Abstract
We investigated the molecular epidemiology and population dynamics of HCV infection among indigenes of two semi-isolated communities in North-Central Nigeria. Despite remoteness and isolation, ~15 % of the population had serological or molecular markers of hepatitis C virus (HCV) infection. Phylogenetic analysis of the NS5b sequences obtained from 60 HCV-infected residents showed that HCV variants belonged to genotype 1 (n=51; 85 %) and genotype 2 (n=9; 15 %). All sequences were unique and intermixed in the phylogenetic tree with HCV sequences from people infected from other West African countries. The high-throughput 454 pyrosequencing of the HCV hypervariable region 1 and an empirical threshold error correction algorithm were used to evaluate intra-host heterogeneity of HCV strains of genotype 1 (n=43) and genotype 2 (n=6) from residents of the communities. Analysis revealed a rare detectable intermixing of HCV intra-host variants among residents. Identification of genetically close HCV variants among all known groups of relatives suggests a common intra-familial HCV transmission in the communities. Applying Bayesian coalescent analysis to the NS5b sequences, the most recent common ancestors for genotype 1 and 2 variants were estimated to have existed 675 and 286 years ago, respectively. Bayesian skyline plots suggest that HCV lineages of both genotypes identified in the Nigerian communities experienced epidemic growth for 200–300 years until the mid-20th century. The data suggest a massive introduction of numerous HCV variants to the communities during the 20th century in the background of a dynamic evolutionary history of the hepatitis C epidemic in Nigeria over the past three centuries.
INTRODUCTION
Hepatitis C virus (HCV) infection is a serious public health problem worldwide. HCV causes chronic infection in 70–80 % of infected people. Chronic HCV infection may progress to chronic hepatitis and cirrhosis, leading in many cases to severe complications including hepatocellular carcinoma (HCC) and death (Itskowitz, 2007). An estimated 170 million people or ~3 % of the world’s population are chronically infected with HCV; and 3–4 million people are newly infected each year, with most of these cases occurring in Africa (EASL, 1999; WHO, 1999).
HCV is a positive-strand, ssRNA virus classified in the genus Hepacivirus of the family Flaviviridae (Shepard et al., 2005). The HCV genome consists of ~9600 nt and encodes a single long polyprotein with the following gene order: 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′ (Lindenbach & Rice, 2005). The HCV genome displays considerable sequence divergence. HCV has been classified into six genotypes, 1–6, with each genotype further subdivided into subtypes displaying different geographical distributions worldwide (Simmonds et al., 2005). For example, HCV subtype 5a and all subtypes of genotype 6 are mainly restricted to South Africa and South East Asia, respectively (Simmonds et al., 1993), while subtypes 1a, 1b, 2b and 3a are distributed globally (Smith et al., 1997). A new genotype 7 has been recently registered in the HCV databases, but the full report on this isolate has not been published (Nakano et al., 2012).
In Africa, divergent strains of HCV genotypes 1 and 2 were found in Ghana, Guinea Conakry, Burkina Faso, Benin Republic and Guinea Bissau, suggesting that these strains have been endemic in the West African subregion (Candotti et al., 2003; Jeannel et al., 1998; Markov et al., 2009; Ruggieri et al., 1996; Wansbrough-Jones et al., 1998). Recently, analysis of the epidemic history of HCV infections traced the modern HCV lineages in West Africa back to the 17th–20th centuries (Markov et al., 2009; Pouillot et al., 2008b). This estimate, however, does not take into account Nigeria, the country with more than three-quarters of the population of West Africa, which occupies a geographically central position within this African subregion. Identifying HCV circulating in Nigeria is important for guiding HCV infection control initiatives, understanding the evolutionary dynamics and natural history of HCV infection among indigenous West Africans, and predicting the future burden of HCV-related disease in Africa.
The epidemiology of HCV infection in Nigeria is not well understood. Most studies of hepatitis C in Nigeria have focused on serological characterization of selected population groups, e.g. prison inmates, patients with diabetes mellitus, blood donors, HIV-infected persons, patients with chronic renal failure and those with sickle cell anaemia, for whom risk for HCV infection in urban areas of Nigeria is variable. Previous studies established a broad HCV-seroprevalence rate, ranging from 1.9 % among pregnant women in Benin City to ~14.5 % among apparently healthy individuals with a family history of diabetes in Plateau State or among HIV-positive patients in Lagos (Balogun et al., 2010; Nwankiti et al., 2009; Onakewhor & Okonofua, 2009). HCV infections have been shown to play a significant role in the aetiology of chronic liver disease and HCC in Nigeria (Laraba et al., 2010; Mustapha et al., 2007). Despite HCV endemicity, association with HCC and importance of HCV genotypes for clinical management (Hnatyszyn, 2005; Zeuzem, 2004), only three small-scale studies have been made to genetically characterize HCV isolates from Nigeria (Agwale et al., 2004; Mellor et al., 1995; Oni & Harrison, 1996). As a result, an extremely limited number of HCV sequences from Nigeria are available in any repository of nucleotide sequences. Genotype distribution and genetic characterization of HCV strains circulating in different African countries, including Nigeria, are critical for designing universally efficacious vaccines against HCV infection, predicting the sensitivity of diagnostic assays and understanding the evolutionary history and molecular epidemiology of HCV in Africa.
Here, we present the first comprehensive phylogenetic and evolutionary analysis of HCV variants obtained from 60 residents of two rural, remote communities in North Central Nigeria.
RESULTS
HCV prevalence
Samples (n=519) were collected from the study sites (341 from village 1 and 178 from village 2) and tested for anti-HCV and for HCV RNA by the NS5b real-time PCR. A total of 73 samples (55 from village 1 and 18 from village 2) were found to be anti-HCV-positive and 60 samples were PCR-positive (47 samples from village 1, 13 from village 2), with 55 samples being both anti-HCV- and PCR-positive. Thus, a total of 78 samples were identified that had serological or molecular markers of HCV infection. An overall high prevalence of 15 % of HCV infection in these communities represents one of the highest estimates reported in Nigeria (Balogun et al., 2010; Nwankiti et al., 2009; Onakewhor & Okonofua, 2009). Of the 60 PCR-positive individuals, males (n=33) and females (n=27) were almost equally represented. Although, the anti-HCV seroprevalence was slightly higher in men than women (sex ratio=1.2), this difference is not statistically significant. The majority (63.3 %) of the infection was found in individuals between the ages of 20 and 40 years.
