Abstract
Background
An alternate HIV testing algorithm has been proposed which includes a fourth-generation immunoassay followed by an HIV-1/HIV-2 antibody differentiation supplemental test for reactive specimens and a nucleic acid test (NAT) for specimens with discordant results.
Objective
To evaluate the performance of five rapid tests (Alere Clearview, Bio-Rad Multispot, OraSure OraQuick, MedMira Reveal, and Trinity Biotech Unigold) as the supplemental antibody assay in the algorithm.
Study Design
A total of 3273 serum and plasma specimens that were third- generation immunoassay repeatedly reactive and Western blot (WB) negative or indeterminate were tested with rapid tests and NAT. Specimens were classified by NAT 1) HIV-1 infected (NAT-reactive; n=184, 5.6%), 2) HIV-status unknown (NAT nonreactive; n=3078, 94.2%) or by Multispot 3) HIV-2 positive (n=5), 4) HIV-1 and HIV-2 positive (n=6). Excluding HIV-2 positive specimens, we calculated the proportion of reactive rapid tests among specimens with reactive and nonreactive NAT.
Results
The proportion of infected specimens with reactive rapid test results and negative or indeterminate WB ranged from 30.4% (56) to 47.8% (88) depending on the rapid test. From 1% to 2% of NAT-negative specimens had reactive rapid test results.
Conclusions
In these diagnostically challenging specimens, all rapid tests identified infections that were missed by the Western blot, but only one FDA-approved rapid test could differentiate HIV-1 from HIV-2. Regardless of which rapid test is used as a supplemental test in the alternative algorithm, false-positive algorithm results (i.e., reactive screening and rapid test in uninfected person) may occur, which will need to be resolved during the baseline medical evaluation.
Background
Although HIV-1 Western blots have historically been used as supplemental tests to confirm HIV infection in specimens with repeatedly reactive immunoassay results, they detect HIV infection weeks later than most currently available screening immunoassays, 1–4 are time-consuming, 5 may produce indeterminate results in persons who are infected or uninfected, 6, 7 and misclassify the majority of HIV-2 infections as HIV-1.8, 9 In response to the limitations of the Western blot, an alternate testing algorithm has been proposed by the Centers for Disease Control and Prevention (CDC) and the Association of Public Health Laboratories (APHL) which uses a fourth-generation antigen/antibody screening immunoassay (IA) and supplemental tests that include an HIV-1/HIV-2 antibody differentiation test and an HIV-1 nucleic acid test (NAT) (Figure 1).5, 10 If the specimen is repeatedly reactive using the fourth-generation IA and the supplemental antibody test is reactive for HIV-1 and/or HIV-2 antibodies, the person is considered to have established HIV-1 or HIV-2 infection. If the supplemental antibody test is negative, an HIV-1 RNA NAT is performed and, if positive, that person is considered to have acute HIV-1 infection. As recommended in the Department of Health and Human Services antiretroviral treatment guidelines, persons who test positive for HIV should receive a baseline medical evaluation that includes laboratory tests .11
Figure 1.
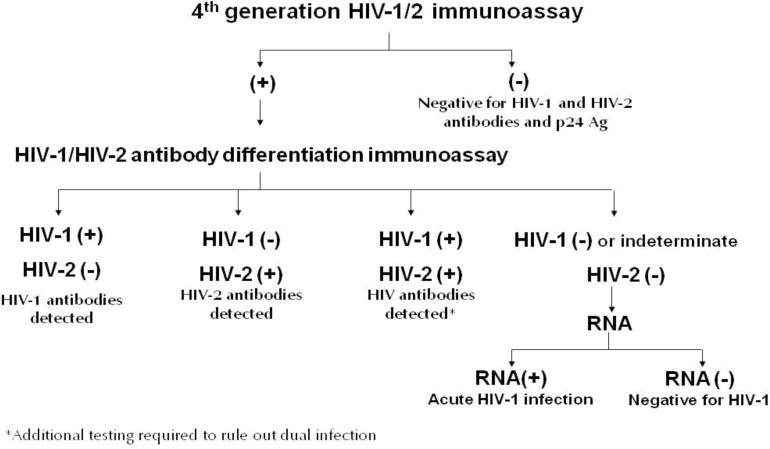
Alternate HIV Laboratory Testing Algorithm
Although an antigen/antibody IA is recommended as the screening assay for this new HIV diagnostic algorithm, third-generation immunoassays are still used in many laboratories.12 The use of a third-generation IA is given as an alternative screening IA in the proposed algorithm and its performance in the algorithm has been evaluated.13,14, 15,4 Most FDA-approved rapid tests can detect both HIV-1 and HIV-2, but only the Multispot HIV-1/HIV-2 Rapid Test (BioRad Laboratories, Redmond, WA) is FDA-approved as a differentiation test for supplemental testing. Although the proposed algorithm provides alternate FDA-approved antibody supplemental tests in place of the differentiation test in the algorithm (e.g., HIV-1 Western blot and immunofluorescence assay),13 the performance of other rapid tests has not been assessed and could inform testing guidance.
