background in figure 2a) under a uPy function which has a unique uPy name (yellow background in figure 2a). These higher-level uPy functions thus support the creation of host-specific graphical objects using a unified syntax (yellow background in figure 2a). The same approach is applied for the creation of GUI widgets and dialogs using the UI adaptor class. Figure 2b illustrates the different API functions called to create a button in each host application and the equivalent single call using uPy (yellow background in Figure 2b). With this approach, a single code can execute the same behavior in every supported host application. For instance, in Figure 3 we depict the plug-in code required to create and scale a sphere. The uPy code will run in the different host application and will generate a GUI dialog exposing a button that triggers the generation of the sphere. The dialog also displays a slider that will scale the generated sphere in real-time.
Figure 2. uPy improves coding efficiency.
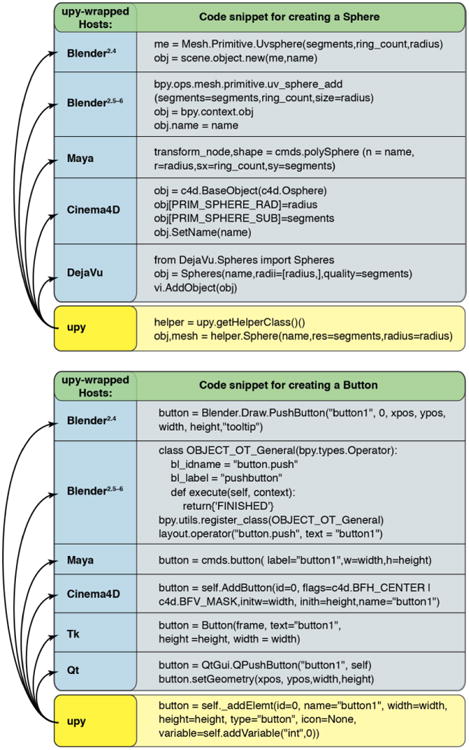
Different hosts require the use of these different lines of code (blue background) to generate a single sphere (a) and to create a GUI button (b) via their APIs. A single uPy call (shown in the yellow fields) can replace all of the native calls in the blue fields to generate a sphere or a button in any and all of the uPy wrapped hosts with the same results that the native code would redundantly produce. In this manner, 5 lines of uPy code effectively replace the 31 lines of code a programmer would have to write to make a sphere and a button in all 5 hosts.
Figure 3. Simple example: create a Sphere-Builder widget in 5 different hosts with a single uPy script file.
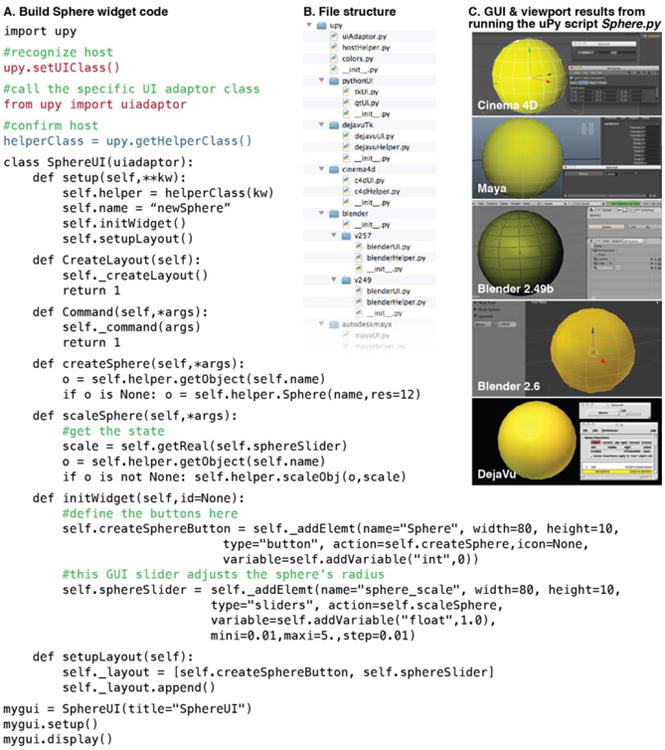
The folder hierarchy of uPy shown in this screengrab represents the Python class hierarchy. Two files, a Modeling Helper and a User Interface Adaptor, exist for each hosting application (B). uPy consists of two main modules (Helper and Adaptor) that automatically recognize the host software as shown in the Sphere.py example code (A). This code produces homogenous results as demonstrated in the screenshots from various hosts shown on the right (C).