Phylogenetic relationships among HCV variants
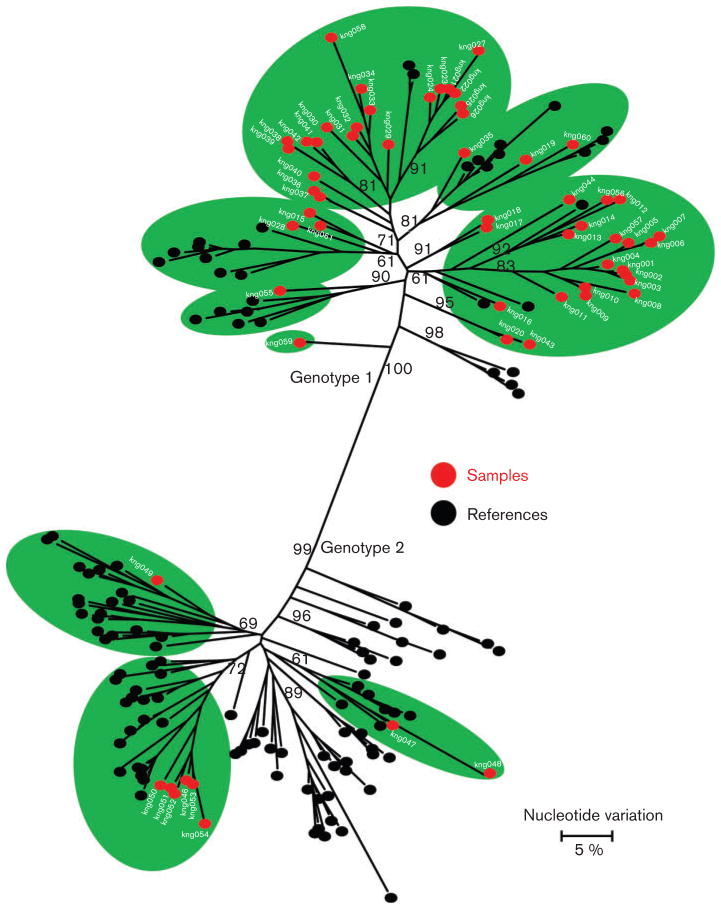
Considering the rural location and relative isolation, it was expected that these two small communities would have limited exposure to HCV and should have had a single or only a few HCV strains circulating among the indigenous residents. However, analysis of the NS5b sequences identified HCV variants belonging to genotypes 1 (n=51; 85 %) and 2 (n=9; 15 %) (Fig. 1). Furthermore, all genotype 1 and 2 variants segregated into subtypes, some of which were not previously characterized (Fig. 1). These findings indicate that the two communities were infected with several HCV strains, suggesting either numerous exposures or exposure to all these HCV lineages in bulk from limited infection events.
Fig. 1.
Phylogenetic maximum-likelihood tree constructed using NS5b sequences (300 nt). Sequences determined in this study are shown in red. HCV genotypes are indicated on the branches. Reference sequences were obtained from the GenBank database and are shown in black. Bootstrap values higher than 60 % are indicated at the major nodes.
The number of HCV genotype 1 variants in these communities was fivefold greater compared with the number of genotype 2 variants. The NS5b sequence analysis did not reveal any community-specific clustering and showed extensive phylogenetic intermixing of these sequences with other HCV isolates from different African countries and strains found worldwide. Consistent with the NS5b phylogenetic analysis, genetic diversity of 17.3 and 16.2 % was observed for the genotype 1 and 2 variants, respectively. The high genetic diversity suggests a long evolutionary history. However, the presence of two genotypes and several subtypes as well as intermixing with HCV variants found worldwide strongly suggest that many, if not all, of these variants were introduced rather than evolved in the two communities. Although many variants could not be classified into any known subtype, there was a trend for the variants to form clusters.
HCV variants from West and Central Africa
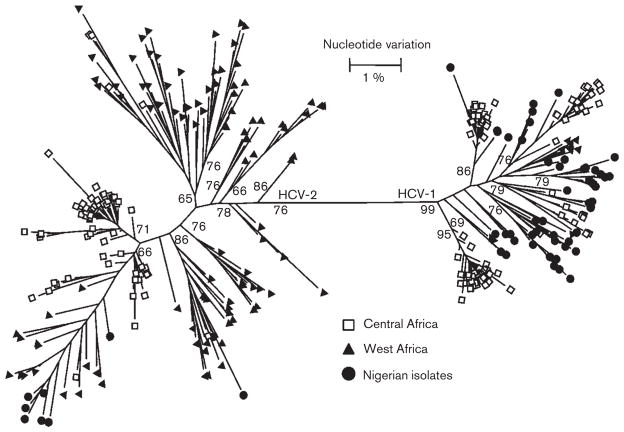
The diversity of the HCV variants observed in the two communities was compared to HCV genotype 1 and 2 strains identified in other West and Central African countries (Fig. 2). HCV NS5b sequences from Central Africa had the tendency to form compact and distinct clusters. A few of these sequences intermixed with sequences from West Africa (Fig. 2). It should be noted, however, that among 141 sequences from Central Africa, 136 were obtained from Cameroon, with only five sequences being from two immediate neighbouring countries (Table 1). Conversely, 107 sequences from West Africa originated from six countries within the West African subregion (Table 1). These sequences were more diverse and grouped with many of the variants determined in this study, thus indicating a genetic relatedness of HCV variants identified among the two communities and HCV strains broadly circulating in West Africa. However, the distribution of HCV variants between genotypes 1 and 2 in these communities in Nigeria is very different from the other West African countries. In the Nigerian communities, 85 % of HCV variants belong to genotype 1, while only 15 % of HCV variants recovered from the other West African countries belong to this genotype.
Fig. 2.
Phylogenetic tree (maximum-likelihood) constructed using NS5b sequences from West and Central Africa. Sequences from Central Africa are shown by unfilled squares, while those from West Africa are shown by black triangles. The sequences from this study are marked by black circles. Bootstrap values higher than 60 % are indicated at the major nodes.
Table 1.
GenBank sequences used in this study
HCV-1, HCV genotype 1, HCV-2, HCV genotype 2.
Intra-host HCV heterogeneity
The presence of two HCV genotypes, several classified/ unclassified subtypes and numerous HCV variants raise questions about the origin and maintenance of HCV diversity in these two communities. To investigate genetic connections among these variants in the population, quasispecies analysis of HVR1 was conducted using high-throughput pyrosequencing of 43 HCV genotype 1 and six genotype 2 samples. The small size of the communities and circulation of many HCV variants suggests a potential for frequent mixed infections. However, analysis of quasi-species revealed a very limited intermixing of HCV quasi-species among residents (Figs 3 and 4).