Objective
To evaluate the performance of five rapid antibody tests as supplemental assays in the alternate HIV testing algorithm using diagnostically challenging specimens that were repeatedly reactive with an initial third generation immunoassay, but not Western blot positive.
Study Design
From January 1, 2009 to September 9, 2010, over 6 million HIV tests were conducted at Quest Diagnostics laboratory facilities in the United States. The HIV prevalence in this population is estimated to be 1.3% among non-pregnant persons. 16 The company identified candidate specimens as all serum or plasma specimens tested in their United States laboratories with repeatedly reactive third-generation immunoassay results (GS HIV-1/HIV-2 Plus O, BioRad Laboratories, Redmond, WA) and negative or indeterminate Western blot results (GS HIV-1 Western blot, BioRad Laboratories, Redmond, WA). These specimens were sent to a central laboratory where links to personally identifying information were removed.
Specimens with sufficient volume were tested with 5 FDA-approved rapid antibody tests according to package insert instructions: Clearview HIV-1/2 StatPak (Clearview, Alere, Orlando, FL), Multispot HIV-1/HIV-2 Rapid Test (Multispot, BioRad Laboratories, Redmond, WA), OraQuick Advance Rapid HIV-1/2 Antibody Test (OraQuick, Orasure Technologies, Bethlehem, PA), Reveal G3 Rapid HIV-1 Antibody Test (Reveal, MedMira Laboratories, Halifax, Nova Scotia, Canada), and Uni-Gold Recombigen HIV Test (Uni-Gold, Trinity BioTech, Wicklow, Ireland). Four rapid tests are FDA-approved for use with serum or plasma. OraQuick, which is approved for use on plasma, was also used with serum. Reveal and Uni-gold are FDA-approved to detect HIV-1 only. Clearview and OraQuick can detect HIV-1 and HIV-2, and Multispot can detect and differentiate HIV-1 and HIV-2.
All specimens with sufficient volume were tested with the APTIMA HIV-1 RNA Qualitative Assay (APTIMA, GEN-PROBE, San Diego, CA), which is FDA approved for use on serum and plasma. Specimens with negative results on all rapid tests were tested with APTIMA in pools of 16 at the Florida Bureau of Laboratories, 17 with the exception that reactive pools were repeated. Specimens in pools that were non-reactive initially or on repeat testing were considered to be APTIMA negative. Pools testing repeatedly reactive on the APTIMA test were deconstructed, and specimens were tested individually with APTIMA. Specimens with one or more positive rapid test were tested individually with APTIMA at the CDC laboratory using 100μL of serum or plasma supplemented with 400μL of IA and APTIMA-negative plasma. It is possible that some specimens may not have been handled according to APTIMA package insert specifications with regard to holding temperatures or freeze-thaw cycles because they were not originally collected with the intent of being tested with that assay.
Specimens were categorized by HIV infection status as: 1) HIV-1 infected (reactive by APTIMA; n=184, 5.6%) ; 2) HIV status unknown (negative APTIMA; n=3078, 94.0%) ; 3) HIV-2 antibody positive (HIV-1 antibody negative and HIV-2 antibody reactive using Multispot, regardless of APTIMA results; n=5,0.2%) ; and 4) HIV-infected, undifferentiated (HIV-1 and HIV-2 antibody reactive using Multispot, regardless of APTIMA results; n=6, 0.2%). Due to volume constraints, specimens with undifferentiated results did not undergo testing according to the Multispot dilution protocol.