Figure 4 illustrates the different geometries currently accessible from the Modeling Helper. These geometries include: (i) text objects, (ii) simple primitives (planes, spheres, cones, cylinders, and cubes), (iii) platonic solids, (iv) point based geometry (points, metaballs, particles), (v) lines (mesh lines, curve, extruded curve and bones), (vi) complex polyhedral meshes, and (vii) object instances. These common geometries are the basis for most scientific model visualizations. For example, meshes can be used to represent an isosurface computed from volumetric data, or to visualize different molecule representations such as surfaces, or ribbon diagrams. For non-scientific applications, meshes may be used most commonly to skin a character or generate a landscape. While the basic geometry objects are common in any scientific or domain-competent 3D software, more advanced objects such as bones/armatures (traditionally used in character animation and robotics to create and control joints using inverse/forward kinematics), particles system (used for rendering clouds or fire), metaballs (blobby spheres), and instances are less common in theses scientific or domain-competent 3D software but are ubiquitous in general professional 3D software and have proven to be useful additions to domain-specific geometry types for building efficient models native to each host. Furthermore, these host features can extend domain-specific functionalities. For example, we have used armatures to model and explore protein conformation and flexibility, and particle systems to efficiently display a grid of points or volumetric data. Instances allow the efficient rendering of multiple copies of a given geometry. Instances are defined by a list of 3D transformation matrices or directly translated and rotated onto another object's model data, e.g., onto the vertex positions of a mesh. Instances can be used to represent atoms of a molecule (instances of sphere primitives), or redundant subunits of viral capsids (instances of mesh models). Furthermore, any geometry can be associated with a material to apply color and texture. For example, a textured plane can display results produced by a graph plotting Python module such as matplotlib (http://matplotlib.sourceforge.net/) or any plotting library.
Figure 4. uPy supports common geometry types.
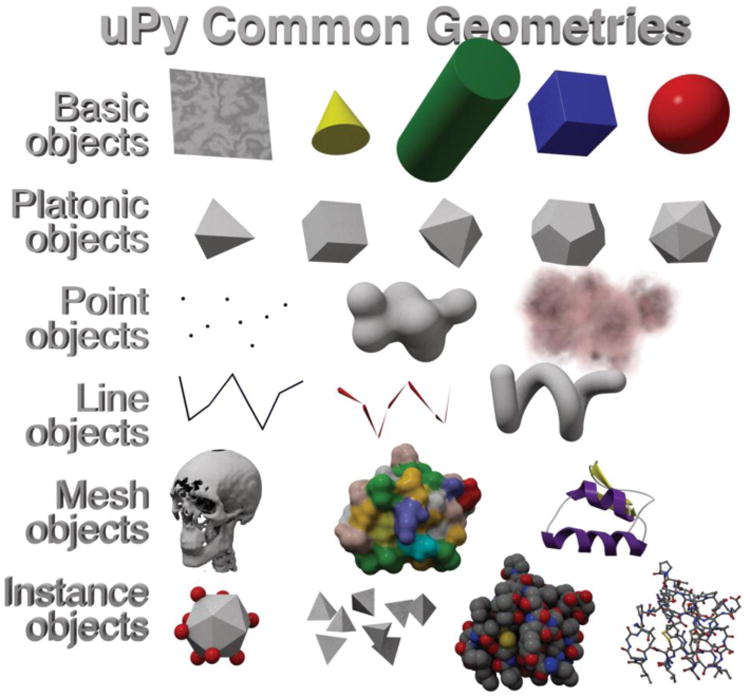
A screenshot from Cinema 4D shows the current common denominator (CD) geometries, i.e. geometries supported by the Modeling Helper available for all host applications. These geometries include: (i) Text Objects, (ii) Basic Objects (planes, spheres, cones, cylinders, and cubes), (iii) Platonic Objects, (iv) Point Objects (points, metaballs, particles), (v) Lines Objects (mesh lines, curves, extruded curves and bones), (vi) Mesh Objects (volume isosurfaces, molecular surfaces, and molecular ribbons), and (vii) Instances Objects (sphere instances at geometry vertices to decorate, platonic instances at vertex coordinates, sphere instances at molecular atomic coordinates, spheres and cylinders instanced at molecular atoms and bond coordinates).