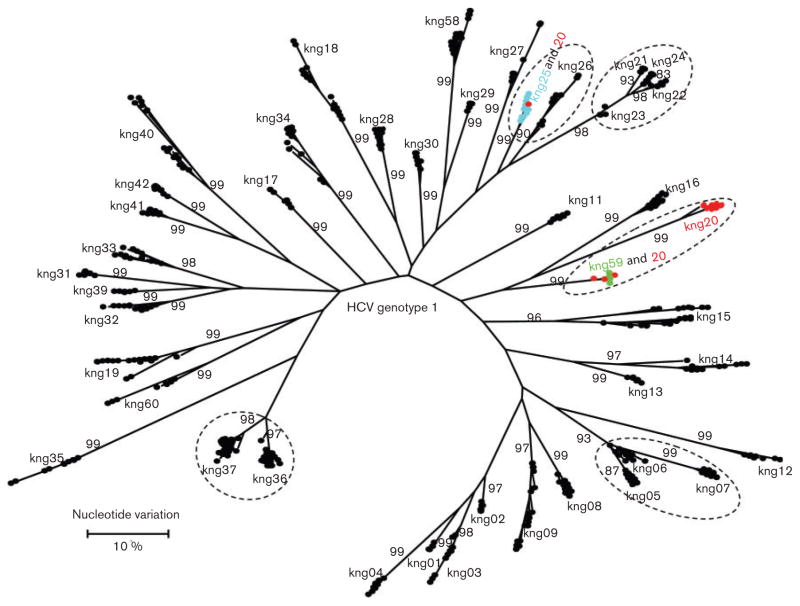
Fig. 3.
Maximum-likelihood tree of intra-host HVR1 variants identified in 43 individuals infected with HCV genotypes 1. All sequences from a single individual are shown using the sample identification code. The three samples that share variants have been identified with same colours. Dotted circles represent members of the same family. Bootstrap values higher than 60 % are indicated at the major nodes.
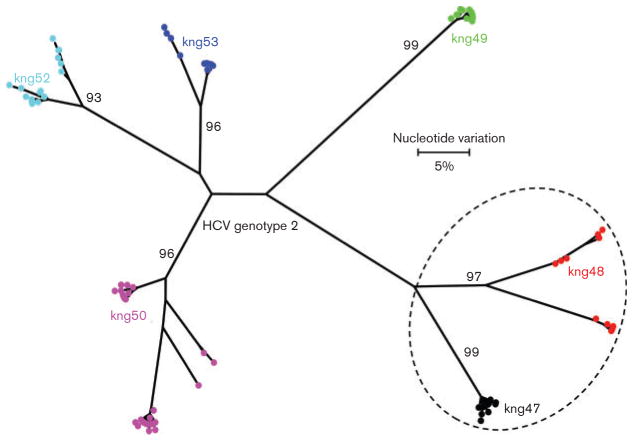
Fig. 4.
Maximum-likelihood tree of intra-host HVR1 variants identified in six individuals infected with HCV genotypes 1. All sequences from a single individual are shown using the sample identification code and same colour. Dotted circle represent members of the same family. Bootstrap values higher than 60 % are indicated at the major nodes.
There were six groups of relatives included in the analysis: group 1 – kng005 (20 years, female), kng006 (40 years, male) and kng007 (33 years, female); group 2 – kng021 (22 years, male), kng022 (45 years, male), kng23 (14 years, male) and kng024 (12 years, male); group 3 – kng036 (60 years, male) and kng037 (4 years, male); group 4 –kng020 (30 years, female) and kng59 (32 years, male); group 5 – kng025 (40 years, female) and kng026 (50 years, female); and group 6 – kng047 (44 years, male) and kng048 (35 years, male). HCV intra-host variants were found clustered together among relatives within groups 1, 2, 3, 5 and 6 (Figs 3 and 4). Although HCV variants identified from two relatives of group 4, kng020 and kng059, segregated into two distinct branches (Fig. 1), some intra-host variants found in kng020 intermixed with variants from kng059 (Fig. 3). Additionally, HCV intra-host variants from kng020 were also found to be intermixed with variants from kng025 (Fig. 3). These findings indicate a probable intra-familial HCV transmission or infections of relatives from a common source. However, clusters of genetically close intra-host variants were also identified among unrelated residents. For example, clusters of HCV variants from kng001 (15 years, male), kng002 (10 years, female), kng003 (28 years, female) and kng004 (28 years, male), or kng031 (60 years, female), kng032 (45 years, female), kng033 (30 years, female) and kng039 (50 years, male) were found among non-relatives. It is important to note that kng039 did not reside in the same community with kng031, kng032 and kng033. The identification of genetically close HCV variants among males and females of a broad age range (4–60 years) residing in different communities suggests a complex pattern of HCV dissemination within the communities, with the intra-familial transmission being common.
Population dynamics
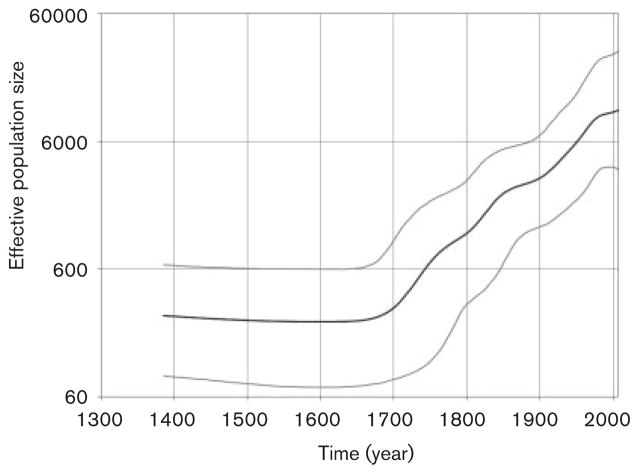
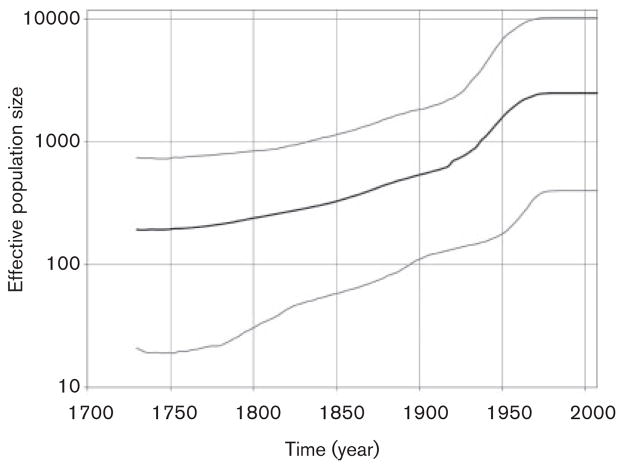
In order to investigate the origin and spread of HCV genotype 1 and 2 variants in these Nigerian communities we used the Bayesian coalescent framework implemented in BEAST. Each genotype was analysed separately. The inferred dates for the most recent common ancestor (tMRCA) for genotypes 1 and 2 were 1335 Common Era (C.E.) [95 % highest posterior density (95 % HPD): 1127–1529] and 1721 C.E. (95 % HPD: 1645–1791), respectively. Both genotypes experienced epidemic growth (Figs 5 and 6). The effective population size of genotype 1 increased progressively during the 18th–20th centuries and reached a plateau by the mid-20th century (Fig. 5). The genotype 2 expansion began during the mid-18th century and stabilized in the mid-20th century (Fig. 6). Genotype 2 showed a rapid exponential expansion between ~1920 and 1960. In support of these observations, Fu’s Fs test rejected the null hypothesis of neutrality and constant population for HCV genotype 1 (P=0.0001) and genotype 2 (P= 0.0006) variants. Collectively, the data suggest that HCV variants of both genotypes identified in the Nigerian communities experienced population expansion for about 200–350 years.