We excluded the specimens with reactive HIV-2 results and undifferentiated results from analysis. Among the HIV-infected specimens, for each rapid test, we calculated the number and proportion of specimens with reactive rapid results stratified by Western blot results. Among those with negative APTIMA results, for each rapid test, we calculated the number and proportion of specimens with positive rapid test results by Western blot results. We also calculated the proportion with negative results on rapid tests. Statistical analysis was conducted using SAS version 9.3 (Cary, NC).
Results
Of the 6934 specimens with IA-repeatedly reactive and Western blot negative or indeterminate results identified during the study period, funding permitted the acquisition of 3999 specimens (57.7%) with sufficient volume for testing with the five rapid tests. Of those, 3273 (81.8%) had sufficient volume to conduct APTIMA and were included in further analyses. Among the 726 specimens not included in analyses, 659 had insufficient volume for APTIMA testing, 60 had reactive pooled APTIMA testing, but insufficient volume for individual testing, and 7 had an invalid pooled result with insufficient volume for repeat testing. Among the specimens not included in the analyses (n=726), the number with reactive rapid tests ranged from 13 (1.8%) (Reveal) to 28 (3.9%) (Multispot).
Of the 3273 total specimens that were GS IA repeatedly reactive and Western blot negative or indeterminate, 184 (5.6%) were HIV-1 infected based on reactive APTIMA results. Of these, 93 (50.5%) had an indeterminate Western blot result. Between 40 (43.0%) and 68 (73.1%) of these were detected by at least one rapid test, and 60 (64.5%) were detected by more than one rapid test (Table 1). There were 91 (49.5%) infected specimens with a negative Western blot result. Between 14 (15.4%) and 20 (22.0%) of these were positive on at least one rapid test, and 18 (19.8%) were positive on more than one rapid test. In most cases, the 95% confidence intervals for the proportion of infections identified by a rapid test overlapped with that of the other rapid tests. In addition, Multispot identified 5 (0.2%) specimens with HIV-2 antibodies and 6 (0.2%) specimens with both HIV-1 and HIV-2 antibodies.
Table 1.
HIV rapid test results among specimens with repeatedly reactive HIV 1/2 IA, negative or indeterminate Western blot, and positive APTIMA results
| Rapid HIV Test | Indeterminate WB (n=93) and positive RT N (%) | Negative WB (n=91) and positive RT N (%) | Total HIV-infecteda (n=184) with positive RT | Proportion HIV-infected with positive RTb (95% Confidence Interval) |
|---|---|---|---|---|
| Clearview | 48 (51.6%) | 16 (17.6%) | 64 | 34.8% (27.9%–42.1%) |
| Multispot | 68 (73.1%) | 20 (22.0%) | 88 | 47.8% (40.4%–55.3%) |
| OraQuick | 44 (47.3%) | 14 (15.4%) | 58 | 31.5% (24.9%–38.8%) |
| Reveal | 40 (43.0%) | 16 (17.6%) | 56 | 30.4% (23.9%–37.6%) |
| Unigold | 59 (63.4%) | 20 (22.0%) | 79 | 42.9% (35.7%–50.4%) |
| >1 reactive rapid test | 60 (64.5%) | 18 (19.8%) | n/a | not applicable |
WB=Western blot; RT=rapid test
HIV-infected=GS IA repeatedly reactive and APTIMA (nucleic acid test)-positive
Specimens with reactive rapid test results/# GS IA repeatedly reactive and APTIMA positive specimens
There were 3078 (94.0%) specimens with non-reactive APTIMA results (Table 2), 735 (23.9%) of which had an indeterminate Western blot. Between 16 (2.2%) and 37 (5.0%) of these were positive by a rapid test, and 33 (4.5%) were positive by more than one rapid test. There were 2343 (76.1%) specimens with negative Western blot results. Between 9 (0.4%) and 26 (1.1%) of these were positive by a rapid test, and 11 (0.5%) were positive by more than one rapid test. The proportion of APTIMA-negative specimens with negative rapid test results ranged from 98.0% to 99.0%, and the 95% confidence intervals of these proportions overlapped for most tests.