The Modeling Helper class provides support for generating geometries as well as support for selecting and updating geometries (modify vertices/points, edges and faces) and for transformations (position, rotation and scale).
Figure 5, demonstrates the different widgets and layouts supported in the UI adaptor, for all host applications. The UI adaptor translates most of the basic widgets commonly used for designing a modern GUI. The UI adaptor currently supports: menu bars, check boxes, buttons, labels, string/integer/float input fields, text area input fields, integer and float sliders, color chooser fields that trigger system-native color choosers, and pull down menus. The UI adaptor also provides access to message and file dialogs that allow for console reporting and native file browsing. These widgets can be arranged in row layouts, and grouped into collapsible or tabbed subsections.
Figure 5. The same uPy code generates a common UI across all supported hosts.

The dialog windows show the lowest common denominator widgets and layouts as provided by the UI Adaptor. The widgets are labeled according their respective type i.e. data input, buttons and menus. The title of each dialog window labels the particular host.
The list of supported features can easily be extended. To add support for a new feature, a programmer creates an empty function in the uPy Modeling Helper base-class to declare the feature name in uPy. The actual implementation of the feature is provided by overriding this new empty function in each host-specific Modeling Helper subclass with the appropriate snippet of host-specific code. If a feature cannot be implemented within a given host API the empty function will be called and no operation will be performed. Rather than limiting development to the lowest-common-denominator hosts, this architecture enables programmers to write many types of scripts for high/specific-functioning hosts and automatically culls the scripts for hosts with limited functionality. Once created, the new function can then be called across all host applications with a single line of code.
Applications
Here we describe several software plugins that we have developed using uPy in order to make them available in a wide range of 3DCGS. Software plugins described below include: (i) ePMV, an embedded Python molecular viewer, (ii) GAMer a mesh improvement tool, (iii) Tetra, a tetrahedral mesh visualization tool, and (iv) autoFill/autoCell, a biological mesoscale modeling and visualization tool. While these plugins relate to our research interests, uPy can be used more generally to create plugins that expose any Python-callable computational method inside any of the supported hosts, or to generate uPy scripts and plugins from scratch that reach a broad user-base across multiple hosts without redundant effort.
ePMV – embedded Molecular Viewer
The Python Molecular Viewer3 (PMV) is a suite of Python packages for visualization and analysis of molecular structure. PMV's core functionality for reading, writing, and manipulating molecular data (adding charges, radii, hydrogen atoms etc.) and generating geometrical objects from molecular data (CPK, ribbons, surfaces, etc.) can be accessed independently of PMV's native GUI in any program providing a Python interpreter.
Users in the molecular illustration community have explored the creation of 3DCGS-specific extensions for dealing with molecular data, leading to the re-implementation of many features, long-existing in PMV. We used uPy to create a PMV plugin: ePMV4 that provides a richer and more easily extensible set of capabilities than any previous molecular plugin for 3DCGS. Moreover, ePMV is usable within any uPy supported host.
ePMV uses the uPy Modeling Helper to translate PMV's molecular representations into host-native geometric objects to be displayed in the host's 3D viewport, leveraging all of the host's rendering capabilities (i.e. materials, textures, lights, animations, etc). Moreover, ePMV retains the correspondence between the host's geometric object and the underlying atomic and molecular representation. Figure 6a shows the ePMV GUI, which provides an easy and intuitive interface to ePMV's capabilities and which was developed with the uPy UI adaptor in order to be usable across all host applications supported by uPy. More details about ePMV can be found at http://epmv.scripps.edu/.
Figure 6. uPy allows deep plugins like ePMV to support hundreds of functions and GUI options.