Fig. 5.
BSP showing the epidemic history estimated for HCV genotype 1 variants from the Nigerian communities. The middle black line is the estimated mean of effective population size (logarithmic scale), while the grey lines show the limits of the 95 % highest posterior probability density for this estimate. The most recent time is the time of collection for the Nigerian sequences (2007) and the leftmost extent of the estimated effective population size is set at the estimated mean for the tMRCA for all the sequences. Time is shown using common era dating.
Fig. 6.
BSP showing the epidemic history estimated for HCV genotype 2 variants from the Nigerian communities. The middle black line is the estimated mean of effective population size (logarithmic scale), while the grey lines show the limits of the 95 % highest posterior probability density for this estimate. The most recent time is the time of collection for the Nigerian sequences (2007) and the leftmost extent of the estimated effective population size is set at the estimated mean for the tMRCA for all the sequences. Time is shown using common era dating.
DISCUSSION
Nigeria has a high burden of chronic hepatitis C and HCC associated with HCV infection (Balogun et al., 2010; Laraba et al., 2010; Mustapha et al., 2007; Nwankiti et al., 2009). The rate of HCV infection was estimated to range from 1.9 to 14.5 % in the country (Balogun et al., 2010; Nwankiti et al., 2009; Onakewhor & Okonofua, 2009). This study was carried out in two remote communities in North Central Nigeria. Despite the relative isolation of these communities, ~15 % of the population experienced HCV infection. The observation of the high HCV prevalence in the villages lends additional support to high level of endemicity of HCV in Nigeria.
Owing to the isolation of these small communities, it was expected that only a few HCV strains would have been introduced and transmitted within the population. However, phylogenetic analysis of the NS5b sequences from 60 isolates obtained in this study revealed the presence of many distinct HCV variants and subtypes belonging to genotypes 1 and 2 (Fig. 1). HCV genotype 1 was detected in 85 % and genotype 2 in 15 % of all PCR-positive specimens. This finding is in agreement with three previous studies from Nigeria that showed predominance of HCV genotype 1, with variants of genotypes 2 and 4 being reported as minority genotypes (Agwale et al., 2004; Mellor et al., 1995; Oni & Harrison, 1996).
Although isolates in this study were collected from individuals living in closed communities, all HCV NS5b sequences were different from each other. Phylogenetic analysis showed extensive intermixing of the HCV variants identified among residents of the Nigerian communities and reported from other countries of West Africa (Fig. 2), indicating their common genetic history. These findings suggest that the heterogeneous HCV variants did not independently evolve within these remote communities but rather were introduced. Due to limited contacts with the general population, it is conceivable that few opportunities have been available for extraneous transmission, thus indicating that the introduction of the many HCV strains to the communities occurred in bulk. Alternatively, there is a remote possibility of the incremental introduction and accumulation of HCV variants but that would take a long period of time.
Analysis of HVR1 quasispecies showed that over a half of 49 HCV-infected members of the communities shared closely related HCV variants (Figs 3 and 4), thus revealing a complex pattern of potential HCV transmissions. The high HCV prevalence in these closed communities and the presence of many HCV lineages should have generated frequent opportunities for mixed infections. However, with the exception of kng020 sharing HCV variants with kng025 and kng059 (Fig. 3), no intermixing was observed among HCV quasispecies from different residents. Consistent with this finding, mixed HCV infections were rarely observed among such highly exposed groups as injection drug users, who experience only transient infections with more than one HCV variant (Viazov et al., 2000, 2010). These observations suggest a significant interference among HCV lineages that thwarts their co-existence in the same host. HCV superinfection exclusion is a possible molecular mechanism contributing to such interference (Tscherne et al., 2007).
The principal mode of transmission resulting in such a complex pattern of transmission is not clear. Findings of genetically close HCV quasispecies among relatives indicate that intra-familial transmission is a common route of HCV spread in the communities. It was previously suggested that sharing of equipment used for parenteral practices is an important route of HCV transmission in rural regions of Africa (Ndong-Atome et al., 2009; Njouom et al., 2003). However, such parenteral modes as body scarification, clitodectomy using unsterilized instruments, circumcision, shaving by local barbers, that are known to be responsible for HCV transmission in Africa (Adewole et al., 2009; Ocama & Seremba, 2011; Pépin et al., 2010), could not be responsible for the introduction of HCV genotypes 1 and 2 strains to these villages but could play a role in maintaining modern lineages of the virus. We can only speculate about the possible introduction and transmission events that occurred many years ago.
The existence of heterogeneous HCV variants reflects a long-term presence and diversifying evolution of this virus, and may be associated with a particularly extensive transmission mode in West Africa. A high prevalence of HCV genotype 4 lineages in Egypt had been linked to massive treatment campaigns against schistosomiasis between 1920 and 1980 (Frank et al., 2000). Studies from other African countries have argued that the HCV epidemic was driven iatrogenically, mostly by campaigns against trypanosomiasis, yaws and syphilis (Njouom et al., 2007; Pépin, Labbé, 2008; Pouillot et al., 2008b). Given this scenario, massive iatrogenic interventions could have jump-started the HCV epidemic in the Nigerian population. However, our data provide no direct indication on the events that could have led to such high prevalence of divergent HCV variants in the two communities.
Bayesian coalescence analysis indicated that the tMRCA for HCV genotype 1 and 2 lineages in the two Nigerian populations existed approximately 675 and 286 years ago, respectively. The tMRCA for genotype 1 variants identified in this study predates the tMRCA for HCV genotype 1 reported globally (Ferraro et al., 2008; Lampe et al., 2010; Magiorkinis et al., 2009; Pépin et al., 2010). This finding is consistent with many suggestions that HCV genotype 1 originated in Africa and spread to other parts of the world (Markov et al., 2009; Simmonds et al., 1993, 2005). The tMRCA for HCV genotype 2 variants found in the Nigerian communities is similar to the reported estimate for other countries in West Africa (Pouillot et al., 2008a). It should be noted, however, that analysis of sequences linked to a single date of collection used in this study could result in estimating tMRCA inaccurately. The timescale approximation for HCV epidemic may vary considerably depending on the estimation of evolutionary rate (Lampe et al., 2010). The rate of 5×10−4 substitutions per site per year used in the present analysis represents the best estimate of HCV NS5b evolutionary rates consistent with estimates made in several studies (Pouillot et al., 2008a; Pybus et al., 2001; Tanaka et al., 2002).