Table 2.
HIV rapid test results among specimens with repeatedly reactive HIV 1/2 IA, negative or indeterminate Western blot, and negative APTIMA results
| Rapid HIV test | Indeterminate WB, negative APTIMA (n=735) and positive RT N (%) | Negative WB, negative APTIMA (n=2343) and positive RT N (%) | Total with positive RT and negative APTIMA (n=3078) | Proportion with negative APTIMA and negative RT (95% Confidence Interval) |
|---|---|---|---|---|
| Clearview | 23 (3.1%) | 10 (0.4%) | 33 | 98.9% (98.5%–99.3%) |
| Multispot | 37 (5.0%) | 26 (1.1%) | 63 | 98.0% (97.4%–98.4%) |
| Oraquick | 26 (3.5%) | 10 (0.4%) | 36 | 98.8% (98.4%–99.2%) |
| Reveal | 16 (2.2%) | 14 (0.6%) | 30 | 99.0%a (98.6%–99.3%) |
| Unigold | 29 (4.0%) | 9 (0.4%) | 38 | 98.8% (98.3%–99.1%) |
| >1 reactive rapid test | 33 (4.5%) | 11 (0.5%) | n/a | not applicable |
WB=Western blot; RT=Rapid test
7 specimens had invalid results with Reveal, and were not included in analyses.
Discussion
In this evaluation of 3273 specimens that were third-generation HIV 1/2 IA reactive but Western blot negative or indeterminate, one-third to one-half of specimens with detectable RNA were classified as antibody-positive by the rapid tests. When the alternate HIV diagnostic algorithm is used and infections are missed by rapid supplemental tests, they are likely to be detected by an HIV-1 NAT which can identify the presence of viral RNA within two weeks of exposure.3, 4, 15 These specimens from HIV-infected individuals, which were not detected by Western blot, likely represent early infections. Other investigations have shown an even lower proportion of early infections detected by rapid tests. 18, 19 It is unknown whether persons misclassified as uninfected based on Western blot ultimately learned of their infection, which highlights the need for the alternate algorithm.
Using the alternate algorithm,5 specimens that are reactive using the screening IA, non-reactive using the rapid antibody test, and reactive by a NAT are flagged as early infections. There is utility in following the highly sensitivity initial IA with an assay that is less sensitive, such as a rapid test, so that early infections can be identified as such. Persons in the early stage of infection are likely to have high viral loads and, thus, pose a greater likelihood of disease transmission.20–22 Maximizing the public health impact of identifying early infections requires a timely response to link persons with care, treatment and partner services.23 Identifying early infections requires that persons present to testing venues shortly after an exposure event, which may necessitate education about the window period of new tests in communities impacted by HIV.23
Only one FDA-approved rapid HIV test, Multispot, distinguishes HIV-1 from HIV-2 and it captured 5 potential HIV-2 infections. It has been validated to perform well in the proposed algorithm, and it had the highest sensitivity among the rapid tests evaluated.4, 14, 15 Although HIV-2 infections are rare in the United States,24 identifying them is important because they do not respond to non-nucleoside reverse transcriptase inhibitors and some protease inhibitors that are used as first-line antiretroviral treatment for HIV-1.25, 26 With wider implementation and use of the algorithm, increases in the number of HIV-2 cases detected in the United States might be observed.