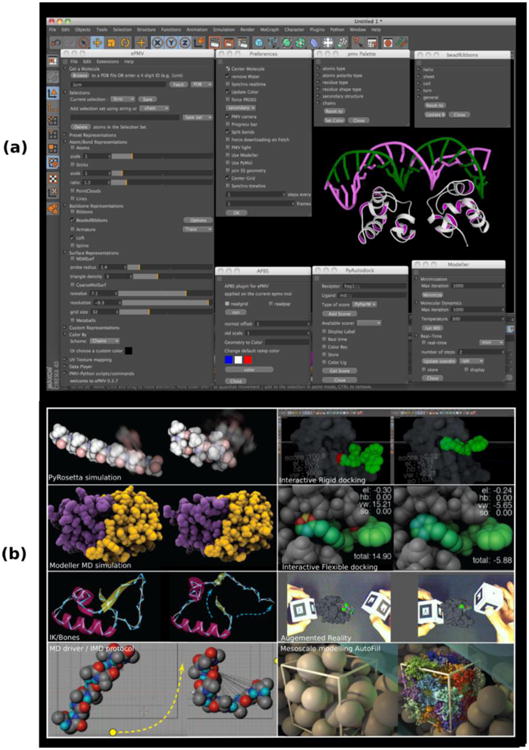
(a) View of ePMV UI windows and sub-windows inside the host Cinema4D. The main window controls the current molecular representation. The sub-windows expose options, detailed-commands, and extensions. (b) ePMV interoperates a large variety of host and external algorithms on the same data set. The left images show the initial state of an imported or constructed model, and the right images show a change in the model state as induced by each labeled method.
When ePMV is instantiated in a particular host, this host's computational capabilities become available for manipulating ePMV's molecular data. For instance, we leveraged Cinema 4D's bullet-based (http://bulletphysics.org) fast collision detection mechanism to implement interactive manual ligand-receptor docking. Here, ePMV provides the docking mechanism where the user, guided by an “energy score,” moves the ligand molecule relative the receptor. The host provides the method for handling collisions of rigid and/or soft bodies. When the molecules are treated as soft bodies, they can deform for an induced fit. Since ePMV interfaces with our Augmented Reality software component5, the user can position the receptor and ligand via handheld markers tracked by a camera (Figure 6b).
Developing ePMV as a uPy plugin (i.e. using uPy to create GUI and geometry objects, and to handle user interactions) allows ePMV to run in a wide range of hosts, thus enabling a larger number of potential users to access ePMV in the 3D software host they are most comfortable with, already own/use, or that they simply prefer. uPy also facilitates the maintenance and extension of ePMV, as the code duplication associated with a host-specific ePMV-type plugin approach does not occur.
GAMer– Mesh Improvement Software
GAMer (Geometry-preserving Adaptive MeshER) is a mesh generation library that produces high-quality simplex meshes of surfaces and volumes6. GAMer is used for: (i) improving meshes (coarsening and smoothing); and (ii) generating quality tetrahedral meshes (using TetGen, http://tetgen.berlios.de/), for use in finite element simulations. GAMer can be used as a stand-alone command line tool. However, visual feedback during mesh improvement has proven very helpful. Originally, GAMer was wrapped to become callable from Python and integrated as a dedicated plug-in for Blender 2.49b. With the new Blender 2.6 arriving along with an increase in GAMer demand by non-Blender users, the plug-in was re-written only once using uPy to facilitate migration to Blender 2.5 and 2.6, and to make GAMer available in the other supported hosts.
Figure 7 shows how the GAMer plug-in can be used on geometry acquired from manual segmentation of an electron tomography data set from a mouse heart cell11. In Figure 7a we see portions of two subcellular features from the segmented surface mesh after it has been imported into Blender. On the left we see a part of the sarcoplasmic reticulum and on the right we see part of a T-tubule. In Figure 7b we have applied the mesh improvement algorithms, both coarsening and smoothing, and in Figure 7c we have merged the surface mesh with a surrounding box mesh. The latter is a non-trivial feature provided by Blender. In Figure 7d we have used tools exposed by the uPy-based GAMer plug-in to mark and color certain regions on the surface mesh. When a volumetric mesh is generated, these regions become boundary domains, which can be used to declare boundary conditions in a finite element computation (see calcium simulation below). The uPy-based GAMer plugin communicates data (meshes) between the hosts and the GAMer module via the Modeling Helper. This process is exposed to the end user by the UI Adaptor interface to provide a comprehensive GUI that looks the same across all supported hosts. Figure 7e shows the uPy-based GAMer plug-in used for the mesh improvements and the boundary selection in Blender 2.6 and Blender 2.49b respectively.