The Bayesian skyline plot (BSP) analysis showed that both genotype 1 and 2 populations experienced expansion (Figs 5 and 6). Fu’s statistical test supports this finding. HCV genotype 1 expansion steadily progressed for over 350 years starting in approximately 1700; the HCV genotype 1 population finally stabilized about the mid-20th century (Fig. 5). Genotype 2 expansion began in the mid-18th century and ended approximately during the mid-20th century (Fig. 6). A similar observation was made for genotype 2 lineages in Guinea-Bissau in West Africa, where the effective population size was shown to have been increasing from the mid-18th century. However, this growth had stabilized by approximately 1900 (Markov et al., 2009). A trend to stabilization was also observed in the genotype 1 and 2 BSPs at about the same time (Figs 5 and 6), indicating potential mitigation of the epidemic growth by the end of the 19th century. However, different from the genotype 2 expansion in Guinea-Bissau, both genotypes 1 and 2 in the Nigerian communities showed expansion before the mid-20th century (Figs 5 and 6). A similarly rapid spread of genotypes 1 and 2 from 1920 to 1950 was also observed in neighbouring Cameroon in Central Africa (Markov et al., 2009). However, this spread was not preceded by epidemic growth as seen in this study. Thus, the Nigerian expansion seems to combine the growth patterns observed in West and Central Africa. During the colonial period, in French speaking areas in Central Africa, unsafe injections practised mostly by mobile medical units for the prevention of sleeping disease early in 20th century were most probably responsible for the rapid HCV expansion in Cameroon (Njouom et al., 2007; Pouillot et al., 2008b). In English-speaking areas in West Africa including Nigeria, medical services have been broadly provided preferentially in stationary facilities since the 19th century (Millar & Foege, 1969) and could have contributed to the expansion during the 19th and mid-20th centuries in Nigeria through unsafe injection practices.
We have recently detected a population expansion of HBV genotype E lineages in these Nigerian communities (Forbi et al., 2010). Strikingly, this expansion occurred during approximately 1970–1980 or approximately 20 years later than the HCV expansion detected here. The tMRCA for the modern HBV genotype E was predicted to have existed approximately in the mid-20th century. The cause for this difference in time of expansion between two viral pathogens that share modes of transmission and co-circulate in the same communities is unknown. The difference in tMRCA indicates that the HCV genotype 1 and 2 lineages are much older than HBV genotype E (Forbi et al., 2010). It is conceivable that the epidemiological processes responsible for the origination of the modern HBV genotype E lineages (Forbi et al., 2010) could have contributed to the continuation of expansion of the already abundant HCV variants during the mid-20th century.
Six of the nine HCV genotype 2 variants identified in the Nigerian communities share a single branch of the phylogenetic tree with the major cluster of the variants from Cameroon, indicating a strong genetic connection between the genotype 2 strains circulating in both countries. These variants represent a subset of all genotype 2 lineages identified in West Africa (Figs 1 and 2). Recently, it was suggested that the genotype originated in West Africa and spread from the most western part of this region toward the centre of the continent (Markov et al., 2009; Ndjomou et al., 2003; Pouillot et al., 2008b). The detection of genetic similarity between the predominant genotype 2 lineages in Nigeria and Cameroon is consistent with this hypothesis and further suggests that the genotype 2 strains were filtered through Nigeria before reaching Cameroon. Nigeria contributes over three-quarters of the population in West Africa and is a convergence point and centre for active population mixing in Africa. Owing to its geographical location and population density, Nigeria may have critically facilitated HCV transmission from West to Central Africa.
The data suggest that these two Nigerian communities have experienced a massive introduction of multiple HCV variants, which probably occurred around the mid-20th century. The extensive genetic variation among HCV variants observed in this study indicates that the introduction was rather recent because genetic drift would have reduced the number of HCV variants circulating in these small communities over an extended period of time spanning more than one human generation. The prevalence of ~15 % is among the highest reported in West and Central Africa (Bekondi et al., 2010; Nagalo et al., 2011). Whether the prevalence and HCV genetic composition in these two communities represents the HCV population circulating in Nigeria as a whole needs further enquiry. This study is the first comprehensive investigation of the epidemic history of HCV in Nigeria revealing significant variation in the evolutionary histories of HCV genotypes 1 and 2 in two remote Nigerian villages that could be critical to understanding the dynamics of the diversity and spread of HCV in West and Central Africa.
METHODS
Ethics statement
Written informed consent was obtained from all participants involved in this study. The use of specimens in this study was approved by the Institutional Review Board (IRB) of the US Centers for Disease Control and Prevention and the Nigerian Federal Ministry of Health.
Serum samples and serological testing
A total of 519 blood samples were randomly obtained in 2007 from asymptomatic indigenes of two small rural communities in Keffi local government area of Nasarawa state in Nigeria, with 341 samples being from village 1 and 178 from village 2. These two villages with a total population of ~2000 inhabitants are ~2 miles apart. The residents have limited access to basic healthcare facilities.
All the studied samples were anonymous with a coding number for analysis. Samples were screened in the field for the presence of HCV antibodies using an immunochromatographic rapid assay (Shantha Biotechnics Ltd) following the manufacturer’s instructions. This assay uses recombinant antigen NS3, NS4 and NS5 to detect anti-HCV activity. Blood samples were centrifuged at 11 620 g for 10 min to separate serum from the cells. The serum samples were stored at − 20 °C until further testing was carried out.
Nucleic acid extraction and cDNA synthesis
Total nucleic acid was extracted from all serum specimens using the Roche MagNA Pure LC instrument and MagNA Pure LC Total Nucleic Acid Isolation kit (Roche Diagnostics), and eluted with 50 μl buffer according to the manufacturer’s instructions. RNA was precipitated and reverse-transcribed using both random and specific primers as described previously (Alter et al., 1999). Reverse transcription was carried out for 60 min at 42 °C in a total volume of 20 μl 5× PCR buffer (Roche), 200 pmol of each deoxynucleotide triphosphate, 1 μg random primers μl−1, 25 U AMV reverse transcriptase (Roche) and 40 U RNase inhibitor (Roche), followed by heating at 95 °C for 5 min.
Molecular detection of NS5b gene by real-time PCR
To identify HCV genotypes and subtypes, a segment of HCV NS5b encompassing positions 8275–8616 was amplified using real-time nested PCR. All amplification reactions were performed using the Fast-start DNA Master plus SYBR Green 1 kit and the Stratagene MX3005p QPCR machine (Agilent Technologies). First-round PCR was conducted using primers NS5B-K1 (5′-TGGGGATCCCGTATGATACCCGC-TGCTTTGA-3′) and NS5B-K2 (5′-GGCGGAATTCCTGGTCATA-GCCTCCGTGAA-3′). The nested reaction was performed with primers NS5B-122 (5′-CTCAACCGTCACTGAGAGAGACAT-3′) and NS5B-R1 (5′-GCTCTCAGGCTCGCCGCGTCCTC-3′). PCR included incubation at 94 °C for 2 min and 30 s, followed by 50 cycles of 94 °C for 20 s, 45 °C for 25 s and 72 °C for 45 s. Nested PCRs with 45 cycles were performed using the same conditions with 2 μl of the first-round product diluted 1 : 100.