Over 90% of the specimens analyzed had negative HIV-1 RNA APTIMA results. Many may be from uninfected individuals (i.e., a false positive HIV-1/2 antibody screening test result with negative rapid and NAT results). Based on the broader population of 6 million specimens, the specificity given in the package insert for the GS IA would predict that 6600 specimens would be false-positive on this screening test. This number is close to the 6934 total specimens identified as third-generation repeatedly reactive and Western blot negative or indeterminate, once those that were infected were removed. Assuming that all APTIMA non-reactive specimens were uninfected, the rapid test specificity would range from 98 to 99%, and Multispot would have been concurrently false-positive with the screening test on more specimens than the other rapid tests. In two studies, Multispot specificity was 99.4%, an estimate which is higher than in this study because they did not include IA-reactive specimens. 14, 27 Those studies also showed Multispot specificity to be slightly lower than that for other rapid tests evaluated. It is conceivable that more cross-reactivity occurred between the GS screening IA and Multispot than other rapid tests because the tests are produced by the same manufacturer, and may use some of the same antigens. In other studies, concordant false positives between a GS screening IA and Multispot were rare.15, 28 If concordant false-positive screening and supplemental rapid test results occur with the alternate algorithm, persons will have negative nucleic acid test results following their baseline evaluation in care 11 and will require further diagnostic testing. The false positive rate of HIV-1 Western blot in blood donors, based on a negative NAT result, was 4.8%, which is higher than the observed false-positive rate for any of the rapid tests evaluated here.29
One limitation of this study is that some of the specimens with negative APTIMA results might be from infected individuals, particularly those exhibiting viral bands on the Western blot and more than one reactive rapid test, but follow-up specimens were not available. Occasional false-negative NAT results have been observed in HIV antibody-positive specimens.3, 17 Those with infections detected by the third-generation immunoassay but not APTIMA may have had this result due to antiretroviral use, dilution of specimens, specimen handling30 or because serum samples, which constituted most of this specimen set, have reduced RNA relative to plasma samples.31 An additional study limitation is that some APTIMA-reactive specimens could have been cross- contaminated during handling, especially those that simultaneously exhibited low APTIMA signal to cut-off values and rapid test non-reactivity (up to 7% of those classified as infected). If true, the rapid-test sensitivity determinations presented here would be underestimates.
In conclusion, although all of the rapid tests identified infections that were missed by the Western blot, only one FDA-approved rapid test differentiates HIV-1 from HIV-2. Regardless of which rapid test is used, false-positive algorithm results may occur, which will need to be resolved during the baseline medical evaluation. For individuals with established HIV infections that will be resolved by the algorithm without NAT testing, the time to test result receipt should be reduced when a rapid test is used as a supplemental test in place of a Western blot. Although currently available FDA-approved rapid tests are not as sensitive as third- or fourth-generation IA for the detection of early HIV infection, they function as well as or better than the Western Blot for detecting early HIV infection.3,4 Their use as a supplemental test in the alternate algorithm in conjunction with a nucleic acid test will allow healthcare personnel to more effectively identify recently infected individuals and to provide a timelier response to interrupt further transmission.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The use of trade names and commercial sources is for identification only and does not imply endorsement by the U.S. Department of Health and Human Services or by Quest Diagnostics.
Acknowledgments
We wish to acknowledge the following persons for their logistical and technical assistance in the conduct of this project: Association of Public Health Laboratories, Steve Ethridge and Dollene Hemmerlein from the Centers for Disease Control and Prevention, Emeka Oraka from ICF International at the Centers for Disease Control and Prevention, Sally Fordan, Olanike David, and Petrice Stephens from the Florida Bureau of Laboratories, and Lynn Collins, Roger Frye, Lawrence Hirsch, James Hong, Annie Howden, Xiaohua Huang, Brenda Kolb, and Constance Rhett, from Quest Diagnostics.
Funding: This project was funded by the Centers for Disease Control and Prevention
Abbreviations
- NAT
nucleic acid test
- IA
immunoassay
- WB
Western blot
Footnotes
Competing Interests: None declared.
Ethical Approval: This study was determined to be research not involving identifiable human subjects by the National Center for HIV, Viral Hepatitis, STD and TB Prevention at the Centers for Disease Control and Prevention.
References
- 1.Association of Public Health Laboratories. HIV Diagnostics Survey. Public Health Laboratory Issues in Brief. 2012 Available from: http://www.aphl.org/AboutAPHL/publications/Documents/ID_2012Dec_HIV-Diagnostics-Survey-Issue-Brief.pdf.