Figure 7. GAMer improves polyhedral meshes.
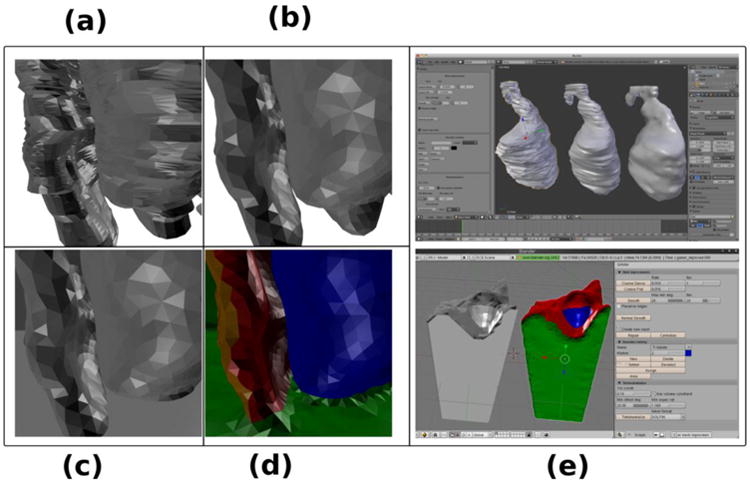
A roughly segmented surface (a) can be smoothed (b), merged with a bounding box (c), and used for selecting a boundary region (d). GAMer segments different surfaces (e) inside of Blender 2.6 (top view) and blender 2.49b (bottom view).
CSMOL– Cardiac Myocyte Calcium Dynamics
Meshs and boundaries generated from the uPy-based GAMer plug-in are used as the domain for simulating calcium (Ca2+) diffusion in cardiac ventricular myocytes. With the recent availability of sub-micron resolution myocyte structural data from confocal and electron microscopy7, a simulation tool, CSMOL, was developed for modeling reaction-diffusion models for calcium in rat and rabbit myocytes. CSMOL was used to model half-sarcomere sub-domains of both rat and rabbit ventricular myocytes and to identify inhomogeneities in calcium flux that are dependent on the geometric and topological features of the cell.
CSMOL results are usually analyzed using volumetric visualizers such as GMViewer or as projected data via MATLAB. These applications provide graphical insight into the Ca2+ concentration as a function of time and position. However, they are not directly integrated into a common platform, complicating the workflow for the average scientific user. We developed a standalone application with uPy to facilitate the visualization of CSMOL simulation results. The core of the application: (i) accesses the raw data from CSMOL (gmv); (ii) produces the tetrahedral mesh; and (iii) colors the mesh according to the compartment and the different Ca2+ concentration and flux values. uPy enabled us to rapidly create a visualization tool for this data. This prototype consists of a Python module that handles the CSMOL data (i.e. parsing) and communicates it to the Modeling Helper, which generates the colored tetrahedral representation of the simulated volume (parsed CSMOL data). This workflow is exposed to the user through a dialog developed using the UI Adaptor. The dialog enables the user to load different tetrahedral meshes, color them according to various properties, and split them based on properties.
We found that for this application, using uPy was particularly advantageous, as it allowed us to switch between hosts at different stages of the analysis to take advantage of the strengths each could offer. The initial, interactive analysis was performed in DejaVu3, where the availability of arbitrary 3D clipping planes and the ability of animating vertex-based colors greatly facilitated the analysis of the change of Ca flux over time in the different compartments (see Figure 8 and movie 1). High quality rendering could then be generated in other hosts to produce more effective images and advanced animations for publication and communication.
Figure 8. CSMOL visualizes calcium dynamics simulations with tetrahedral meshing from GAMer.