Sequencing of the NS5b region
Sequencing was performed using the second-round PCR products and nested primers. Sequencing reactions were conducted using the BigDye v3.1 chemistry sequencing kit (Applied Biosystems), and products were sequenced using an automated sequencer (3130xl Genetic Analyzer; Applied Biosystems). Sequencing PCR involved 25 cycles, each cycle consisting of 96 °C for 10 s, 50 °C for 5 s and 60 °C for 4 min.
Deep pyrosequencing of the HVR1 region
Forty-three samples from patients infected with HCV genotype 1 (HCV-1) and six from patients with HCV genotype 2 (HCV-2) were used for quasispecies analysis. This subset was analysed using the next-generation sequencing technology (454/Roche GS FLX platform) on the HVR1 region of the genome. The junction E1/E2 region (309 nt), which contains the HVR1 region, was amplified using the nested PCR protocol described in Ramachandran et al. (2008). Each sample was amplified independently with fusion primers including the 454-primer key (A and B for forward and reverse primers, respectively), a different multiple identifier (MID) for each sample and HCV-specific sequence. The PCR products were purified using Agentcourt AmPure XP (Beckman Coulter). The quality of the amplicons was assessed using 3100 bioanalyser (Agilent Technologies). Purified amplicons were quantified using the Quan-iT PicoGreen dsDNA assay kit (Invitrogen). PCR amplicons were mixed at equimolar concentrations and diluted to a final concentration of 107 molecules ml−1 prior to being subjected to emulsion (em)PCR, which was performed following the instructions supplied with the kit and enriched beads were subjected to pyrosequencing (titanium chemistry) using the 454/ Roche GS FLX instrument. The original sequence reads (raw data) were processed using the SFFFILE tools. Sequence reads belonging to each sample were identified and separated using MID. Low quality reads were removed. The pyrosequencing files were post-processed with the empirical threshold (ET) error correction algorithm (Skums et al., 2011), which shows very high accuracy in finding true haplotypes, removing false haplotypes and estimating the frequency of true ones. The ET algorithm includes a calibration step using sequence reads from single-clone HVR1 samples, estimating an empirical frequency threshold for indels and haplotypes, and also correcting homopolymer errors using sequence alignment (Skums et al., 2011).
Phylogenetic analysis
Preliminary sequence analysis was conducted using SeqMan and MEGALIGN programs from the Lasergene DNA and Protein analysis software (version 8.0, DNASTAR Inc.). The Accelrys GCG Package (Genetic Computer Group, version 11.1-UNIX, Accelrys Inc.) was used for further analysis. Nucleotide sequences were aligned using the GCG multiple alignment program PILEUP. HCV genotypes/subgenotypes were classified based on the NS5b sequence (Smith et al., 1997) and by comparing each sequence with published reference sequences from GenBank. Initial phylogenetic trees were built using the Kimura two-parameter model of nucleotide substation (Tamura et al., 2004). Phylogenetic trees were constructed using the maximum-likelihood algorithm implemented in DNAML (PHYLIP package, v.3.6). Bootstrap values were calculated using 1000 replicates.
All sequences generated in this study were further subjected to phylogenetic analysis together with published sequences (NS5b region) from West and Central Africa retrieved from GenBank (Table 1).
Estimation of evolutionary dates and demographic history
The date of the MRCA of HCV strains from Nigeria was calculated using 362 nt NS5b sequences from the two villages. These sequences belong to genotypes 1 (n=51) and 2 (n=9). Divergence times were calculated using BEAST (ver. 1.6.0) (Drummond & Rambaut, 2007). The GTR substitution model was used with four gamma categories and invariant sites. Codons were partitioned into three partitions with unlinked substitution model and unlinked rate heterogeneity model across the codon partitions. Each sequence alignment was analysed with a strict or relaxed clock with an initial estimate for the rate of substitution as 5×10−4 (Njouom et al., 2007). Constant size, exponential and expansion growth priors were used. All models were run until the effective sample size for each was greater than 200. In order to compare the models the Bayes factor was estimated using importance sampling of the posterior probability (Newton et al., 1994). The exponential substitution model with an expansion growth prior was chosen because it had the largest Bayes factor; however, none of the models tested was found to be superior to any other model.
BSP analysis was done by segregating the NS5b sequences into the genotypes 1 (n=51) and 2 (n=9) groups. Each dataset was analysed separately using BEAST (ver. 1.6.0) (Drummond & Rambaut, 2007) with a strict clock, a substitution rate of 5×10−4 and a piecewise-constant skyline model with six groups.
Population dynamics of HCV in West and Central Africa
Unbiased estimates of nucleotide diversity were calculated according to Nei (1987) using the program ARLEQUIN (Excoffier et al., 2005). Fu’s Fs test for detecting population growth, was calculated using ARLEQUIN (Excoffier et al., 2005). A P-value of 0.05 or less was considered statistically significant.
Nucleotide sequence accession numbers
Sequences from Nigeria reported in this study have been deposited in the National Center for Biotechnology Information database and can be retrieved under accession numbers JQ679028–JQ679087.
Acknowledgments
The authors sincerely thank Dr Teo Chong-Gee for providing archived documents relevant to this study and for critically reviewing the manuscript. Disclaimer: this information is distributed solely for the purpose of pre-dissemination peer review under applicable information quality guidelines. It has not been formally disseminated by the Centers for Disease Control and Prevention/Agency for Toxic Substances and Disease Registry. It does not represent and should not be construed to represent any agency determination or policy.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences reported in this study are JQ679028–JQ679087.