- 2.Centers for Disease Control. Interpretation and use of the western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb Mortal Wkly Rep. 1989;38(Suppl 7):1–7. [PubMed] [Google Scholar]
- 3.Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, et al. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol. 2008;46(5):1588–95. doi: 10.1128/JCM.02196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(Suppl 1):S17–22. doi: 10.1016/j.jcv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 5.CLSI. CLSI Document M53A. Clinical and Laboratory Standards Institute; Wayne, PA: 2011. Criteria for Laboratory Testing and Diagnosis of Human Immunodeficiency Virus Infection; Approved Guideline. [Google Scholar]
- 6.Vardinon N, Yust I, Katz O, Iaina A, Katzir Z, Modai D, et al. Anti-HIV indeterminate western blot in dialysis patients: a long-term follow-up. Am J Kidney Dis. 1999;34(1):146–9. doi: 10.1016/s0272-6386(99)70121-6. [DOI] [PubMed] [Google Scholar]
- 7.Guan M. Frequency, causes, and new challenges of indeterminate results in Western blot confirmatory testing for antibodies to human immunodeficiency virus. Clin Vaccine Immunol. 2007;14(6):649–59. doi: 10.1128/CVI.00393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasrullah M, Ethridge SF, Delaney KP, Wesolowski LG, Granade TC, Schwendemann J, et al. Comparison of alternative interpretive criteria for the HIV-1 Western blot and results of the Multispot HIV-1/HIV-2 Rapid Test for classifying HIV-1 and HIV-2 infections. J Clin Virol. 2011;52(Suppl 1):S23–7. doi: 10.1016/j.jcv.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Torian LV, Eavey JJ, Punsalang AP, Pirillo RE, Forgione LA, Kent SA, et al. HIV type 2 in New York City, 2000–2008. Clin Infect Dis. 2010;51(11):1334–42. doi: 10.1086/657117. [DOI] [PubMed] [Google Scholar]
- 10.Pandori MW, Branson BM. 2010 HIV Diagnostics Conference. Expert Rev Anti Infect Ther. 2010;8(6):631–3. doi: 10.1586/eri.10.48. [DOI] [PubMed] [Google Scholar]
- 11.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents: Baseline Evaluation. 2012 2012 June 14]; Available from: http://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-treatment-guidelines/36/baseline-evaluation.
- 12.Association of Public Health Laboratories. Public Health Laboratory Issues in Brief: 2012 HIV Diagnostics Survey. 2012 Available from: http://www.aphl.org/aphlprograms/infectious/hiv/Documents/HIV_2012_Survey.pdf.
- 13.Branson BM. Draft Recommendations: Diagnostic Laboratory Testing for HIV Infection. Centers for Disease Control and Prevention: 2012 HIV Diagnostics Conference Website 2012 [Google Scholar]
- 14.Wesolowski LG, Delaney KP, Hart C, Dawson C, Owen SM, Candal D, et al. Performance of an alternative laboratory-based algorithm for diagnosis of HIV infection utilizing a third generation immunoassay, a rapid HIV-1/HIV-2 differentiation test and a DNA or RNA-based nucleic acid amplification test in persons with established HIV-1 infection and blood donors. J Clin Virol. 2011;52(Suppl 1):S45–9. doi: 10.1016/j.jcv.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 15.Delaney KP, Heffelfinger JD, Wesolowski LG, Owen SM, Meyer WA, III, Kennedy S, et al. Performance of an alternative laboratory-based algorithm for HIV diagnosis in a high-risk population. J Clin Virol. 2011;52(Suppl 1):S5–10. doi: 10.1016/j.jcv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Wesolowski LG, Delaney KP, Lampe MA, Nesheim SR. False-positive human immunodeficiency virus enzyme immunoassay results in pregnant women. PLoS One. 2011;6(1):e16538. doi: 10.1371/journal.pone.0016538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel P, Mackellar D, Simmons P, Uniyal A, Gallagher K, Bennett B, et al. Detecting acute human immunodeficiency virus infection using 3 different screening immunoassays and nucleic acid amplification testing for human immunodeficiency virus RNA, 2006–2008. Arch Intern Med. 2010;170(1):66–74. doi: 10.1001/archinternmed.2009.445. [DOI] [PubMed] [Google Scholar]