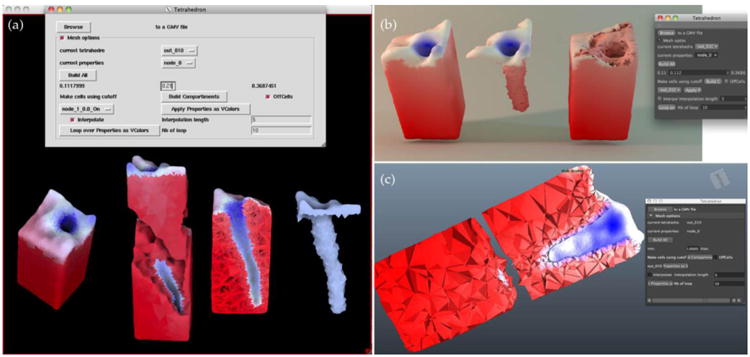
A cylindrical membrane feature (t-tubule) from muscle cells is colored red to indicate high calcium as compared to initial concentrations in blue. (a) View in DejaVu of the cell interior using splitting tools and real time cutting planes. (b) View in Cinema 4D of the same cell surface and interior more usable for publication of a static image thanks to enhanced rendering features. (c) View of a two perpendicular plane slices in Maya.
autoFill– Mesoscale Modeling and Visualization
The autoFill software program synthesizes 3D models of densely filled volumes and surfaces with arbitrarily shaped geometries (including procedurally defined structures). A specialization of autoFill, called autoCell, models the biological mesoscale, an intermediate scale (10-7–10-8m) between molecular and cellular biology that is difficult to visualize with experimental methods. autoCell works by packing molecular shapes into larger cellular frameworks while satisfying chemical and biological constraints. The autoFill project can be followed at http://autofill.scripps.edu.
In general, autoFill positions ingredient objects, into, onto, and around larger volumes/compartments with various methods and degrees of control according to autoFill recipe files. The most common type of ingredient preparation requires a user to generate a specific filetype called a sphereTree (.sph file). autoFill uses this sphereTree for efficient collision detection between ingredients and containers. The sphereTree algorithm identifies a set of spheres approximating an object's shape. We have used uPy to create a plugin that expands the original sphereTree clustering algorithm's functionality to be available from any host. An advantage of using uPy is that the sphereTree clustering algorithm can be used on any 3D model type available in any host application, including, for instance, models generated by: the host (Figure 9a); plugins such as ePMV (Figure 9b); or imported from other formats that are highly useful, but not traditionally accessible to molecular modeling software.
Figure 9. uPy exposes the capabilities of a Python SphereTree generator.
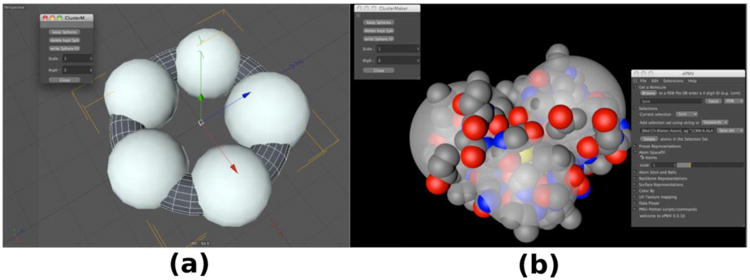
(a) A tree of five Spheres (one trunk with 5 branches) generated to approximate a torus geometry clustered stochastically about the coordinates of the torus's vertices. (b) Tree of three Spheres (one trunk with 3 branches) clustered about the atomic coordinates provided in the protein databank file 1crn to approximate the molecule's shape in a coarse but efficient manner suitable for use in generating a coarse mesoscale model with autoCell (see figure 10).
With all ingredients and recipes prepared and defined (shape, concentration, behavior, etc.), autoFill packs the ingredients into a specified volume. During packing, autoFill produces a list of transformation matrices for each placed ingredient, which becomes the output file. To efficiently visualize the results, we developed a Python script that calls uPy's Modeling Helper functions to generate the master geometry for each unique ingredient and deposit visible instances of each ingredient's geometry according to the autoFill output transformation matrices. Ingredient geometries can be molecular meshes, simplified representations (e.g. primitive spheres, cylinders, and boxes), curve points for fibrous molecules, and grids points for volumetric data (see examples in Figure 10A).
Figure 10. uPy interfaces with autoFill to construct, visualize, and analyse multiscale models of complex 3D volumes including biological cellscapes.