References
- Adewole OO, Anteyi E, Ajuwon Z, Wada I, Elegba F, Ahmed P, Betiku Y, Okpe A, Eze S, et al. Hepatitis B and C virus co-infection in Nigerian patients with HIV infection. J Infect Dev Ctries. 2009;3:369–375. doi: 10.3855/jidc.245. [DOI] [PubMed] [Google Scholar]
- Agwale SM, Tanimoto L, Womack C, Odama L, Leung K, Duey D, Negedu-Momoh R, Audu I, Mohammed SB, et al. Prevalence of HCV coinfection in HIV-infected individuals in Nigeria and characterization of HCV genotypes. J Clin Virol. 2004;31 (Suppl 1):S3–S6. doi: 10.1016/j.jcv.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- Balogun TM, Emmanuel S, Wright KO. Hepatitis C virus co infection in HIV positive patients. Nig Q J Hosp Med. 2010;20:117–120. [PubMed] [Google Scholar]
- Bracho MA, Carrillo-Cruz FY, Ortega E, Moya A, Gonzalez-Candelas F. A new subtype of hepatitis C virus genotype 1: complete genome and phylogenetic relationships of an Equatorial Guinea isolate. J Gen Virol. 2006;87:1697–1702. doi: 10.1099/vir.0.81666-0. [DOI] [PubMed] [Google Scholar]
- Bekondi C, Mobima T, Ouavènè JO, Koffi B, Konamna X, Béré A, Le Faou A. Etiopathological factors of hepatocellular carcinoma in Bangui, Central African Republic: clinical, biological characteristics and virological aspects of patients. Pathol Biol (Paris) 2010;58:152–155. doi: 10.1016/j.patbio.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Candotti D, Temple J, Sarkodie F, Allain JP. Frequent recovery and broad genotype 2 diversity characterize hepatitis C virus infection in Ghana, West Africa. J Virol. 2003;77:7914–7923. doi: 10.1128/JVI.77.14.7914-7923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EASL. EASL International Consensus Conference on hepatitis C. Paris, 26–27 February 1999. Consensus statement. J Hepatol. 1999;31 (Suppl 1):3–8. [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Ferraro D, Genovese D, Argentini C, Giordano V, Pizzillo P, Stroffolini T, Craxì A, Rapicetta M, Di Stefano R. Phylogenetic reconstruction of HCV genotype 1b dissemination in a small city centre: the Camporeale model. J Med Virol. 2008;80:1723–1731. doi: 10.1002/jmv.21276. [DOI] [PubMed] [Google Scholar]
- Forbi JC, Vaughan G, Purdy MA, Campo DS, Xia GL, Ganova-Raeva LM, Ramachandran S, Thai H, Khudyakov YE. Epidemic history and evolutionary dynamics of hepatitis B virus infection in two remote communities in rural Nigeria. PLoS ONE. 2010;5:e11615. doi: 10.1371/journal.pone.0011615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C, Mohamed MK, Strickland GT, Lavanchy D, Arthur RR, Magder LS, El Khoby T, Abdel-Wahab Y, Aly Ohn ES, et al. The role of parenteral antischistosomal therapy in the spread of hepatitis C virus in Egypt. Lancet. 2000;355:887–891. doi: 10.1016/s0140-6736(99)06527-7. [DOI] [PubMed] [Google Scholar]
- Hnatyszyn HJ. Chronic hepatitis C and genotyping: the clinical significance of determining HCV genotypes. Antivir Ther. 2005;10:1–11. [PubMed] [Google Scholar]
- Itskowitz MS. Hepatitis C: epidemiology, diagnosis, and management. Compr Ther. 2007;33:87–93. doi: 10.1007/s12019-007-8005-8. [DOI] [PubMed] [Google Scholar]
- Jeannel D, Fretz C, Traore Y, Kohdjo N, Bigot A, PêGamy E, Jourdan G, Kourouma K, Maertens G, et al. Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J Med Virol. 1998;55:92–97. [PubMed] [Google Scholar]
- Lampe E, Espirito-Santo MP, Martins RM, Bello G. Epidemic history of Hepatitis C virus in Brazil. Infect Genet Evol. 2010;10:886–895. doi: 10.1016/j.meegid.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Laraba A, Wadzali G, Sunday B, Abdulfatai O, Fatai S. Hepatitis C virus infection in Nigerians with chronic liver disease. The Internet Journal of Gastroenterology. 2010;9 [Google Scholar]
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, Pybus OG, Allain JP, Hatzakis A. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med. 2009;6:e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov PV, Pepin J, Frost E, Deslandes S, Labbé AC, Pybus OG. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol. 2009;90:2086–2096. doi: 10.1099/vir.0.011569-0. [DOI] [PubMed] [Google Scholar]
- Mellor J, Holmes EC, Jarvis LM, Yap PL, Simmonds P The International HCV Collaborative Study Group. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. J Gen Virol. 1995;76:2493–2507. doi: 10.1099/0022-1317-76-10-2493. [DOI] [PubMed] [Google Scholar]
- Millar JD, Foege WH. Status of eradication of smallpox (and control of measles) in West and Central Africa. J Infect Dis. 1969;120:725–732. doi: 10.1093/infdis/120.6.725. [DOI] [PubMed] [Google Scholar]
- Mustapha SK, Bolori MT, Ajayi NA, Nggada HA, Pindiga UH, Gashau W, Khalil MIA. Hepatitis C virus antibodies in Nigerians with hepatocellular carcinoma. The Internet Journal of Oncology. 2007;4 [Google Scholar]
- Nagalo MB, Sanou M, Bisseye C, Kaboré MI, Nebie YK, Kienou K, Kiba A, Dahourou H, Ouattara S, et al. Seroprevalence of human immunodeficiency virus, hepatitis B and C viruses and syphilis among blood donors in Koudougou (Burkina Faso) in 2009. Blood Transfus. 2011;4:1–6. doi: 10.2450/2011.0112-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Lau GM, Lau GM, Sugiyama M, Mizokami M. An updated analysis of hepatitis C virus genotypes and subtypes based on the complete coding region. Liver Int. 2012;32:339–345. doi: 10.1111/j.1478-3231.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- Ndjomou J, Pybus OG, Matz B. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J Gen Virol. 2003;84:2333–2341. doi: 10.1099/vir.0.19240-0. [DOI] [PubMed] [Google Scholar]
- Ndong-Atome GR, Njouom R, Padilla C, Bisvigou U, Makuwa M, Kazanji M. Absence of intrafamilial transmission of hepatitis C virus and low risk for sexual transmission in rural central Africa indicate a cohort effect. J Clin Virol. 2009;45:349–353. doi: 10.1016/j.jcv.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Newton MA, Raftery AE, Davison AC, Bacha M, Celeux G, Carlin BP, Clifford P, Lu C, Sherman M, et al. Approximate Bayesian-inference with the weighted likelihood bootstrap. J Roy Stat Soc B Met. 1994;56:3–48. [Google Scholar]
- Njouom R, Pasquier C, Ayouba A, Gessain A, Froment A, Mfoupouendoun J, Pouillot R, Dubois M, Sandres-Sauné K, et al. High rate of hepatitis C virus infection and predominance of genotype 4 among elderly inhabitants of a remote village of the rain forest of South Cameroon. J Med Virol. 2003;71:219–225. doi: 10.1002/jmv.10473. [DOI] [PubMed] [Google Scholar]