- 18.Patel P, Bennett B, Sullivan T, Parker MM, Heffelfinger JD, Sullivan PS, et al. Rapid HIV screening: missed opportunities for HIV diagnosis and prevention. J Clin Virol. 2012;54(1):42–7. doi: 10.1016/j.jcv.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louie B, Wong E, Klausner JD, Liska S, Hecht F, Dowling T, et al. Assessment of rapid tests for detection of human immunodeficiency virus-specific antibodies in recently infected individuals. J Clin Microbiol. 2008;46(4):1494–7. doi: 10.1128/JCM.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daar ES, Moudgil T, Meyer RD, Ho DD. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324(14):961–4. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- 21.Morrison CS, Demers K, Kwok C, Bulime S, Rinaldi A, Munjoma M, et al. Plasma and cervical viral loads among Ugandan and Zimbabwean women during acute and early HIV-1 infection. AIDS. 2010;24(4):573–82. doi: 10.1097/QAD.0b013e32833433df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilcher CD, Joaki G, Hoffman IF, Martinson FE, Mapanje C, Stewart PW, et al. Amplified transmission of HIV-1: comparison of HIV-1 concentrations in semen and blood during acute and chronic infection. AIDS. 2007;21(13):1723–30. doi: 10.1097/QAD.0b013e3281532c82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly JA, Morin SF, Remien RH, Steward WT, Higgins JA, Seal DW, et al. Lessons learned about behavioral science and acute/early HIV infection. The NIMH Multisite Acute HIV Infection Study: V. AIDS Behav. 2009;13(6):1068–74. doi: 10.1007/s10461-009-9579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease Control and Prevention. HIV-2 Infection Surveillance–United States, 1987–2009. MMWR Morb Mortal Wkly Rep. 2011;60(29):985–8. [PubMed] [Google Scholar]
- 25.Ntemgwa ML, d’Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob Agents Chemother. 2009;53(9):3611–9. doi: 10.1128/AAC.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hizi A, Tal R, Shaharabany M, Currens MJ, Boyd MR, Hughes SH, et al. Specific inhibition of the reverse transcriptase of human immunodeficiency virus type 1 and the chimeric enzymes of human immunodeficiency virus type 1 and type 2 by nonnucleoside inhibitors. Antimicrob Agents Chemother. 1993;37(5):1037–42. doi: 10.1128/aac.37.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaney KP, Branson BM, Uniyal A, Phillips S, Candal D, Owen SM, et al. Evaluation of the performance characteristics of 6 rapid HIV antibody tests. Clin Infect Dis. 2011;52(2):257–63. doi: 10.1093/cid/ciq068. [DOI] [PubMed] [Google Scholar]
- 28.Nasrullah M, Wesolowski LG, Meyer WA, 3rd, Owen SM, Masciotra S, Vorwald C, et al. Performance of a fourth-generation HIV screening assay and an alternative HIV diagnostic testing algorithm. AIDS. 2012 doi: 10.1097/QAD.0b013e32835bc535. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinman S, Busch MP, Hall L, Thomson R, Glynn S, Gallahan D, et al. False-positive HIV-1 test results in a low-risk screening setting of voluntary blood donation. Retrovirus Epidemiology Donor Study. JAMA. 1998;280(12):1080–5. doi: 10.1001/jama.280.12.1080. [DOI] [PubMed] [Google Scholar]
- 30.Gessoni G, Barin P, Valverde S, Giacomini A, Di Natale C, Orlandini E, et al. Biological qualification of blood units: considerations about the effects of sample’s handling and storage on stability of nucleic acids. Transfus Apher Sci. 2004;30(3):197–203. doi: 10.1016/j.transci.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 31.Griffith BP, Rigsby MO, Garner RB, Gordon MM, Chacko TM. Comparison of the Amplicor HIV-1 monitor test and the nucleic acid sequence-based amplification assay for quantitation of human immunodeficiency virus RNA in plasma, serum, and plasma subjected to freeze-thaw cycles. J Clin Microbiol. 1997;35(12):3288–91. doi: 10.1128/jcm.35.12.3288-3291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