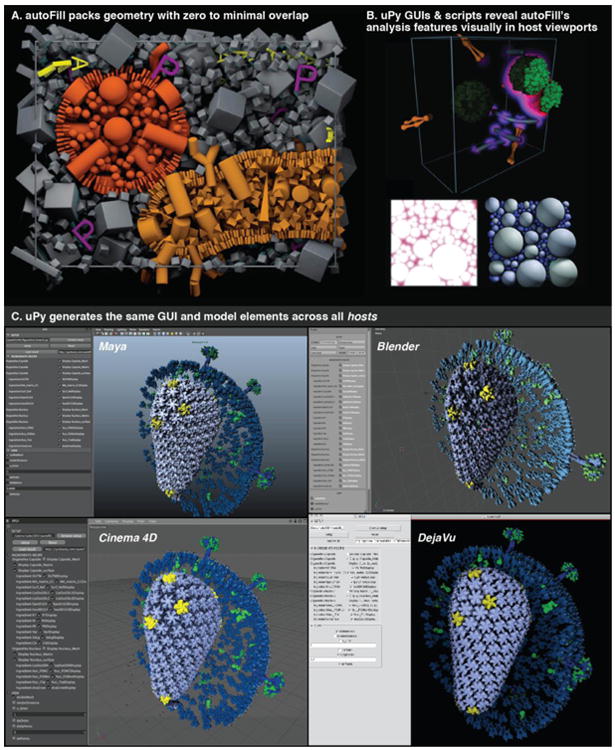
(A) Cylinders populate the surfaces of a red-orange and a yellow-orange mesh. Sphereoids fill the orange mesh and pyramids the green while cuboids fill the space outside either volume while avoiding existing “P” meshes. AutoFill quickly positions these objects with zero to minimal overlap as allowed by the user. (B) uPy enables many of autoFill's result analysis output and analysis tools to display interactively. (C) A lightweight autoFill Result Viewer builds, hides, reveals, and replaces various components of a multiscale autoFill model (here the HIV virus), directly from a result file, using the most efficient instancing systems available to each host to speed up viewport interaction and to reduce filesize.
To help the visualization and the development of autoFill analysis, we implemented uPy scripts for: the visualization of packing at run-time and different object coloring modes for debugging; the visualization of size-dependent available volumes, distance heat-mapping, packing order, and graphs of distribution and randomness statistics for analysis of results (see examples in Figure 10B).
Using uPy to develop autoFill result visualization tools enabled us to exploit the capabilities offered by different hosts. For instance, the DejaVu host optimized for viewport interactivity allows for realtime manipulation of a data-heavy model of HIV in full detail (see figure 10C and movie 2 of the rotation of an HIV virus). Other hosts, proved useful to set up autoFill recipes, e.g., to model certain ingredients, and to produce high quality ray-traced images and animations.
High quality host rendering options produce massive yet detailed images for large and dense volume such as blood serum. In order to illustrate such large volumes, difficult to visualize in real time, we used Cinema 4D to render a high resolution Gigapan image viewable at http://gigapan.org/gigapans/85568 This zoomable web interface shows a 1μm2 cuboid filled with molecular detail from a blood serum recipe to enable easier study, communication, and iterative improvement of the model by experts in the subject matter without having to produce multiscale viewers de novo.
Discussion and Conclusions
The modular nature of the Python programming language is conducive to the creation of re-usable software components. Moreover, the rapidly growing number of large applications and frameworks, either developed in Python or that embed a Python interpreter, creates a significant opportunity to re-use software components and make software interoperate in unprecedented ways. uPy supports this evolution by providing a unified API for writing scripts and plug-ins for a variety of host 3DCGS, creating new domain-competent applications.
uPy has proven to be a useful module for: (i) prototyping, developing, and maintaining complete plug-ins for all of the different supported hosts with reduced redundant effort, (ii) exploring the advantages, drawbacks and possible enhancements of various hosts, and (iii) expanding the functionality of individual Python wrapped applications by uniting them to the now standard host functionalities such as physics, particles, inverse kinematics, and Hollywood caliber animation and rendering. uPy enables software from two distinct communities, scientific and computer graphics, to interoperate in ways that can enhance the efforts of both.
Nevertheless, using uPy has raised some issues that have to be considered for any project. While every host application comes with its own set of strengths and weaknesses we found that all of them run into problems when displaying large numbers of spheres or polygonal meshes. This problem is well known in the computer graphics community and solutions exist, including: distance-dependent level-of-detail adjustment, field-of-view culling, employing particles with billboard imposters, low-polygon objects with normal textures applied, etc. While these techniques are not typically available in domain-specific modeling and visualization applications, the professional 3D software hosts already support many of these solutions and are likely to be among the first to integrate new techniques as they become available that will reduce these and other issues, minimizing the need for workarounds.