- Njouom R, Pasquier C, Ayouba A, Tejiokem MC, Vessiere A, Mfoupouendoun J, Tene G, Eteki N, Lobe MM, Izopet J, et al. Low risk of mother-to-child transmission of hepatitis C virus in Yaounde, Cameroon: the ANRS 1262 study. Am J Trop Med Hyg. 2005;73:460–466. [PubMed] [Google Scholar]
- Njouom R, Nerrienet E, Dubois M, Lachenal G, Rousset D, Vessière A, Ayouba A, Pasquier C, Pouillot R. The hepatitis C virus epidemic in Cameroon: genetic evidence for rapid transmission between 1920 and 1960. Infect Genet Evol. 2007;7:361–367. doi: 10.1016/j.meegid.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Njouom R, Frost E, Deslandes S, Mamadou-Yaya F, Labbe AC, Pouillot R, Mbelesso P, Mbadingai S, Rousset D, Pepin J. Predominance of hepatitis C virus genotype 4 infection and rapid transmission between 1935 and 1965 in the Central African Republic. J Gen Virol. 2009;90:2452–2456. doi: 10.1099/vir.0.011981-0. [DOI] [PubMed] [Google Scholar]
- Nwankiti OO, Ndako JA, Echeonwu GO, Olabode AO, Nwosuh CI, Onovoh EM, Okeke LA, Akinola JO, Duru BN, et al. Hepatitis C virus infection in apparentenly healthy individuals with family history of diabetes in Vom, Plateau State Nigeria. Virol J. 2009;6:110. doi: 10.1186/1743-422X-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocama P, Seremba E. Management of HIV and Hepatitis C Virus Infections in Resource-Limited Settings. Curr Opin HIV AIDS. 2011;6:539–545. doi: 10.1097/COH.0b013e32834bd23f. [DOI] [PubMed] [Google Scholar]
- Onakewhor JU, Okonofua FE. Seroprevalence of Hepatitis C viral antibodies in pregnancy in a tertiary health facility in Nigeria. Niger J Clin Pract. 2009;12:65–73. [PubMed] [Google Scholar]
- Oni AO, Harrison TJ. Genotypes of hepatitis C virus in Nigeria. J Med Virol. 1996;49:178–186. doi: 10.1002/(SICI)1096-9071(199607)49:3<178::AID-JMV4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Pasquier C, Njouom R, Ayouba A, Dubois M, Sartre MT, Vessière A, Timba I, Thonnon J, Izopet J, Nerrienet E. Distribution and heterogeneity of hepatitis C genotypes in hepatitis patients in Cameroon. J Med Virol. 2005;77:390–398. doi: 10.1002/jmv.20468. [DOI] [PubMed] [Google Scholar]
- Pépin J, Labbé AC. Noble goals, unforeseen consequences: control of tropical diseases in colonial Central Africa and the iatrogenic transmission of blood-borne viruses. Trop Med Int Health. 2008;13:744–753. doi: 10.1111/j.1365-3156.2008.02060.x. [DOI] [PubMed] [Google Scholar]
- Pépin J, Lavoie M, Pybus OG, Pouillot R, Foupouapouognigni Y, Rousset D, Labbé AC, Njouom R. Risk factors for hepatitis C virus transmission in colonial Cameroon. Clin Infect Dis. 2010;51:768–776. doi: 10.1086/656233. [DOI] [PubMed] [Google Scholar]
- Pouillot R, Lachenal G, Pybus OG, Rousset D, Njouom R. Variable epidemic histories of hepatitis C virus genotype 2 infection in West Africa and Cameroon. Infect Genet Evol. 2008a;8:676–681. doi: 10.1016/j.meegid.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Pouillot R, Lachenal G, Pybus OG, Rousset D, Njouom R. Variable epidemic histories of hepatitis C virus genotype 2 infection in West Africa and Cameroon. Infect Genet Evol. 2008b;8:676–681. doi: 10.1016/j.meegid.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Charleston MA, Gupta S, Rambaut A, Holmes EC, Harvey PH. The epidemic behavior of the hepatitis C virus. Science. 2001;292:2323–2325. doi: 10.1126/science.1058321. [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Xia GL, Ganova-Raeva LM, Nainan OV, Khudyakov Y. End-point limiting-dilution real-time PCR assay for evaluation of hepatitis C virus quasispecies in serum: performance under optimal and suboptimal conditions. J Virol Methods. 2008;151:217–224. doi: 10.1016/j.jviromet.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Ruggieri A, Argentini C, Kouruma F, Chionne P, D’Ugo E, Spada E, Dettori S, Sabbatani S, Rapicetta M. Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry) J Gen Virol. 1996;77:2073–2076. doi: 10.1099/0022-1317-77-9-2073. [DOI] [PubMed] [Google Scholar]
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Skums P, Dimitrova Z, Campo DS, Vaughan G, Rossi L, Forbi JC, Yokosawa J, Zelikovsky A, Khudyakov Y. Efficient error correction for deep sequencing of viral amplicons. BMC Bioinformatics. 2012 doi: 10.1186/1471-2105-13-S10-S6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, Yap PL, Simmonds P. The origin of hepatitis C virus genotypes. J Gen Virol. 1997;78:321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- Stuyver L, Wyseur A, van Arnhem W, Lunel F, Laurent-Puig P, Pawlotsky JM, Kleter B, Bassit L, Nkengasong J, van Doorn L-J, et al. Hepatitis C virus genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 1995;38:137–157. doi: 10.1016/0168-1702(95)00052-r. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Hanada K, Mizokami M, Yeo AET, Shih JWK, Gojobori T, Alter HJ. A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc Natl Acad Sci U S A. 2002;99:15584–15589. doi: 10.1073/pnas.242608099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne DM, Evans MJ, von Hahn T, Jones CT, Stamataki Z, McKeating JA, Lindenbach BD, Rice CM. Superinfection exclusion in cells infected with hepatitis C virus. J Virol. 2007;81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viazov S, Widell A, Nordenfelt E. Mixed infection with two types of hepatitis C virus is probably a rare event. Infection. 2000;28:21–25. doi: 10.1007/s150100050005. [DOI] [PubMed] [Google Scholar]
- Viazov S, Ross SS, Kyuregyan KK, Timm J, Neumann-Haefelin C, Isaeva OV, Popova OE, Dmitriev PN, El Sharkawi F, et al. Hepatitis C virus recombinants are rare even among intravenous drug users. J Med Virol. 2010;82:232–238. doi: 10.1002/jmv.21631. [DOI] [PubMed] [Google Scholar]
- Wansbrough-Jones MH, Frimpong E, Cant B, Harris K, Evans MR, Teo CG. Prevalence and genotype of hepatitis C virus infection in pregnant women and blood donors in Ghana. Trans R Soc Trop Med Hyg. 1998;92:496–499. doi: 10.1016/s0035-9203(98)90887-2. [DOI] [PubMed] [Google Scholar]
- WHO. World Health Organisation Fact sheet no. 164. 1999. Hepatitis C: Weekly Epidemiological Record. No. 49, 10 December 1999. [Google Scholar]
- Zeuzem S. Heterogeneous virologic response rates to interferon-based therapy in patients with chronic hepatitis C: who responds less well? Ann Intern Med. 2004;140:370–381. doi: 10.7326/0003-4819-140-5-200403020-00033. [DOI] [PubMed] [Google Scholar]