In the examples that we have described the uPy API has been restricted to the set of features common to all supported host applications. The number of common features is likely to shrink as the list of supported host applications grows. As described in the Methods paragraph, uPy currently supports basic types of geometric objects and a finite set of widgets, but it is possible to extend uPy beyond theses common primitives. For instance, Platonic solids are only natively supported in MAXON Cinema4D. To overcome this limitation, we implemented a Platonic solid modeler in the Modeling Helper base-class, thus making it available in all host-specific Modeling Helper classes. The Cinema4D subclass overrides this generic implementation to use its native one. This approach can be applied for a variety of functions missing in one or more of the supported host applications. Another example is the lack of Noise functions in the Maya Python API, while Blender and Cinema4D provide such functions. Again, the Modeling Helper class can provide a generic implementation of a missing function like this that can be overridden by the native versions. These implementations can be coded from scratch, or can be based on third party packages such as the Python Computer Graphics kit (cgkit http://cgkit.sourceforge.net/), which provides a complete noise module. As the core functionality of some hosts such as Maya are available as 3rd party Python packages, uPy's approach makes it possible to embed Maya functionality into any other host.
Currently uPy cannot be used to develop plugins that require the execution of OpenGL code or plugins that rely on reacting to mouse interactions in the host's 3D viewport beyond identifying currently selected objects and object positions. However, adding support for handling such events can be achieved, since all currently supported host applications provide the necessary APIs, with the exception, that the Cinema4D Python API does not currently support OpenGL code execution.
With regard to performance, while uPy is written in pure Python, it is used to encapsulate the hosts' native calls and thus does not reduce performance when compared to using the host's Python APIs directly.
The main advantage of using uPy is that uPy plugins are usable in a variety of host applications. This has several beneficial consequences: 1) users can utilize their domain specific plugins in the host that they are most familiar with; 2) the same plugin can run in different hosts at different stages of a project pipeline (e.g. for large studios with multiple hosts) or for scientific projects, at different stages of the analysis (for instance one host may work better when interactivity is critical verses when high-quality rendering is the goal); and finally, 3) the plugin is usable by a broader audience.
From the domain-specific methods-development point of view, uPy exposes computational methods in more environments without having to write more than one plugin. From the 3D graphics software development point of view, uPy extends capabilities across the set of supported hosts (i.e. implementing a feature in the uPy base class). From the application end-user point of view, uPy offers a choice among the hosts with the possibility to reproduce work in any of the other supported host applications as well.
We believe that uPy can help strengthen and broaden the connections between the computer graphics community and the information-visualization, scientific-visualization, visual-analytics, and other scientific communities by facilitating the development of new plugins and applications.
Acknowledgments
We would like to thank Peter Kekenes-Huskey for providing data and his help during the development of the Ca Simulation View plugins. This project was supported by grants from the National Center for Research Resources (5P41RR008605-19), the National Institute of General Medical Sciences (8 P41 GM103426-19) from the National Institutes of Health, the National Biomedical Computational Resource NIH grant 8P41GM103426-19 and partly supported by Centre of Excellence grant from the Research Council of Norway to the Centre for Biomedical Computing at Simula Research Laboratory.
References
- 1.McGill G. Molecular movies… coming to a lecture near you. Cell. 2008;133(7):1127–1132. doi: 10.1016/j.cell.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 2.The Python Papers Monograph. 2006;2 ISSN 1837-7092. [Google Scholar]
- 3.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 4.Johnson GT, Autin L, et al. ePMV Embeds Molecular Modeling into Professional Animation Software Environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillet A, et al. Tangible interfaces for structural molecular biology. Structure. 2005;13:483–491. doi: 10.1016/j.str.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Yu Z, et al. Feature-Preserving Adaptive Mesh Generation for Molecular Shape Modeling and Simulation. Journal of Molecular Graphics and Modeling. 2008;26(8):1370–1380. doi: 10.1016/j.jmgm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi T, et al. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J Cell Sci. 2009;122(7):1005–1013. doi: 10.1242/jcs.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]


